

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0023.16
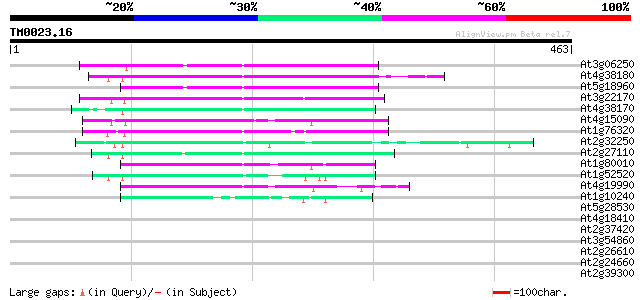
(463 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g06250 unknown protein 102 3e-22
At4g38180 unknown protein (At4g38180) 102 6e-22
At5g18960 FAR1 - like protein 100 3e-21
At3g22170 far-red impaired response protein, putative 90 2e-18
At4g38170 hypothetical protein 88 1e-17
At4g15090 unknown protein 82 6e-16
At1g76320 putative phytochrome A signaling protein 78 9e-15
At2g32250 Mutator-like transposase 75 1e-13
At2g27110 Mutator-like transposase 72 6e-13
At1g80010 hypothetical protein 66 4e-11
At1g52520 F6D8.26 63 4e-10
At4g19990 putative protein 60 2e-09
At1g10240 unknown protein 46 5e-05
At5g28530 far-red impaired response protein (FAR1) - like 38 0.010
At4g18410 MuDR transposable element - like protein 32 0.97
At2g37420 putative kinesin heavy chain 31 1.3
At3g54860 vacuolar protein sorting protein 33 (VPS33) 30 2.2
At2g26610 unknown protein 30 2.8
At2g24660 putative retroelement pol polyprotein 30 2.8
At2g39300 hypothetical protein 29 6.3
>At3g06250 unknown protein
Length = 764
Score = 102 bits (255), Expect = 3e-22
Identities = 72/257 (28%), Positives = 119/257 (46%), Gaps = 14/257 (5%)
Query: 58 FDVTEKYLCAWHLLRNANQNLGN-PLLTK-GFKNCMFRKK--------WDELVLNLGLQD 107
F T AW + +NL + P K ++ C+++ + W LV GL+D
Sbjct: 454 FPGTHHRFSAWQIRSKERENLRSFPNEFKYEYEKCLYQSQTTVEFDTMWSSLVNKYGLRD 513
Query: 108 NCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNC 167
N W++E YEKR WV A+LR FF G + F G ++S +L +F+ + +
Sbjct: 514 NMWLREIYEKREKWVPAYLRASFFGGIHVDGTFDPF---YGTSLNSLTSLREFISRYEQG 570
Query: 168 VKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDC 227
++ R E ++D +S + P ++T E +E + +T +F +F+ L + L
Sbjct: 571 LEQRREEERKEDFNSYNLQPFLQTK-EPVEEQCRRLYTLTIFRIFQSELAQSYNYLGLKT 629
Query: 228 TDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEE 287
+ + V K ++ V+ S+ N + SC E GL C HI+ V L I E
Sbjct: 630 YEEGAISRFLVRKCGNENEKHAVTFSASNLNASCSCQMFEYEGLLCRHILKVFNLLDIRE 689
Query: 288 LSECFIFKRWSKGAKDG 304
L +I RW+K A+ G
Sbjct: 690 LPSRYILHRWTKNAEFG 706
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 102 bits (253), Expect = 6e-22
Identities = 74/307 (24%), Positives = 128/307 (41%), Gaps = 33/307 (10%)
Query: 66 CAWHLLRNANQNLGN-----PLLTKGFKNCM--------FRKKWDELVLNLGLQDNCWVK 112
C WH+L+ + L + P F C+ F + W L+ L+D+ W++
Sbjct: 386 CKWHILKKCQEKLSHVFLKHPSFESDFHKCVNLTESVEDFERCWFSLLDKYELRDHEWLQ 445
Query: 113 ETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHMR 172
Y R WV +LR FFA T R + S ++++ NL+ F + + ++
Sbjct: 446 AIYSDRRQWVPVYLRDTFFADMSLTHRSDSINSYFDGYINASTNLSQFFKLYEKALESRL 505
Query: 173 FREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAVT 232
+E++ D +++ P +KT +E A++ +T +F+ F+ L D
Sbjct: 506 EKEVKADYDTMNSPPVLKTP-SPMEKQASELYTRKLFMRFQEELVGTLTFMASKADDDGD 564
Query: 233 CLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECF 292
+ Y VAK K V + SC E G+ C HI+ V + L +
Sbjct: 565 LVTYQVAKYGEAHKAHFVKFNVLEMRANCSCQMFEFSGIICRHILAVFRVTNLLTLPPYY 624
Query: 293 IFKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNTAADFNDATQKA 352
I KRW++ AK V I +D+ +L+ YL S+T +N KA
Sbjct: 625 ILKRWTRNAKSSV-----IFDDY-------------NLHAYANYLESHTVR-YNTLRHKA 665
Query: 353 SELIEQS 359
S ++++
Sbjct: 666 SNFVQEA 672
Score = 30.8 bits (68), Expect = 1.6
Identities = 11/60 (18%), Positives = 31/60 (51%)
Query: 2 LKNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
++ + + + ++ ++ + + N+F + S+ ++FW D + + +GD + FD T
Sbjct: 246 IEGEIQLLLDYLRQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDFTHFGDTVTFDTT 305
>At5g18960 FAR1 - like protein
Length = 788
Score = 99.8 bits (247), Expect = 3e-21
Identities = 63/213 (29%), Positives = 102/213 (47%), Gaps = 4/213 (1%)
Query: 92 FRKKWDELVLNLGLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHV 151
F W L+ GL+D+ W++E YE+R WV A+LR FFAG E F G +
Sbjct: 522 FDSVWSALINKYGLRDDVWLREIYEQRENWVPAYLRASFFAGIPINGTIEPF---FGASL 578
Query: 152 HSRYNLTDFLQQLQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLL 211
+ L +F+ + + ++ R E ++D +S + P ++T E +E + +T VF +
Sbjct: 579 DALTPLREFISRYEQALEQRREEERKEDFNSYNLQPFLQTK-EPVEEQCRRLYTLTVFRI 637
Query: 212 FRPVLQHAPLMKVLDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGL 271
F+ L + L + + V K +++ V+ S+ N + SC E GL
Sbjct: 638 FQNELVQSYNYLCLKTYEEGAISRFLVRKCGNESEKHAVTFSASNLNSSCSCQMFEHEGL 697
Query: 272 ACEHIVDVLAHLRIEELSECFIFKRWSKGAKDG 304
C HI+ V L I EL +I RW+K A+ G
Sbjct: 698 LCRHILKVFNLLDIRELPSRYILHRWTKNAEFG 730
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 90.1 bits (222), Expect = 2e-18
Identities = 61/265 (23%), Positives = 115/265 (43%), Gaps = 15/265 (5%)
Query: 58 FDVTEKYLCAWHLLRNANQNLGNPL-----LTKGFKNCMFR--------KKWDELVLNLG 104
F T L WH+L ++NLG + F+ C+++ +KW + + G
Sbjct: 363 FPNTRHCLFLWHVLMKVSENLGQVVKQHDNFMPKFEKCIYKSGKDEDFARKWYKNLARFG 422
Query: 105 LQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQL 164
L+D+ W+ YE R W ++ AG T+ R + + K++H + ++ +F++
Sbjct: 423 LKDDQWMISLYEDRKKWAPTYMTDVLLAGMSTSQRADSINAFFDKYMHKKTSVQEFVKVY 482
Query: 165 QNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKV 224
++ E + D + PAMK+ E S ++ +T VF F+ + A
Sbjct: 483 DTVLQDRCEEEAKADSEMWNKQPAMKSP-SPFEKSVSEVYTPAVFKKFQIEVLGAIACSP 541
Query: 225 LDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLR 284
+ TC + V +++ V+ + + C E +G C H ++VL
Sbjct: 542 REENRDATCSTFRVQDFE-NNQDFMVTWNQTKAEVSCICRLFEYKGYLCRHTLNVLQCCH 600
Query: 285 IEELSECFIFKRWSKGAKDGVYSGK 309
+ + +I KRW+K AK +SG+
Sbjct: 601 LSSIPSQYILKRWTKDAKSRHFSGE 625
Score = 30.0 bits (66), Expect = 2.8
Identities = 18/76 (23%), Positives = 34/76 (44%), Gaps = 3/76 (3%)
Query: 5 DSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT--- 61
D + + F+ + S + N+F ++++FW D R +Y + DV++ D T
Sbjct: 234 DFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFWVDAKSRHNYGSFCDVVSLDTTYVR 293
Query: 62 EKYLCAWHLLRNANQN 77
KY + NQ+
Sbjct: 294 NKYKMPLAIFVGVNQH 309
>At4g38170 hypothetical protein
Length = 531
Score = 87.8 bits (216), Expect = 1e-17
Identities = 63/259 (24%), Positives = 104/259 (39%), Gaps = 17/259 (6%)
Query: 52 YGDVLAFDVTEKYLCAWHLLRNANQNLGNPLLTKGFKNCM--------FRKKWDELVLNL 103
+ L F+ TE+ L H+ + +P F NC+ F WD +V
Sbjct: 136 FSQPLIFEETEEKLA--HVFQ------AHPTFESEFINCVTETETAAEFEASWDSIVRRY 187
Query: 104 GLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQ 163
++DN W++ Y R WV +R F+ T S V + + ++Q
Sbjct: 188 YMEDNDWLQSIYNARQQWVRVFIRDTFYGELSTNEGSSILNSFFQGFVDASTTMQMLIKQ 247
Query: 164 LQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMK 223
+ + R +E++ D + + P MKT +E A +T F+ F+ +
Sbjct: 248 YEKAIDSWREKELKADYEATNSTPVMKTP-SPMEKQAASLYTRAAFIKFQEEFVETLAIP 306
Query: 224 VLDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHL 283
+D+ T Y VAK + K VS S SC E G+ C HI+ V +
Sbjct: 307 ANIISDSGTHTTYRVAKFGEVHKGHTVSFDSLEVKANCSCQMFEYSGIICRHILAVFSAK 366
Query: 284 RIEELSECFIFKRWSKGAK 302
+ L ++ +RW+K AK
Sbjct: 367 NVLALPSRYLLRRWTKEAK 385
Score = 31.6 bits (70), Expect = 0.97
Identities = 12/25 (48%), Positives = 16/25 (64%)
Query: 37 HLFWRDGVGRLSYQVYGDVLAFDVT 61
++FW D RL+Y +GD L FD T
Sbjct: 31 NVFWADPTCRLNYTYFGDTLVFDTT 55
>At4g15090 unknown protein
Length = 768
Score = 82.0 bits (201), Expect = 6e-16
Identities = 65/270 (24%), Positives = 113/270 (41%), Gaps = 25/270 (9%)
Query: 61 TEKYLCAWHLLRNANQNLGNPL------LTKGFKNCMFRK--------KWDELVLNLGLQ 106
T WH+L + + + L K F C+FR +W ++V GL+
Sbjct: 298 TRHCFALWHVLEKIPEYFSHVMKRHENFLLK-FNKCIFRSWTDDEFDMRWWKMVSQFGLE 356
Query: 107 DNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQN 166
++ W+ +E R WV + F AG T+ R E S K++H + L +FL+Q
Sbjct: 357 NDEWLLWLHEHRQKWVPTFMSDVFLAGMSTSQRSESVNSFFDKYIHKKITLKEFLRQYGV 416
Query: 167 CVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLD 226
+++ E D + H PA+K+ + AT +T+ +F F+ ++ V+
Sbjct: 417 ILQNRYEEESVADFDTCHKQPALKSPSPWEKQMATT-YTHTIFKKFQ-----VEVLGVVA 470
Query: 227 CTDAVTCLNYTVAKLSFLAKE----WHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAH 282
C + +A E + V+ S + C E +G C H + +L
Sbjct: 471 CHPRKEKEDENMATFRVQDCEKDDDFLVTWSKTKSELCCFCRMFEYKGFLCRHALMILQM 530
Query: 283 LRIEELSECFIFKRWSKGAKDGVYSGKNIE 312
+ +I KRW+K AK GV +G+ +
Sbjct: 531 CGFASIPPQYILKRWTKDAKSGVLAGEGAD 560
Score = 35.4 bits (80), Expect = 0.067
Identities = 15/59 (25%), Positives = 31/59 (52%)
Query: 3 KNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
+ DS + +++ + ++ +F ++ L++LFW D R Y + DV++FD T
Sbjct: 164 EGDSQVLLEYFKRIKKENPKFFYAIDLNEDQRLRNLFWADAKSRDDYLSFNDVVSFDTT 222
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 78.2 bits (191), Expect = 9e-15
Identities = 63/267 (23%), Positives = 112/267 (41%), Gaps = 21/267 (7%)
Query: 61 TEKYLCAWHLLRNANQNLG------NPLLTKGFKNCMFR--------KKWDELVLNLGLQ 106
T C WH+L +NL + + K FK C++R ++W +L+ L+
Sbjct: 294 TRHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFK-CIYRSWSEEEFDRRWLKLIDKFHLR 352
Query: 107 DNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQN 166
D W++ YE+R W +RG FAG R E S ++VH +L +FL+
Sbjct: 353 DVPWMRSLYEERKFWAPTFMRGITFAGLSMRCRSESVNSLFDRYVHPETSLKEFLEGYGL 412
Query: 167 CVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFR-PVLQHAPLMKVL 225
++ E + D + H P +K+ E +++ +F F+ VL A
Sbjct: 413 MLEDRYEEEAKADFDAWHEAPELKSP-SPFEKQMLLVYSHEIFRRFQLEVLGAAACHLTK 471
Query: 226 DCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRI 285
+ + T Y+V +++ V D SC E +G C H + VL +
Sbjct: 472 ESEEGTT---YSVKDFDD-EQKYLVDWDEFKSDIYCSCRSFEYKGYLCRHAIVVLQMSGV 527
Query: 286 EELSECFIFKRWSKGAKDGVYSGKNIE 312
+ ++ +RW+ A++ +N+E
Sbjct: 528 FTIPINYVLQRWTNAARNRHQISRNLE 554
Score = 32.3 bits (72), Expect = 0.57
Identities = 19/85 (22%), Positives = 42/85 (49%), Gaps = 3/85 (3%)
Query: 5 DSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT--- 61
D+ + ++F++ + ++ +F + + L+++FW D G Y+ + DV++F+ +
Sbjct: 162 DAEILLEFLMRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFV 221
Query: 62 EKYLCAWHLLRNANQNLGNPLLTKG 86
KY L N ++ LL G
Sbjct: 222 SKYKVPLVLFVGVNHHVQPVLLGCG 246
>At2g32250 Mutator-like transposase
Length = 684
Score = 74.7 bits (182), Expect = 1e-13
Identities = 86/401 (21%), Positives = 162/401 (39%), Gaps = 55/401 (13%)
Query: 55 VLAFDVTEKYLCAWHLLRNANQNLGNPLLTK------GFKNCM--------FRKKWDELV 100
V F C W +L ++ L NP +++ F NC+ F ++W ++
Sbjct: 295 VEVFPDVRHIFCLWSVLSKISEML-NPFVSQDDGFMESFGNCVASSWTDEHFERRWSNMI 353
Query: 101 LNLGLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDF 160
L +N WV+ + R WV + G AG R S K+++S DF
Sbjct: 354 GKFELNENEWVQLLFRDRKKWVPHYFHGICLAGLSGPERSGSIASHFDKYMNSEATFKDF 413
Query: 161 LQQLQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFR---PVLQ 217
+ +++ E +DD P ++++L + E + +T+ F F+ P +
Sbjct: 414 FELYMKFLQYRCDVEAKDDLEYQSKQPTLRSSL-AFEKQLSLIYTDAAFKKFQAEVPGVV 472
Query: 218 HAPLMKVL-DCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHI 276
L K D T A+ + + +F V+++++ D SC E +G C+H
Sbjct: 473 SCQLQKEREDGTTAIFRIEDFEERQNFF-----VALNNELLDACCSCHLFEYQGFLCKHA 527
Query: 277 VDVLAHLRIEELSECFIFKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALGY 336
+ VL + + +I KRWSK G N E+ K+ +C+ +
Sbjct: 528 ILVLQSADVSRVPSQYILKRWSK-------KGNNKED------KNDKCATI--------- 565
Query: 337 LNSNTAADFNDATQKASEL-IEQSKAKKSCQREGSLSLSQT--NILNLKDPIRLRRKGRG 393
N A F+D ++ +L + S + ++C+ L L +T + +++ + + +
Sbjct: 566 --DNRMARFDDLCRRFVKLGVVASLSDEACKTALKL-LEETVKHCVSMDNSSKFPSEPDK 622
Query: 394 TKNKSSLGLTVKHVINCL--IFKTPGHNKLTGTYCSQQGHT 432
S+GL + V++C + K K YC + T
Sbjct: 623 LMTGGSIGLENEGVLDCASKVSKKKKIQKKRKVYCGPEDAT 663
Score = 30.4 bits (67), Expect = 2.2
Identities = 13/57 (22%), Positives = 27/57 (46%)
Query: 3 KNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFD 59
+ D + ++ +E+ K +F D ++++FW D + Y + DV+ FD
Sbjct: 167 EEDLKLLLEHFMEMQDKQPGFFYAVDFDSDKRVRNVFWLDAKAKHDYCSFSDVVLFD 223
>At2g27110 Mutator-like transposase
Length = 851
Score = 72.0 bits (175), Expect = 6e-13
Identities = 60/263 (22%), Positives = 99/263 (36%), Gaps = 16/263 (6%)
Query: 68 WHLLRNANQNLGN-----PLLTKGFKNCM--------FRKKWDELVLNLGLQDNCWVKET 114
W +LR + L + P NC+ F W ++ L + W+
Sbjct: 335 WDVLREGQEKLAHVCLAYPSFQVELYNCINFTETIEEFESSWSSVIDKYDLGRHEWLNSL 394
Query: 115 YEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHMRFR 174
Y R+ WV + R FFA + G S +V+ + L F + + ++
Sbjct: 395 YNARAQWVPVYFRDSFFAAVFPSQGYSG--SFFDGYVNQQTTLPMFFRLYERAMESWFEM 452
Query: 175 EIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAVTCL 234
EIE D +++ P +KT +E A FT +F F+ L D T
Sbjct: 453 EIEADLDTVNTPPVLKTP-SPMENQAANLFTRKIFGKFQEELVETFAHTANRIEDDGTTS 511
Query: 235 NYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECFIF 294
+ VA K + V+ SC E G+ C H++ V I L +I
Sbjct: 512 TFRVANFENDNKAYIVTFCYPEMRANCSCQMFEHSGILCRHVLTVFTVTNILTLPPHYIL 571
Query: 295 KRWSKGAKDGVYSGKNIENDFWD 317
+RW++ AK V +++ + D
Sbjct: 572 RRWTRNAKSMVELDEHVSENGHD 594
Score = 34.3 bits (77), Expect = 0.15
Identities = 14/62 (22%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query: 5 DSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVTEKY 64
D+ +++ + +++ +F D+ N + ++FW D R++Y +GD + D +Y
Sbjct: 196 DAHNLLEYFKRMQAENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDT--RY 253
Query: 65 LC 66
C
Sbjct: 254 RC 255
>At1g80010 hypothetical protein
Length = 696
Score = 66.2 bits (160), Expect = 4e-11
Identities = 49/224 (21%), Positives = 95/224 (41%), Gaps = 21/224 (9%)
Query: 92 FRKKWDELVLNLGLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCE-GFQSELGKH 150
F W+E+++ G+ +N +++ ++ R +W +L+ F AG T +
Sbjct: 417 FETAWEEMIIRFGMTNNETIRDMFQDRELWAPVYLKDTFLAGALTFPLGNVAAPFIFSGY 476
Query: 151 VHSRYNLTDFLQQLQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFL 210
VH +L +FL+ ++ + RE D S+ +P +KT E K FT +F
Sbjct: 477 VHENTSLREFLEGYESFLDKKYTREALCDSESLKLIPKLKTT-HPYESQMAKVFTMEIFR 535
Query: 211 LFRPVLQHAPLMKVLDCTDAVTCLNYTVAKLSFLAKE------------WHVSVSSDNKD 258
F+ + + VT ++ + S++ KE + S ++ +
Sbjct: 536 RFQDE------VSAMSSCFGVTQVHSNGSASSYVVKEREGDKVRDFEVIYETSAAAQVRC 589
Query: 259 FKYSCMKMESRGLACEHIVDVLAHLRIEELSECFIFKRWSKGAK 302
F C G C H++ +L+H ++E+ +I +RW K K
Sbjct: 590 F-CVCGGFSFNGYQCRHVLLLLSHNGLQEVPPQYILQRWRKDVK 632
Score = 37.7 bits (86), Expect = 0.013
Identities = 20/50 (40%), Positives = 31/50 (62%), Gaps = 2/50 (4%)
Query: 12 FMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
F ++L+S + Y + +AD SL+++FW D R +Y +GDVL FD T
Sbjct: 251 FQIQLSSPNFLYLMD-LADD-GSLRNVFWIDARARAAYSHFGDVLLFDTT 298
>At1g52520 F6D8.26
Length = 703
Score = 62.8 bits (151), Expect = 4e-10
Identities = 61/265 (23%), Positives = 104/265 (39%), Gaps = 43/265 (16%)
Query: 69 HLLRNANQNLGN----PLLTKGFKNCM--------FRKKWDELVLNLGLQDNCWVKETYE 116
H++R + LG + K F + F W +V N G+ +N W++ YE
Sbjct: 386 HIMRKIPEKLGGLHNYDAVRKAFTKAVYETLKVVEFEAAWGFMVHNFGVIENEWLRSLYE 445
Query: 117 KRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHMRFREI 176
+R+ W +L+ FFAG E + ++VH + L +FL + + ++ E
Sbjct: 446 ERAKWAPVYLKDTFFAGIAAAHPGETLKPFFERYVHKQTPLKEFLDKYELALQKKHREET 505
Query: 177 EDDCHS-IHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAVTCLN 235
D S +KT S E ++ +T ++F F+ ++ + +C +
Sbjct: 506 LSDIESQTLNTAELKTKC-SFETQLSRIYTRDMFKKFQ-----------IEVEEMYSCFS 553
Query: 236 YTVAKLS-----FLAKEWHVSVSS--DNKDF-----------KYSCMKMESRGLACEHIV 277
T + FL KE SS + +DF + C G C H +
Sbjct: 554 TTQVHVDGPFVIFLVKERVRGESSRREIRDFEVLYNRSVGEVRCICSCFNFYGYLCRHAL 613
Query: 278 DVLAHLRIEELSECFIFKRWSKGAK 302
VL +EE+ +I RW K K
Sbjct: 614 CVLNFNGVEEIPLRYILPRWRKDYK 638
Score = 32.0 bits (71), Expect = 0.74
Identities = 19/88 (21%), Positives = 37/88 (41%), Gaps = 3/88 (3%)
Query: 3 KNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFD--- 59
+ DS + + + N+F + L+++FW D ++S +GDV+ D
Sbjct: 245 RGDSAAIYNYFCRMQLTNPNFFYLMDVNDEGQLRNVFWADAFSKVSCSYFGDVIFIDSSY 304
Query: 60 VTEKYLCAWHLLRNANQNLGNPLLTKGF 87
++ K+ N + LL+ GF
Sbjct: 305 ISGKFEIPLVTFTGVNHHGKTTLLSCGF 332
>At4g19990 putative protein
Length = 672
Score = 60.5 bits (145), Expect = 2e-09
Identities = 55/247 (22%), Positives = 103/247 (41%), Gaps = 35/247 (14%)
Query: 92 FRKKWDELVLNLGLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHV 151
F K W E+V ++DN W++ YE R WV +++ AG T R + S L K++
Sbjct: 333 FEKNWWEVVDRFHMRDNVWLQSLYEDREYWVPVYMKDVSLAGMCTAQRSDSVNSGLDKYI 392
Query: 152 HSRYNLTDFLQQLQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLL 211
+ FL+Q + ++ E + + +++ P +K+ + +T +F
Sbjct: 393 QRKTTFKAFLEQYKKMIQERYEEEEKSEIETLYKQPGLKSP-SPFGKQMAEVYTREMFKK 451
Query: 212 FRPVLQHAPLMKVLDCTDAVTCLNYTVAKLSFLAKEWH------VSVSSDNKDFKYSCMK 265
F+ ++ + C V K +F +++ V +S++ + SC
Sbjct: 452 FQ-----VEVLGGVACHPKKESEEDGVNKRTFRVQDYEQNRSFVVVWNSESSEVVCSCRL 506
Query: 266 MESRGLACEHIVDVLAHLRIEELS--ECFIFKRWSKGAKDGVYSGKNIENDFWDFQKSSR 323
E +G ELS ++ KRW+K AK S + +E+D D + S++
Sbjct: 507 FELKG----------------ELSIPSQYVLKRWTKDAK----SREVMESDQTDVE-STK 545
Query: 324 CSALMDL 330
DL
Sbjct: 546 AQRYKDL 552
Score = 32.7 bits (73), Expect = 0.43
Identities = 12/30 (40%), Positives = 19/30 (63%)
Query: 32 INSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
+ SL+++FW D GR Y + DV++ D T
Sbjct: 183 MQSLRNIFWVDAKGRFDYTCFSDVVSIDTT 212
>At1g10240 unknown protein
Length = 680
Score = 45.8 bits (107), Expect = 5e-05
Identities = 43/218 (19%), Positives = 86/218 (38%), Gaps = 26/218 (11%)
Query: 92 FRKKWDELVLNLGLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHV 151
F W ++V + GL N + Y RS+W +LR F AG T R + + + + +
Sbjct: 403 FELGWRDMVNSFGLHTNRHINNLYASRSLWSLPYLRSHFLAGMTLTGRSKAINAFIQRFL 462
Query: 152 HSRYNLTDFLQQLQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLL 211
++ L F++Q+ V + +D M+ NL+++ + ++ +
Sbjct: 463 SAQTRLAHFVEQVAVVV------DFKDQATE---QQTMQQNLQNISLKTGAPMESHAASV 513
Query: 212 FRPVLQHAPLMKVLDCTDAVTCLNYTVAKL--SFLAKEWHVSVSSDNKDF--------KY 261
P + L + L V +Y ++ +L + H + K +
Sbjct: 514 LTP-FAFSKLQEQL-----VLAAHYASFQMDEGYLVRH-HTKLDGGRKVYWVPQEGIISC 566
Query: 262 SCMKMESRGLACEHIVDVLAHLRIEELSECFIFKRWSK 299
SC E G C H + VL+ ++ + ++ RW +
Sbjct: 567 SCQLFEFSGFLCRHALRVLSTGNCFQVPDRYLPLRWRR 604
Score = 35.0 bits (79), Expect = 0.087
Identities = 14/61 (22%), Positives = 32/61 (51%)
Query: 3 KNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVTE 62
+++++ ++ + KD N+ E D + L+++ W SY+++GD + FD T
Sbjct: 225 EDENIDFLRMCQSIKEKDPNFKFEFTLDANDKLENIAWSYASSIQSYELFGDAVVFDTTH 284
Query: 63 K 63
+
Sbjct: 285 R 285
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 38.1 bits (87), Expect = 0.010
Identities = 44/206 (21%), Positives = 79/206 (37%), Gaps = 7/206 (3%)
Query: 92 FRKKWDELVLNLGLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHV 151
F ++WD LV GL + Y R+ W+ +R F A T+ S L + V
Sbjct: 433 FEQQWDLLVTRFGLVPDRHAALLYSCRASWLPCCIREHFVAQTMTSEFNLSIDSFLKRVV 492
Query: 152 HSRYNLTDFLQQLQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLL 211
+ L++ + ++ + P++KT + +E A T F
Sbjct: 493 DGATCMQLLLEE--SALQVSAAASLAKQILPRFTYPSLKTCMP-MEDHARGILTPYAF-- 547
Query: 212 FRPVLQHAPLMKVLDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGL 271
VLQ+ ++ V + + V + E V + +N++ + SC + E G+
Sbjct: 548 --SVLQNEMVLSVQYAVAEMANGPFIVHHYKKMEGECCVIWNPENEEIQCSCKEFEHSGI 605
Query: 272 ACEHIVDVLAHLRIEELSECFIFKRW 297
C H + VL + E + RW
Sbjct: 606 LCRHTLRVLTVKNCFHIPEQYFLLRW 631
Score = 32.3 bits (72), Expect = 0.57
Identities = 13/59 (22%), Positives = 33/59 (55%)
Query: 3 KNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
++D++ ++ LA +D ++ + +D+ ++++ W G Y ++GDV+ FD +
Sbjct: 255 ESDTLELLECCKGLAERDMDFVYDCTSDENQKVENIAWAYGDSVRGYSLFGDVVVFDTS 313
>At4g18410 MuDR transposable element - like protein
Length = 633
Score = 31.6 bits (70), Expect = 0.97
Identities = 17/68 (25%), Positives = 31/68 (45%), Gaps = 5/68 (7%)
Query: 262 SCMKMESRGLACEHIVDVLAHLRIEE---LSECFIFKRWSKGAKDGVYSGKNIENDFWDF 318
+C+K + G+ CEH V+ H +++ + + F W K DG++ + + FW
Sbjct: 491 TCIKWQICGIPCEHAYGVILHKKLQPEDFVCQWFRTAMWKKNYTDGLFPQRGPK--FWPE 548
Query: 319 QKSSRCSA 326
R A
Sbjct: 549 SNGPRVFA 556
>At2g37420 putative kinesin heavy chain
Length = 1022
Score = 31.2 bits (69), Expect = 1.3
Identities = 27/86 (31%), Positives = 38/86 (43%), Gaps = 7/86 (8%)
Query: 303 DGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNT-AADFNDATQKASELIEQSKA 361
D Y KNI+N QK S+ L DLY L + + AA + A E Q +
Sbjct: 382 DYAYRAKNIKNKPEANQKLSKAVLLKDLYLELERMKEDVRAARDKNGVYIAHERYTQEEV 441
Query: 362 KKSC------QREGSLSLSQTNILNL 381
+K Q E L+LS++N +L
Sbjct: 442 EKKARIERIEQLENELNLSESNFRDL 467
>At3g54860 vacuolar protein sorting protein 33 (VPS33)
Length = 592
Score = 30.4 bits (67), Expect = 2.2
Identities = 30/150 (20%), Positives = 67/150 (44%), Gaps = 10/150 (6%)
Query: 141 EGFQSELGKHVHSRYNLTDFLQQLQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSA 200
E Q+E K V+ + F++ + + +++ + I+ D + ++ VP E +
Sbjct: 70 EPVQTECTKVVYLVRSQLSFMKFIASHIQNDIAKAIQRDYY-VYFVPRRSVACEKILEQE 128
Query: 201 TKH--FTNNVFLLFRPVLQHAPLMKVLDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKD 258
H T F L+ L + L+ ++ ++ V+ L WH++ + +
Sbjct: 129 KVHNLVTVKEFPLYMVPLDEDVISFELELSEKDCLVDGDVSSL------WHIAKAIHELE 182
Query: 259 FKYSCM-KMESRGLACEHIVDVLAHLRIEE 287
F + + KM ++G A + D+L +++EE
Sbjct: 183 FSFGVISKMRAKGKASVRVADILNRMQVEE 212
>At2g26610 unknown protein
Length = 852
Score = 30.0 bits (66), Expect = 2.8
Identities = 29/102 (28%), Positives = 45/102 (43%), Gaps = 17/102 (16%)
Query: 112 KETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHM 171
KE EK SI + + L + C E F +G V S LT + QL + ++++
Sbjct: 317 KECAEKFSIHLCSSLGEQVHVLCAI----EEFMPTIGNSVPSPSLLTLLISQLDSTLQNI 372
Query: 172 RFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFR 213
R +IH L+S E+ + F N+ FLLF+
Sbjct: 373 R---------TIHS----DALLDSSELEISFDFNNDAFLLFK 401
>At2g24660 putative retroelement pol polyprotein
Length = 1156
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/27 (48%), Positives = 17/27 (62%)
Query: 157 LTDFLQQLQNCVKHMRFREIEDDCHSI 183
L L++L N V H R + IE DCHS+
Sbjct: 1052 LRKLLKELANPVFHERTKHIESDCHSV 1078
>At2g39300 hypothetical protein
Length = 768
Score = 28.9 bits (63), Expect = 6.3
Identities = 29/110 (26%), Positives = 48/110 (43%), Gaps = 11/110 (10%)
Query: 321 SSRCSALMDLYRALGYLN---SNTAADFNDATQKASELIEQSKAKKSCQREGSLSLSQTN 377
SSRC ++ G ++ SN ++ D E +E+SK K SLS S+
Sbjct: 78 SSRCLTPERQFKEYGSMSTCSSNVSSQVLDRYIDGEEHLERSKQKSGSLHSSSLSGSRRR 137
Query: 378 I-------LNLKDPIRLRRKGRGTKNKSSLGLTVKHVINCLIFKTPGHNK 420
+ L + + +RK +G ++ S+ L + VI L T G +K
Sbjct: 138 LPPRAQSPSPLSESGKDKRKSKGLRDASARSL-ARSVIERLSHNTQGKSK 186
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,550,607
Number of Sequences: 26719
Number of extensions: 454076
Number of successful extensions: 1063
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1017
Number of HSP's gapped (non-prelim): 38
length of query: 463
length of database: 11,318,596
effective HSP length: 103
effective length of query: 360
effective length of database: 8,566,539
effective search space: 3083954040
effective search space used: 3083954040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0023.16


