

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0026.11
(461 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
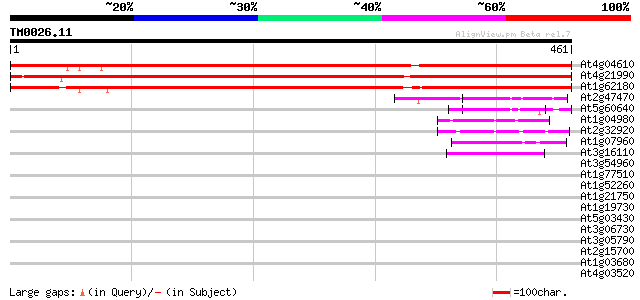
Score E
Sequences producing significant alignments: (bits) Value
At4g04610 5'-adenylylsulfate reductase 684 0.0
At4g21990 PRH26 683 0.0
At1g62180 putative adenosine-5'-phosphosulfate reductase 660 0.0
At2g47470 putative protein disulfide-isomerase 60 3e-09
At5g60640 protein disulfide isomerase precursor - like 53 4e-07
At1g04980 isulfide isomerase-related like protein 52 9e-07
At2g32920 putative protein disulfide isomerase 47 2e-05
At1g07960 unknown protein 44 1e-04
At3g16110 disulfide isomerase like protein 43 4e-04
At3g54960 protein disulfide-isomerase-like protein 40 0.002
At1g77510 putative thioredoxin 40 0.003
At1g52260 protein disulfide isomerase precursor like protein 40 0.003
At1g21750 putative disulfide isomerase 39 0.005
At1g19730 thioredoxin 37 0.018
At5g03430 unknown protein 36 0.039
At3g06730 thioredoxin, putative 36 0.051
At3g05790 putative mitochondrial LON ATP-dependent protease 35 0.11
At2g15700 copia-like retroelement pol polyprotein 35 0.11
At1g03680 putative thioredoxin-m 35 0.11
At4g03520 putative M-type thioredoxin 33 0.25
>At4g04610 5'-adenylylsulfate reductase
Length = 465
Score = 684 bits (1766), Expect = 0.0
Identities = 342/471 (72%), Positives = 400/471 (84%), Gaps = 16/471 (3%)
Query: 1 MALAVTCSISISSSSSSSSTFQSSQPKFSQIASIRVSEIPHGGGVN---ASQRRSFVKP- 56
MA++V S S SS +S S +PK SQI S+R+ + H V+ + +R S VKP
Sbjct: 1 MAMSVNVSSSSSSGIINSRFGVSLEPKVSQIGSLRLLDRVHVAPVSLNLSGKRSSSVKPL 60
Query: 57 --PQRSRDSIAPLAATIVAS---DVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNHI 111
+++DS+ PLAAT+VA +V E +++++LA LENASPL+IMD ALEK+GN I
Sbjct: 61 NAEPKTKDSMIPLAATMVAEIAEEVEVVEIEDFEELAKKLENASPLEIMDKALEKYGNDI 120
Query: 112 AIAFSGAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAV 171
AIAFSGAEDVALIEYA LTGRPFRVFSLDTGRLNPETYRFFD VEKHYGI IEYMFPD+V
Sbjct: 121 AIAFSGAEDVALIEYAHLTGRPFRVFSLDTGRLNPETYRFFDAVEKHYGIRIEYMFPDSV 180
Query: 172 EVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVV 231
EVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGL+AWITGQRKDQSPGTRSE+PVV
Sbjct: 181 EVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPVV 240
Query: 232 QVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRA 291
QVDPVFEG+DGGVGSLVKWNPVANV+G+D+W+FLRTM+VPVN+LH+ GYISIGCEPCT+A
Sbjct: 241 QVDPVFEGLDGGVGSLVKWNPVANVEGNDVWNFLRTMDVPVNTLHAAGYISIGCEPCTKA 300
Query: 292 VLPGQHEREGRWWWEDAKAKECGLHKGNVKQDA-EAELNGNGVANTNGTATVADIFNTQN 350
VLPGQHEREGRWWWEDAKAKECGLHKGNVK+++ +A++NG + VADIF ++N
Sbjct: 301 VLPGQHEREGRWWWEDAKAKECGLHKGNVKENSDDAKVNG------ESKSAVADIFKSEN 354
Query: 351 VVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRA 410
+V+LSR GIENL KLENRKEPW+VVLYAPWCP+CQAME SY +LADKLA SG+KV KFRA
Sbjct: 355 LVTLSRQGIENLMKLENRKEPWIVVLYAPWCPFCQAMEASYDELADKLAGSGIKVAKFRA 414
Query: 411 DGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVNALR 461
DG+QKEFAK+EL LGSFPTI+ FPK+SSRPIKYPSE RDV+SL +F+N +R
Sbjct: 415 DGDQKEFAKQELQLGSFPTILVFPKNSSRPIKYPSEKRDVESLTSFLNLVR 465
>At4g21990 PRH26
Length = 458
Score = 683 bits (1763), Expect = 0.0
Identities = 338/463 (73%), Positives = 391/463 (84%), Gaps = 7/463 (1%)
Query: 1 MALAVTCSISISSSSSSSSTFQSSQPKFSQIASIRVSEIPH--GGGVNASQRRSFVKPPQ 58
MALA+ S S SSS+ SSS+F SS K ++I S+R+ + ++ S +RS VK
Sbjct: 1 MALAINVSSS-SSSAISSSSFPSSDLKVTKIGSLRLLNRTNVSAASLSLSGKRSSVKALN 59
Query: 59 RSRDSIAPLAATIVASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNHIAIAFSGA 118
+ + A+ V + E +++++LA LENASPL+IMD ALEKFGN IAIAFSGA
Sbjct: 60 VQSITKESIVASEVTEKLDVVEVEDFEELAKRLENASPLEIMDKALEKFGNDIAIAFSGA 119
Query: 119 EDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEVQGLVR 178
EDVALIEYA LTGRP+RVFSLDTGRLNPETYR FD VEKHYGI IEYMFPDAVEVQ LVR
Sbjct: 120 EDVALIEYAHLTGRPYRVFSLDTGRLNPETYRLFDTVEKHYGIRIEYMFPDAVEVQALVR 179
Query: 179 SKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVDPVFE 238
+KGLFSFYEDGHQECCR+RKVRPLRRALKGLRAWITGQRKDQSPGTRSE+PVVQVDPVFE
Sbjct: 180 NKGLFSFYEDGHQECCRIRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFE 239
Query: 239 GVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHE 298
G+DGGVGSLVKWNPVANV+G+D+W+FLRTM+VPVN+LH+ GY+SIGCEPCTRAVLPGQHE
Sbjct: 240 GLDGGVGSLVKWNPVANVEGNDVWNFLRTMDVPVNTLHAAGYVSIGCEPCTRAVLPGQHE 299
Query: 299 REGRWWWEDAKAKECGLHKGNVKQDAEAELNGNGVANTNGTATVADIFNTQNVVSLSRTG 358
REGRWWWEDAKAKECGLHKGN+K++ NGN AN NGTA+VADIFN++NVV+LSR G
Sbjct: 300 REGRWWWEDAKAKECGLHKGNIKENT----NGNATANVNGTASVADIFNSENVVNLSRQG 355
Query: 359 IENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFA 418
IENL KLENRKE W+VVLYAPWCP+CQAME S+ +LADKL SGVKV KFRADG+QK+FA
Sbjct: 356 IENLMKLENRKEAWIVVLYAPWCPFCQAMEASFDELADKLGGSGVKVAKFRADGDQKDFA 415
Query: 419 KRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVNALR 461
K+EL LGSFPTI+ FPK+SSRPIKYPSE RDVDSL +F+N +R
Sbjct: 416 KKELQLGSFPTILVFPKNSSRPIKYPSEKRDVDSLTSFLNLVR 458
>At1g62180 putative adenosine-5'-phosphosulfate reductase
Length = 454
Score = 660 bits (1704), Expect = 0.0
Identities = 334/467 (71%), Positives = 388/467 (82%), Gaps = 19/467 (4%)
Query: 1 MALAVTCSISISSSSSSSSTFQSSQPKFSQIASIRVSEIPHGGGVNASQRRSFVKP---P 57
MALAVT S + S SS S + SS+ K QI SIR+S+ H SQRR +KP
Sbjct: 1 MALAVTSSSTAISGSSFSRSGASSESKALQICSIRLSDRTH-----LSQRRYSMKPLNAE 55
Query: 58 QRSR-DSIAPLAATIVASDVAET--EQDNYQQLAVDLENASPLQIMDAALEKFGNHIAIA 114
SR +S A+T++A +V E E ++++QLA LE+ASPL+IMD ALE+FG+ IAIA
Sbjct: 56 SHSRSESWVTRASTLIAPEVEEKGGEVEDFEQLAKKLEDASPLEIMDKALERFGDQIAIA 115
Query: 115 FSGAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEVQ 174
FSGAEDVALIEYARLTG+PFRVFSLDTGRLNPETYR FD VEK YGI IEYMFPDAVEVQ
Sbjct: 116 FSGAEDVALIEYARLTGKPFRVFSLDTGRLNPETYRLFDAVEKQYGIRIEYMFPDAVEVQ 175
Query: 175 GLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVD 234
LVR+KGLFSFYEDGHQECCRVRKVRPLRRALKGL+AWITGQRKDQSPGTRSE+P+VQVD
Sbjct: 176 ALVRNKGLFSFYEDGHQECCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPIVQVD 235
Query: 235 PVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLP 294
PVFEG+DGGVGSLVKWNP+ANV+G D+W+FLRTM+VPVN+LH++GY+SIGCEPCTR VLP
Sbjct: 236 PVFEGLDGGVGSLVKWNPLANVEGADVWNFLRTMDVPVNALHAQGYVSIGCEPCTRPVLP 295
Query: 295 GQHEREGRWWWEDAKAKECGLHKGNVKQDAEAELNGNGVANTNGTATVADIFNTQNVVSL 354
GQHEREGRWWWEDAKAKECGLHKGN+K++ +G A++ A V +IF + NVV+L
Sbjct: 296 GQHEREGRWWWEDAKAKECGLHKGNIKEE-------DGAADSK-PAAVQEIFESNNVVAL 347
Query: 355 SRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQ 414
S+ G+ENL KLENRKE WLVVLYAPWCP+CQAME SY++LA+KLA GVKV KFRADGEQ
Sbjct: 348 SKGGVENLLKLENRKEAWLVVLYAPWCPFCQAMEASYIELAEKLAGKGVKVAKFRADGEQ 407
Query: 415 KEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVNALR 461
KEFAK+EL LGSFPTI+ FPK + R IKYPSE+RDVDSLM+FVN LR
Sbjct: 408 KEFAKQELQLGSFPTILLFPKRAPRAIKYPSEHRDVDSLMSFVNLLR 454
>At2g47470 putative protein disulfide-isomerase
Length = 361
Score = 60.1 bits (144), Expect = 3e-09
Identities = 44/147 (29%), Positives = 69/147 (46%), Gaps = 10/147 (6%)
Query: 317 KGNVK-QDAEAELNGNGVA---NTNGTATVADIFNTQNVVSLSRTGIENLTKLENRKEPW 372
KG+++ Q E N +A N G V QNVV L+ + + +N+
Sbjct: 105 KGSLEPQKYEGPRNAEALAEYVNKEGGTNVKLAAVPQNVVVLTPDNFDEIVLDQNKDV-- 162
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKL-AESGVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
LV YAPWC +C+++ +Y +A E GV + AD + + G+ FPT+
Sbjct: 163 LVEFYAPWCGHCKSLAPTYEKVATVFKQEEGVVIANLDADAHKA--LGEKYGVSGFPTLK 220
Query: 432 FFPKHSSRPIKYPSENRDVDSLMAFVN 458
FFPK + Y RD+D ++F+N
Sbjct: 221 FFPKDNKAGHDYDG-GRDLDDFVSFIN 246
Score = 51.6 bits (122), Expect = 9e-07
Identities = 31/87 (35%), Positives = 45/87 (51%), Gaps = 4/87 (4%)
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAES-GVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
LV YAPWC +C+ + Y L ++ V + K D EQK + G+ +PTI
Sbjct: 44 LVEFYAPWCGHCKKLAPEYEKLGASFKKAKSVLIAKVDCD-EQKSVCTK-YGVSGYPTIQ 101
Query: 432 FFPKHSSRPIKYPSENRDVDSLMAFVN 458
+FPK S P KY R+ ++L +VN
Sbjct: 102 WFPKGSLEPQKYEGP-RNAEALAEYVN 127
>At5g60640 protein disulfide isomerase precursor - like
Length = 597
Score = 52.8 bits (125), Expect = 4e-07
Identities = 29/80 (36%), Positives = 42/80 (52%), Gaps = 2/80 (2%)
Query: 361 NLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKR 420
N T + + LV YAPWC +CQ++ Y A +L E GV + K A E+ E A +
Sbjct: 112 NFTDVIENNQYVLVEFYAPWCGHCQSLAPEYAAAATELKEDGVVLAKIDAT-EENELA-Q 169
Query: 421 ELGLGSFPTIMFFPKHSSRP 440
E + FPT++FF +P
Sbjct: 170 EYRVQGFPTLLFFVDGEHKP 189
Score = 46.6 bits (109), Expect = 3e-05
Identities = 33/91 (36%), Positives = 46/91 (50%), Gaps = 9/91 (9%)
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIMF 432
L+ +YAPWC +CQA+E Y LA L V + DG E K + FPTI+F
Sbjct: 463 LLEVYAPWCGHCQALEPMYNKLAKHLRSIDSLV-ITKMDGTTNEHPKAK--AEGFPTILF 519
Query: 433 FP--KHSSRPIKYPSENRDVDSLMAFVNALR 461
FP +S PI ++ +++AF LR
Sbjct: 520 FPAGNKTSEPITVDTDR----TVVAFYKFLR 546
>At1g04980 isulfide isomerase-related like protein
Length = 443
Score = 51.6 bits (122), Expect = 9e-07
Identities = 29/92 (31%), Positives = 45/92 (48%), Gaps = 5/92 (5%)
Query: 352 VSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRAD 411
V L+ + + L + KE W+V +APWC +C+ + + A+ L + VK+G D
Sbjct: 166 VELNSSNFDEL--VTESKELWIVEFFAPWCGHCKKLAPEWKKAANNL-KGKVKLGHVNCD 222
Query: 412 GEQKEFAKRELGLGSFPTIMFFPKHSSRPIKY 443
EQ K + FPTI+ F S P+ Y
Sbjct: 223 AEQS--IKSRFKVQGFPTILVFGSDKSSPVPY 252
Score = 38.1 bits (87), Expect = 0.010
Identities = 26/94 (27%), Positives = 41/94 (42%), Gaps = 5/94 (5%)
Query: 363 TKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKREL 422
+K+ N LV +APWC +CQ++ ++ +A L D + + ++
Sbjct: 40 SKVLNSNGVVLVEFFAPWCGHCQSLTPTWEKVASTLKGIATVAA---IDADAHKSVSQDY 96
Query: 423 GLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAF 456
G+ FPTI F PI Y RD S+ F
Sbjct: 97 GVRGFPTIKVFVP-GKPPIDYQGA-RDAKSISQF 128
>At2g32920 putative protein disulfide isomerase
Length = 440
Score = 47.0 bits (110), Expect = 2e-05
Identities = 33/110 (30%), Positives = 53/110 (48%), Gaps = 8/110 (7%)
Query: 352 VSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRAD 411
V L+ + ++L N E W+V +APWC +C+ + + A L + VK+G D
Sbjct: 165 VELNASNFDDLVIESN--ELWIVEFFAPWCGHCKKLAPEWKRAAKNL-QGKVKLGHVNCD 221
Query: 412 GEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSEN-RDVDSLMAFVNAL 460
EQ ++ + FPTI+ F S P YP E R ++ +F + L
Sbjct: 222 VEQSIMSR--FKVQGFPTILVFGPDKSSP--YPYEGARSASAIESFASEL 267
Score = 41.2 bits (95), Expect = 0.001
Identities = 32/106 (30%), Positives = 51/106 (47%), Gaps = 7/106 (6%)
Query: 351 VVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRA 410
VV L+ + ++ K+ N LV +APWC +C+A+ ++ +A+ + + V A
Sbjct: 32 VVQLTASNFKS--KVLNSNGVVLVEFFAPWCGHCKALTPTWEKVAN-ILKGVATVAAIDA 88
Query: 411 DGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAF 456
D Q A ++ G+ FPTI F PI Y RD S+ F
Sbjct: 89 DAHQS--AAQDYGIKGFPTIKVFVP-GKAPIDYQGA-RDAKSIANF 130
>At1g07960 unknown protein
Length = 146
Score = 44.3 bits (103), Expect = 1e-04
Identities = 26/95 (27%), Positives = 45/95 (47%), Gaps = 5/95 (5%)
Query: 364 KLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKL-AESGVKVGKFRADGEQKEFAKREL 422
K++ + W V PWC +C+ + + DL + + ++VG+ + K E
Sbjct: 38 KIKEKDTAWFVKFCVPWCKHCKKLGNLWEDLGKAMEGDDEIEVGEVDCGTSRAVCTKVE- 96
Query: 423 GLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFV 457
+ S+PT M F ++ + RDV+SL AFV
Sbjct: 97 -IHSYPTFMLF--YNGEEVSKYKGKRDVESLKAFV 128
>At3g16110 disulfide isomerase like protein
Length = 534
Score = 42.7 bits (99), Expect = 4e-04
Identities = 23/80 (28%), Positives = 39/80 (48%)
Query: 360 ENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAK 419
+N +L + E +V+ YAPWC + + + A L E G V + DGE+
Sbjct: 83 DNTKRLIDGNEYVMVLGYAPWCARSAELMPRFAEAATDLKEIGSSVLMAKIDGERYSKVA 142
Query: 420 RELGLGSFPTIMFFPKHSSR 439
+L + FPT++ F +S+
Sbjct: 143 SQLEIKGFPTLLLFVNGTSQ 162
>At3g54960 protein disulfide-isomerase-like protein
Length = 579
Score = 40.4 bits (93), Expect = 0.002
Identities = 23/63 (36%), Positives = 31/63 (48%), Gaps = 5/63 (7%)
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKL-AESGVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
L+ +YAPWC +CQ+ E Y L L + V K + AK + FPTI+
Sbjct: 459 LLEIYAPWCGHCQSFEPIYNKLGKYLKGIDSLVVAKMDGTSNEHPRAKAD----GFPTIL 514
Query: 432 FFP 434
FFP
Sbjct: 515 FFP 517
Score = 35.4 bits (80), Expect = 0.067
Identities = 19/74 (25%), Positives = 31/74 (41%), Gaps = 3/74 (4%)
Query: 360 ENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAK 419
+N T+ +V YAPWC CQA+ Y A +L + D ++
Sbjct: 107 DNFTEFVGNNSFAMVEFYAPWCGACQALTPEYAAAATELKGLAALA---KIDATEEGDLA 163
Query: 420 RELGLGSFPTIMFF 433
++ + FPT+ F
Sbjct: 164 QKYEIQGFPTVFLF 177
>At1g77510 putative thioredoxin
Length = 508
Score = 40.0 bits (92), Expect = 0.003
Identities = 23/100 (23%), Positives = 49/100 (49%), Gaps = 5/100 (5%)
Query: 361 NLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQ---KEF 417
N T+ ++ + +V YAPWC +CQ + Y A +L+ + + D + KEF
Sbjct: 38 NFTETISKHDFIVVEFYAPWCGHCQKLAPEYEKAASELSSHNPPLALAKIDASEEANKEF 97
Query: 418 AKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFV 457
A E + FPT+ ++ + ++ + R+ + ++ ++
Sbjct: 98 A-NEYKIQGFPTLKIL-RNGGKSVQDYNGPREAEGIVTYL 135
Score = 29.6 bits (65), Expect = 3.6
Identities = 19/79 (24%), Positives = 32/79 (40%), Gaps = 2/79 (2%)
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIMF 432
L+ YAPWC +CQ + ++A ++ V + D + + FPTI +
Sbjct: 394 LIEFYAPWCGHCQKLAPILDEVALSF-QNDPSVIIAKLDATANDIPSDTFDVKGFPTI-Y 451
Query: 433 FPKHSSRPIKYPSENRDVD 451
F S + Y + D
Sbjct: 452 FRSASGNVVVYEGDRTKED 470
>At1g52260 protein disulfide isomerase precursor like protein
Length = 537
Score = 40.0 bits (92), Expect = 0.003
Identities = 22/78 (28%), Positives = 35/78 (44%)
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIMF 432
+V+ YAPWC + + + A L E G V + DG++ EL + FPT++
Sbjct: 98 MVLGYAPWCARSAELMPRFAEAATALKEIGSSVLMAKIDGDRYSKIASELEIKGFPTLLL 157
Query: 433 FPKHSSRPIKYPSENRDV 450
F +S S D+
Sbjct: 158 FVNGTSLTYNGGSSAEDI 175
Score = 29.3 bits (64), Expect = 4.8
Identities = 22/93 (23%), Positives = 40/93 (42%), Gaps = 4/93 (4%)
Query: 367 NRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGS 426
N +E L+ ++ PWC C+A+ + LA K + + R D E K ++
Sbjct: 433 NSRENVLLEVHTPWCVNCEALSKQIEKLA-KHFKGFENLVFARIDASANEHTKLQVD-DK 490
Query: 427 FPTIMFFPK-HSSRPIKYPSENRDVDSLMAFVN 458
+P I+ + +P+K S + F+N
Sbjct: 491 YPIILLYKSGEKEKPLKL-STKLSAKDIAVFIN 522
>At1g21750 putative disulfide isomerase
Length = 501
Score = 39.3 bits (90), Expect = 0.005
Identities = 26/109 (23%), Positives = 51/109 (45%), Gaps = 6/109 (5%)
Query: 351 VVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAES--GVKVGKF 408
V++L T N T N+ + +V YAPWC +C+ + Y A L+ + V + K
Sbjct: 32 VLTLDHT---NFTDTINKHDFIVVEFYAPWCGHCKQLAPEYEKAASALSSNVPPVVLAKI 88
Query: 409 RADGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFV 457
A E + + FPTI F ++ + ++ + R+ + ++ ++
Sbjct: 89 DASEETNREFATQYEVQGFPTIKIF-RNGGKAVQEYNGPREAEGIVTYL 136
Score = 37.7 bits (86), Expect = 0.013
Identities = 25/87 (28%), Positives = 42/87 (47%), Gaps = 5/87 (5%)
Query: 373 LVVLYAPWCPYCQAMEESYVDLA-DKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
L+ YAPWC +CQ + ++A ++S V + K D +F K + FPTI
Sbjct: 396 LLEFYAPWCGHCQKLAPILDEVAVSYQSDSSVVIAKL--DATANDFPKDTFDVKGFPTIY 453
Query: 432 FFPKHSSRPIKYPSENRDVDSLMAFVN 458
F K +S + +R + ++FV+
Sbjct: 454 F--KSASGNVVVYEGDRTKEDFISFVD 478
>At1g19730 thioredoxin
Length = 119
Score = 37.4 bits (85), Expect = 0.018
Identities = 19/71 (26%), Positives = 35/71 (48%), Gaps = 3/71 (4%)
Query: 362 LTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRE 421
L K + + ++ A WCP C+ + + DLA K S + F+ D ++ + +E
Sbjct: 21 LDKAKESNKLIVIDFTASWCPPCRMIAPIFNDLAKKFMSSAI---FFKVDVDELQSVAKE 77
Query: 422 LGLGSFPTIMF 432
G+ + PT +F
Sbjct: 78 FGVEAMPTFVF 88
>At5g03430 unknown protein
Length = 497
Score = 36.2 bits (82), Expect = 0.039
Identities = 20/62 (32%), Positives = 28/62 (44%)
Query: 223 GTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYIS 282
G R P F G ++ NP+ + D+W+FL T V SL+ +GY S
Sbjct: 133 GVRIGDPTAVGQEQFSPSSPGWPPFMRVNPILDWSYRDVWAFLLTCKVKYCSLYDQGYTS 192
Query: 283 IG 284
IG
Sbjct: 193 IG 194
>At3g06730 thioredoxin, putative
Length = 183
Score = 35.8 bits (81), Expect = 0.051
Identities = 30/89 (33%), Positives = 42/89 (46%), Gaps = 4/89 (4%)
Query: 351 VVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRA 410
V LS ++ L K +RK P +V YA WC C M + LA + ES + K
Sbjct: 77 VKKLSAQELQELVK-GDRKVPLIVDFYATWCGPCILMAQELEMLAVEY-ESNAIIVKVDT 134
Query: 411 DGEQKEFAKRELGLGSFPTIMFFPKHSSR 439
D E EFA R++ + PT+ F S+
Sbjct: 135 DDEY-EFA-RDMQVRGLPTLFFISPDPSK 161
>At3g05790 putative mitochondrial LON ATP-dependent protease
Length = 942
Score = 34.7 bits (78), Expect = 0.11
Identities = 50/241 (20%), Positives = 93/241 (37%), Gaps = 25/241 (10%)
Query: 8 SISISSSSSSSSTFQSSQPKFSQIASIRVSEIPHGGGVNASQRRSFVKPP------QRSR 61
S + S + SS+T S++P ++ +PH + +VK P Q SR
Sbjct: 53 SSDLDSDTKSSTTTVSAKPHLDDCLTVIALPLPHKPLIPGFYMPIYVKDPKVLAALQESR 112
Query: 62 DSIAPLAATIVASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNHIAIAFSGAEDV 121
AP A + D A ++ + + LE ++++ + + G I+ E V
Sbjct: 113 RQQAPYAGAFLLKDDASSDSSSSSETENILEKLKGKELIN-RIHEVGTLAQISSIQGEQV 171
Query: 122 ALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYG-----IHIEYMFPDAVEVQGL 176
LI + R R+ + + +P T + +K Y I Y F ++ +
Sbjct: 172 ILIGH-----RQLRITEMVSESEDPLTVKVDHLKDKPYDKDDDVIKATY-FQVMSTLRDV 225
Query: 177 VRSKGLFSFYEDGHQECCRVRKVRPLRR-------ALKGLRAWITGQRKDQSPGTRSEVP 229
+++ L+ + + + C + LR L A I+G K Q+ G E+
Sbjct: 226 LKTTSLWRDHVRTYTQACSLHIWHCLRHIGEFNYPKLADFGAGISGANKHQNQGVLEELD 285
Query: 230 V 230
V
Sbjct: 286 V 286
>At2g15700 copia-like retroelement pol polyprotein
Length = 1166
Score = 34.7 bits (78), Expect = 0.11
Identities = 26/99 (26%), Positives = 43/99 (43%), Gaps = 3/99 (3%)
Query: 12 SSSSSSSSTFQSSQPKFSQIASIRVSEIPHGGGVNASQRRSFVKPPQRSRDSIAPLAATI 71
SSSS SS + S+ + S + +RRS ++PP R D A
Sbjct: 664 SSSSESSENLEESEMNEEVVGSENEQSLDDYLLARDMKRRSNIRPPSRFEDEDFVAYALA 723
Query: 72 VASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNH 110
A D+ E E +Y++ L+++ Q +A E+ +H
Sbjct: 724 TAEDLEEEEPKSYEEA---LKSSKRKQWENAMKEEMDSH 759
>At1g03680 putative thioredoxin-m
Length = 179
Score = 34.7 bits (78), Expect = 0.11
Identities = 19/64 (29%), Positives = 32/64 (49%), Gaps = 3/64 (4%)
Query: 370 EPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPT 429
EP V +APWC C+ ++ +LA K A + ++ + ++ + G+ S PT
Sbjct: 93 EPVFVDFWAPWCGPCKMIDPIVNELAQKYAG---QFKFYKLNTDESPATPGQYGVRSIPT 149
Query: 430 IMFF 433
IM F
Sbjct: 150 IMIF 153
>At4g03520 putative M-type thioredoxin
Length = 186
Score = 33.5 bits (75), Expect = 0.25
Identities = 18/63 (28%), Positives = 31/63 (48%), Gaps = 3/63 (4%)
Query: 371 PWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTI 430
P +V +APWC C+ ++ DLA K+ ++ + ++ + G+ S PTI
Sbjct: 100 PVVVDFWAPWCGPCKMIDPLVNDLAQHYTG---KIKFYKLNTDESPNTPGQYGVRSIPTI 156
Query: 431 MFF 433
M F
Sbjct: 157 MIF 159
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,181,650
Number of Sequences: 26719
Number of extensions: 438579
Number of successful extensions: 1543
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 1495
Number of HSP's gapped (non-prelim): 50
length of query: 461
length of database: 11,318,596
effective HSP length: 103
effective length of query: 358
effective length of database: 8,566,539
effective search space: 3066820962
effective search space used: 3066820962
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0026.11


