

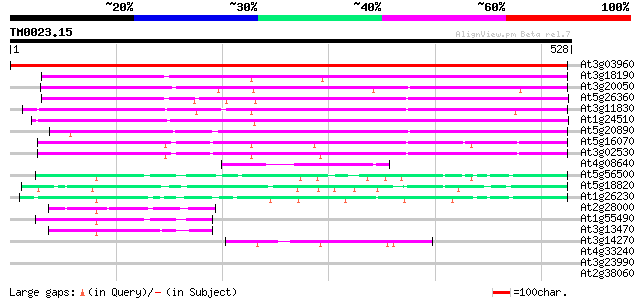
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0023.15
(528 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g03960 putative T-complex protein 1, theta subunit (TCP-1-Theta) 887 0.0
At3g18190 chaperonin subunit, putative 247 1e-65
At3g20050 t-complex polypeptide 1 homologue 234 7e-62
At5g26360 chaperonin gamma chain - like protein 224 7e-59
At3g11830 putative T-complex protein 1, ETA subunit 218 9e-57
At1g24510 T-complex chaperonin protein , epsilon subunit 203 2e-52
At5g20890 T-complex protein 1, beta subunit 192 5e-49
At5g16070 TCP-1 chaperonin-like protein 164 1e-40
At3g02530 putative chaperonin 162 3e-40
At4g08640 hypothetical protein 97 2e-20
At5g56500 RuBisCO subunit binding-protein beta subunit precursor... 61 2e-09
At5g18820 chaperonin 60 alpha chain - like protein 61 2e-09
At1g26230 chaperonin precursor, putative 60 2e-09
At2g28000 putative rubisco subunit binding-protein alpha subunit 52 8e-07
At1g55490 Rubisco subunit binding-protein beta subunit 50 3e-06
At3g13470 chaperonin 60 beta - like protein 47 3e-05
At3g14270 unknown protein 42 8e-04
At4g33240 putative protein 34 0.17
At3g23990 mitochondrial chaperonin hsp60 33 0.39
At2g38060 putative Na+-dependent inorganic phosphate cotransporter 33 0.39
>At3g03960 putative T-complex protein 1, theta subunit (TCP-1-Theta)
Length = 549
Score = 887 bits (2292), Expect = 0.0
Identities = 442/525 (84%), Positives = 493/525 (93%)
Query: 1 MGFNMQSYGIQSMLKEGHKHLSGLDEAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLD 60
+G +MQ YGIQSMLKEG++HLSGLDEAVIKNI+ACK+LSTITRTSLGPNGMNKMVINHLD
Sbjct: 2 VGMSMQPYGIQSMLKEGYRHLSGLDEAVIKNIEACKELSTITRTSLGPNGMNKMVINHLD 61
Query: 61 KLFVTNDAATIVNELEVQHPAAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRM 120
KLFVTNDAATIVNELE+QHPAAK+LVLA KAQQEEIGDGANLTISFAGELLQ AEELIRM
Sbjct: 62 KLFVTNDAATIVNELEIQHPAAKLLVLAAKAQQEEIGDGANLTISFAGELLQNAEELIRM 121
Query: 121 GLHPSEIISGYTKAIAKTIEILDELVEKGSESMDVRDKVQVISRMRAAVASKQFGQEDTI 180
GLHPSEIISGYTKA++K +EIL++LVE GSE+MDVR+K +VISRMRAAVASKQFGQE+ I
Sbjct: 122 GLHPSEIISGYTKAVSKAVEILEQLVETGSETMDVRNKDEVISRMRAAVASKQFGQEEII 181
Query: 181 CSLVADACIQVCPKNPANFNVDNVRVAKLLGGGLHNSTVVRGMVLKSDAVGSIKRIEKAK 240
CSLV DACIQVCPKNP NFNVDNVRV+KLLGGGLHNS +VRGMVLKSDAVGSIKR+EKAK
Sbjct: 182 CSLVTDACIQVCPKNPTNFNVDNVRVSKLLGGGLHNSCIVRGMVLKSDAVGSIKRMEKAK 241
Query: 241 VAVFVSGVDTSATDTKGTVLIHSAEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVG 300
VAVF GVDT+AT+TKGTVLIHSAEQLENY+KTEEAKVEELIKAVA+SGAKVIVSGG++G
Sbjct: 242 VAVFAGGVDTTATETKGTVLIHSAEQLENYAKTEEAKVEELIKAVAESGAKVIVSGGSIG 301
Query: 301 EMALHFCERYKLMVLKISSKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGV 360
EMALHFCERYK+MVLKISSKFELRRFCRT GAVA LKLS+P+P+DLGYVDS+SVEE+GGV
Sbjct: 302 EMALHFCERYKIMVLKISSKFELRRFCRTAGAVAHLKLSRPSPEDLGYVDSISVEEIGGV 361
Query: 361 TVTIVKNEEGGNSVSTVVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIE 420
TVTI +NEEGGNS+STVVLRGSTDSILDDLERAVDDGVNTYKAMC+DSR VPGAAATEIE
Sbjct: 362 TVTIARNEEGGNSISTVVLRGSTDSILDDLERAVDDGVNTYKAMCRDSRIVPGAAATEIE 421
Query: 421 LAKRVKDFSFKETGLDQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKV 480
LA+R+K+++ E GLD+YAI K+AESFE +P+TLA+NAGLNAMEII++LY H SGNTK+
Sbjct: 422 LAQRLKEYANAEIGLDKYAITKYAESFEFVPKTLADNAGLNAMEIIAALYTGHGSGNTKL 481
Query: 481 GIDLEEGVCKDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
GIDLEEG CKDV+ T VWDL TK FALKYA+DA CTVLRVDQ+I
Sbjct: 482 GIDLEEGACKDVSETKVWDLFATKLFALKYASDAACTVLRVDQII 526
>At3g18190 chaperonin subunit, putative
Length = 536
Score = 247 bits (630), Expect = 1e-65
Identities = 145/505 (28%), Positives = 267/505 (52%), Gaps = 14/505 (2%)
Query: 31 NIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILVLAGK 90
NI++ + +S RTSLGP GM+KM+ ++ +TND ATI+N++EV PAAK+LV K
Sbjct: 33 NINSARAVSDAVRTSLGPKGMDKMISTANGEVIITNDGATILNKMEVLQPAAKMLVELSK 92
Query: 91 AQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDELVEKGS 150
+Q GDG + AG LL+ + L+ G+HP+ I KA K I+IL + +
Sbjct: 93 SQDSAAGDGTTTVVVIAGALLKECQSLLTNGIHPTVISDSLHKACGKAIDILTAM----A 148
Query: 151 ESMDVRDKVQVISRMRAAVASKQFGQEDTICS-LVADACIQVC-PKNPANFNVDNVRVAK 208
+++ D+ ++ ++ SK Q T+ + L DA + V P+ P ++ ++++ K
Sbjct: 149 VPVELTDRDSLVKSASTSLNSKVVSQYSTLLAPLAVDAVLSVIDPEKPEIVDLRDIKIVK 208
Query: 209 LLGGGLHNSTVVRGMVLK---SDAVGSIKRIEKAKVAVFVSGVDTSATDTKGTVLIHSAE 265
LGG + ++ V+G+V S A G R+E AK+AV + TD + ++++
Sbjct: 209 KLGGTVDDTHTVKGLVFDKKVSRAAGGPTRVENAKIAVIQFQISPPKTDIEQSIVVSDYT 268
Query: 266 QLENYSKTEEAKVEELIKAVADSGAKVI-----VSGGAVGEMALHFCERYKLMVLKISSK 320
Q++ K E + +IK + +G V+ + AV +++LH+ + K+MV+K +
Sbjct: 269 QMDRILKEERNYILGMIKKIKATGCNVLLIQKSILRDAVTDLSLHYLAKAKIMVIKDVER 328
Query: 321 FELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTVVLR 380
E+ +T + + + + LG+ D V +G + + + ++V++R
Sbjct: 329 DEIEFVTKTLNCLPIANIEHFRAEKLGHADLVEEASLGDGKILKITGIKDMGRTTSVLVR 388
Query: 381 GSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLDQYAI 440
GS +LD+ ER++ D + + + + G A EIEL++++ ++ G++ Y +
Sbjct: 389 GSNQLVLDEAERSLHDALCVVRCLVSKRFLIAGGGAPEIELSRQLGAWAKVLHGMEGYCV 448
Query: 441 AKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKDVTTTHVWDL 500
FAE+ E+IP TLAENAGLN + I++ L +HA G GI++ +G ++ +V
Sbjct: 449 KSFAEALEVIPYTLAENAGLNPIAIVTELRNKHAQGEINAGINVRKGQITNILEENVVQP 508
Query: 501 HVTKFFALKYAADAVCTVLRVDQVI 525
+ A+ A + V +L++D ++
Sbjct: 509 LLVSTSAITLATECVRMILKIDDIV 533
>At3g20050 t-complex polypeptide 1 homologue
Length = 545
Score = 234 bits (598), Expect = 7e-62
Identities = 151/516 (29%), Positives = 271/516 (52%), Gaps = 24/516 (4%)
Query: 30 KNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILVLAG 89
+N+ AC+ +S I +TSLGP G++KM+++ + + +TND ATI+ LEV+HPAAK+LV
Sbjct: 23 QNVMACQAVSNIVKTSLGPVGLDKMLVDDIGDVTITNDGATILRMLEVEHPAAKVLVELA 82
Query: 90 KAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDELVEKG 149
+ Q E+GDG + A ELL+ A +L+R +HP+ IISGY A+ ++ + ++E +
Sbjct: 83 ELQDREVGDGTTSVVIVAAELLKRANDLVRNKIHPTSIISGYRLAMRESCKYIEEKLVTK 142
Query: 150 SESMDVRDKVQVISRMRAAVASKQF-GQEDTICSLVADACIQVCPKN---PANFNVDNVR 205
E + KV +I+ + +++SK G D +LV +A + V N + + +
Sbjct: 143 VEKL---GKVPLINCAKTSMSSKLISGDSDFFANLVVEAVLSVKMTNQRGEIKYPIKGIN 199
Query: 206 VAKLLGGGLHNSTVVRGMVLKSD--AVGSIKRIEKAKVAVFVSGVDTSATDTKGTVLIHS 263
+ K G +S ++ G L + A G R+ AK+A + + V+++
Sbjct: 200 ILKAHGQSARDSYLLNGYALNTGRAAQGMPLRVSPAKIACLDFNLQKTKMQLGVQVVVND 259
Query: 264 AEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSKFEL 323
+LE + E +E I+ + +GA VI++ + +MAL + + ++ K ++
Sbjct: 260 PRELEKIRQREADMTKERIEKLLKAGANVILTTKGIDDMALKYFVEAGAIAVRRVRKEDM 319
Query: 324 RRFCRTTGAVAVLKLSQP------NPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTV 377
R + TGA V + +P LG D V E + V ++K + ++VS +
Sbjct: 320 RHVAKATGATLVTTFADMEGEETFDPAHLGSADEVVEERIADDDVILIKGTKTSSAVS-L 378
Query: 378 VLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLDQ 437
+LRG+ D +LD++ERA+ D + K + + V G A E L+ ++ + +Q
Sbjct: 379 ILRGANDYMLDEMERALHDALCIVKRTLESNTVVAGGGAVESALSVYLEHLATTLGSREQ 438
Query: 438 YAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTK--------VGIDLEEGVC 489
AIA+FA++ +IP+ LA NA +A E+++ L A H + TK +G+DL G
Sbjct: 439 LAIAEFADALLIIPKVLAVNAAKDATELVAKLRAYHHTAQTKADKKHYSSMGLDLVNGTI 498
Query: 490 KDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
++ V + ++K +++A +A T+LR+D +I
Sbjct: 499 RNNLEAGVIEPAMSKVKIIQFATEAAITILRIDDMI 534
>At5g26360 chaperonin gamma chain - like protein
Length = 555
Score = 224 bits (572), Expect = 7e-59
Identities = 146/508 (28%), Positives = 262/508 (50%), Gaps = 19/508 (3%)
Query: 31 NIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILVLAGK 90
NI A K ++ I RT+LGP M KM+++ + VTND I+ EL+V HPAAK ++ +
Sbjct: 24 NIQASKAVADIIRTTLGPRSMLKMLLDAGGGIVVTNDGNAILRELDVAHPAAKSMIELSR 83
Query: 91 AQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDELVEKGS 150
Q EE+GDG I AGE+L AE + HP+ I Y KA+ +I +LD++ +
Sbjct: 84 TQDEEVGDGTTSVIVLAGEMLHVAEAFLEKNYHPTVICRAYIKALEDSIAVLDKI----A 139
Query: 151 ESMDVRDKVQVISRMRAAVASK---QFGQEDTICSLVADACIQV-CPKNPANFNVD---N 203
S+D+ D+ QV+ +++ + +K QFG D I L DA V VD
Sbjct: 140 MSIDINDRSQVLGLVKSCIGTKFTSQFG--DLIADLAIDATTTVGVDLGQGLREVDIKKY 197
Query: 204 VRVAKLLGGGLHNSTVVRGMVLKSDAV--GSIKR-IEKAKVAVFVSGVDTSATDTKGTVL 260
++V K+ GG +S V++G++ D V G +KR I ++ + ++ + +
Sbjct: 198 IKVEKVPGGQFEDSEVLKGVMFNKDVVAPGKMKRKIVNPRIILLDCPLEYKKGENQTNAE 257
Query: 261 IHSAEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSK 320
+ E E K EE +E + + ++++ + ++A H+ + + ++ K
Sbjct: 258 LVREEDWEVLLKLEEEYIENICVQILKFKPDLVITEKGLSDLACHYFSKAGVSAIRRLRK 317
Query: 321 FELRRFCRTTGAVAVLKLSQPNPDDLGY-VDSVSVEEVGGVTVTIVKNEEGGNSVSTVVL 379
+ R + GAV V + + D+G V+++G + + + + + TV+L
Sbjct: 318 TDNNRIAKACGAVIVNRPDELQESDIGTGAGLFEVKKIGDDFFSFIVDCKEPKA-CTVLL 376
Query: 380 RGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLDQYA 439
RG + ++++ER + D ++ + + K+ + VPG ATE+ ++ +K S G++++
Sbjct: 377 RGPSKDFINEVERNLQDAMSVARNIIKNPKLVPGGGATELTVSATLKQKSATIEGIEKWP 436
Query: 440 IAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASG-NTKVGIDLEEGVCKDVTTTHVW 498
A +FE IPRTLA+N G+N + +++L +HA+G N GID G D+ + +W
Sbjct: 437 YEAAAIAFEAIPRTLAQNCGVNVIRTMTALQGKHANGENAWTGIDGNTGAIADMKESKIW 496
Query: 499 DLHVTKFFALKYAADAVCTVLRVDQVIN 526
D + K K A +A C +LR+D +++
Sbjct: 497 DSYNVKAQTFKTAIEAACMLLRIDDIVS 524
>At3g11830 putative T-complex protein 1, ETA subunit
Length = 557
Score = 218 bits (554), Expect = 9e-57
Identities = 141/523 (26%), Positives = 255/523 (47%), Gaps = 17/523 (3%)
Query: 13 MLKEGHKHLSGLDEAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIV 72
+LKEG G + ++ NI+AC + + RT+LGP GM+K++ + + ++ND ATI+
Sbjct: 11 LLKEGTDTSQGKAQ-LVSNINACTAVGDVVRTTLGPRGMDKLIHDDKGSVTISNDGATIM 69
Query: 73 NELEVQHPAAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYT 132
L++ HPAAKILV K+Q E+GDG + A E L+ A+ I G+H +I Y
Sbjct: 70 KLLDIVHPAAKILVDIAKSQDSEVGDGTTTVVLLAAEFLKEAKPFIEDGVHAQNLIRSYR 129
Query: 133 KAIAKTIEILDELVEKGSESMDVRDKVQVISRMRAAVASKQF--GQEDTICSLVADACIQ 190
A I + EL E V +K ++++ A S + G+++ ++V DA +
Sbjct: 130 TASTLAIAKVKELA-VSIEGKSVEEKKGLLAKCAATTLSSKLIGGEKEFFATMVVDAVMA 188
Query: 191 VCPKNPANFNVDNVRVAKLLGGGLHNSTVVRGMVLK-----SDAVGSIKRIEKAKVAVFV 245
+ + N + + K+ GG + +S +V G+ K + K+ K+ +
Sbjct: 189 IGNDDRLNL----IGIKKVPGGNMRDSFLVDGVAFKKTFSYAGFEQQPKKFLNPKILLLN 244
Query: 246 SGVDTSATDTKGTVLIHSAEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALH 305
++ + + + Q ++ E + + + +SGAKV++S A+G++A
Sbjct: 245 IELELKSEKENAEIRLSDPSQYQSIVDAEWNIIYDKLDKCVESGAKVVLSRLAIGDLATQ 304
Query: 306 FCERYKLMVLKISSKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIV 365
+ + ++ +L R G ++ + LG + ++VGG I
Sbjct: 305 YFADRDIFCAGRVAEEDLNRVAAAAGGTVQTSVNNIIDEVLGTCEIFEEKQVGGERFNIF 364
Query: 366 KNEEGGNSVSTVVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRV 425
G + +T+VLRG D +++ ER++ D + + K+S VPG A ++E++K +
Sbjct: 365 SGCPSGRT-ATIVLRGGADQFIEEAERSLHDAIMIVRRAVKNSTVVPGGGAIDMEISKYL 423
Query: 426 KDFSFKETGLDQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHA--SG-NTKVGI 482
+ S G Q I +A++ E+IPR L +NAG +A ++++ L +HA SG G+
Sbjct: 424 RQHSRTIAGKSQLFINSYAKALEVIPRQLCDNAGFDATDVLNKLRQKHAMQSGEGASYGV 483
Query: 483 DLEEGVCKDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
D+ G D VW+ V K A+ A +A C +L VD+ +
Sbjct: 484 DINTGGIADSFANFVWEPAVVKINAINAATEAACLILSVDETV 526
>At1g24510 T-complex chaperonin protein , epsilon subunit
Length = 535
Score = 203 bits (517), Expect = 2e-52
Identities = 140/511 (27%), Positives = 250/511 (48%), Gaps = 8/511 (1%)
Query: 21 LSGLDEAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHP 80
L G+D A NI A K ++ I R+SLGP GM+KM+ + +TND ATI+ +++V +
Sbjct: 24 LRGID-AQKANIAAGKAVARILRSSLGPKGMDKMLQGPDGDITITNDGATILEQMDVDNQ 82
Query: 81 AAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIE 140
AK++V ++Q EIGDG + AG LL+ AE + G+HP I GY A +E
Sbjct: 83 IAKLMVELSRSQDYEIGDGTTGVVVMAGALLEQAERQLDRGIHPIRIAEGYEMASRVAVE 142
Query: 141 ILDELVEKGSESMDVRDKVQVISRMRAAVASKQFGQ-EDTICSLVADACIQVCPKNPANF 199
L+ + +K DV + ++ ++SK + + ++ + A + V +
Sbjct: 143 HLERIAQK--FEFDVNNYEPLVQTCMTTLSSKIVNRCKRSLAEIAVKAVLAVADLERRDV 200
Query: 200 NVDNVRVAKLLGGGLHNSTVVRGMVLKSDA--VGSIKRIEKAKVAVFVSGVDTSATDTKG 257
N+D ++V +GG L ++ ++ G+++ D K+IE A +A+ + TK
Sbjct: 201 NLDLIKVEGKVGGKLEDTELIYGILIDKDMSHPQMPKQIEDAHIAILTCPFEPPKPKTKH 260
Query: 258 TVLIHSAEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKI 317
V I + E+ E K E+ +E+++ D GA +++ + A H L ++
Sbjct: 261 KVDIDTVEKFETLRKQEQQYFDEMVQKCKDVGATLVICQWGFDDEANHLLMHRNLPAVRW 320
Query: 318 SSKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVS-T 376
EL TG V + + P+ LG V + G ++ E NS + T
Sbjct: 321 VGGVELELIAIATGGRIVPRFQELTPEKLGKAGVVREKSFGTTKERMLYIEHCANSKAVT 380
Query: 377 VVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLD 436
V +RG ++++ +R++ D + + + ++ V G A EI + V + K G++
Sbjct: 381 VFIRGGNKMMIEETKRSIHDALCVARNLIRNKSIVYGGGAAEIACSLAVDAAADKYPGVE 440
Query: 437 QYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTK-VGIDLEEGVCKDVTTT 495
QYAI FAE+ + +P LAEN+GL +E +S++ ++ N GID + D+
Sbjct: 441 QYAIRAFAEALDSVPMALAENSGLQPIETLSAVKSQQIKENIPFYGIDCNDVGTNDMREQ 500
Query: 496 HVWDLHVTKFFALKYAADAVCTVLRVDQVIN 526
+V++ + K + A V +L++D VI+
Sbjct: 501 NVFETLIGKQQQILLATQVVKMILKIDDVIS 531
>At5g20890 T-complex protein 1, beta subunit
Length = 527
Score = 192 bits (487), Expect = 5e-49
Identities = 133/493 (26%), Positives = 237/493 (47%), Gaps = 11/493 (2%)
Query: 38 LSTITRTSLGPNGMNKMV--INHLDKLFVTNDAATIVNELEVQHPAAKILVLAGKAQQEE 95
+S + +++LGP GM+K++ + VTND ATI+ L + +PAAK+LV K Q +E
Sbjct: 30 ISDLVKSTLGPKGMDKILQSTGRGHAVTVTNDGATILKSLHIDNPAAKVLVDISKVQDDE 89
Query: 96 IGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDELVEKGSESMDV 155
+GDG + AGELL+ AE+L+ +HP II+GY A L + V ++ +
Sbjct: 90 VGDGTTSVVVLAGELLREAEKLVASKIHPMTIIAGYRMASECARNALLKRVIDNKDNAE- 148
Query: 156 RDKVQVISRMRAAVASKQFGQE-DTICSLVADACIQVCPKNPANFNVDNVRVAKLLGGGL 214
+ + ++ + SK Q+ + + DA ++ + N++ +++ K GG L
Sbjct: 149 KFRSDLLKIAMTTLCSKILSQDKEHFAEMAVDAVFRL----KGSTNLEAIQIIKKPGGSL 204
Query: 215 HNSTVVRGMVL-KSDAVGSIKRIEKAKVAVFVSGVDTSATDTKGT-VLIHSAEQLENYSK 272
+S + G +L K +G KRIE A + V + +DT G V + S ++
Sbjct: 205 KDSFLDEGFILDKKIGIGQPKRIENANILVANTAMDTDKVKIYGARVRVDSMTKVAEIEG 264
Query: 273 TEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSKFELRRFCRTTGA 332
E+ K+++ +K + G V+ + ++ ++ + + R TG
Sbjct: 265 AEKEKMKDKVKKIIGHGINCFVNRQLIYNFPEELFADAGILAIEHADFEGIERLGLVTGG 324
Query: 333 VAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTVVLRGSTDSILDDLER 392
P LG+ + +G + E G + S +VLRG++ +LD+ ER
Sbjct: 325 EIASTFDNPESVKLGHCKLIEEIMIGEDKLIHFSGCEMGQACS-IVLRGASHHVLDEAER 383
Query: 393 AVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLDQYAIAKFAESFEMIPR 452
++ D + D+R + G E+ +AK V + + K G +AI F+ + IP
Sbjct: 384 SLHDALCVLSQTVNDTRVLLGGGWPEMVMAKEVDELARKTAGKKSHAIEAFSRALVAIPT 443
Query: 453 TLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKDVTTTHVWDLHVTKFFALKYAA 512
T+A+NAGL++ E+++ L AEH + GID+ G D+ +++ K L A
Sbjct: 444 TIADNAGLDSAELVAQLRAEHHTEGCNAGIDVITGAVGDMEERGIYEAFKVKQAVLLSAT 503
Query: 513 DAVCTVLRVDQVI 525
+A +LRVD++I
Sbjct: 504 EASEMILRVDEII 516
>At5g16070 TCP-1 chaperonin-like protein
Length = 535
Score = 164 bits (414), Expect = 1e-40
Identities = 132/516 (25%), Positives = 241/516 (46%), Gaps = 26/516 (5%)
Query: 27 AVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILV 86
A+ I+A K L + +++LGP G KM++ + +T D T++ E+++Q+P A ++
Sbjct: 18 ALHMTINAAKGLQDVLKSNLGPKGTIKMLVGGSGDIKLTKDGNTLLKEMQIQNPTAIMIA 77
Query: 87 LAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDEL- 145
AQ + GDG T+ F GEL++ +E I G+HP ++ G+ A T++ LD
Sbjct: 78 RTAVAQDDISGDGTTSTVIFIGELMKQSERCIDEGMHPRVLVDGFEIAKRATLQFLDNFK 137
Query: 146 --VEKGSESMDVRDKVQVISRMRAAVASKQF-GQEDTICSLVADACIQVCPKNPAN-FNV 201
V G E DK + R + +K + G D + +V ++ + C + P ++
Sbjct: 138 TPVVMGDEV----DKEILKMVARTTLRTKLYEGLADQLTDIVVNSVL--CIRKPEEAIDL 191
Query: 202 DNVRVAKLLGGGLHNSTVVRGMVLK--SDAVGSIKRIEKAKVAVFVSGVDTSATDTKGTV 259
V + + ++ +V G+VL S +R E + ++ ++
Sbjct: 192 FMVEIMHMRHKFDVDTRLVEGLVLDHGSRHPDMKRRAENCHILTCNVSLEYEKSEINAGF 251
Query: 260 LIHSAEQLENYSKTEEAKVEELIKAV--------ADSGAKVIVSGGAVGEMALHFCERYK 311
+AEQ E E V+E +K + D+ V+++ + +L R
Sbjct: 252 FYSNAEQREAMVTAERRSVDERVKKIIELKKKVCGDNDNFVVINQKGIDPPSLDLLAREG 311
Query: 312 LMVLKISSKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGG 371
++ L+ + + + R G AV + P+ LG+ V +G T V+ +
Sbjct: 312 IIGLRRAKRRNMERLVLACGGEAVNSVDDLTPESLGWAGLVYEHVLGEEKYTFVEQVKNP 371
Query: 372 NSVSTVVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFK 431
NS T++++G D + ++ AV DG+ + K +D V GA A E+ + + + K
Sbjct: 372 NS-CTILIKGPNDHTIAQIKDAVRDGLRSVKNTIEDECVVLGAGAFEVAARQHLLN-EVK 429
Query: 432 ET--GLDQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVC 489
+T G Q + FA + ++P+TLAENAGL+ ++I SL +EH GN VG++L++G
Sbjct: 430 KTVQGRAQLGVEAFANALLVVPKTLAENAGLDTQDVIISLTSEHDKGNV-VGLNLQDGEP 488
Query: 490 KDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
D ++D + K + +L VD+VI
Sbjct: 489 IDPQLAGIFDNYSVKRQLINSGPVIASQLLLVDEVI 524
>At3g02530 putative chaperonin
Length = 535
Score = 162 bits (411), Expect = 3e-40
Identities = 131/516 (25%), Positives = 238/516 (45%), Gaps = 25/516 (4%)
Query: 27 AVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILV 86
A+ I+A K L + +++LGP G KM++ + +T D T++ E+++Q+P A ++
Sbjct: 18 ALHMTINAAKGLQDVLKSNLGPKGTIKMLVGGSGDIKLTKDGNTLLKEMQIQNPTAIMIA 77
Query: 87 LAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDEL- 145
AQ + GDG T+ F GEL++ +E I G+HP ++ G+ A T++ LD
Sbjct: 78 RTAVAQDDISGDGTTSTVIFIGELMKQSERCIDEGMHPRVLVDGFEIAKRATLQFLDTFK 137
Query: 146 --VEKGSESMDVRDKVQVISRMRAAVASKQF-GQEDTICSLVADACIQVCPKNPAN-FNV 201
V G E DK + R + +K + G D + +V ++ + C + P ++
Sbjct: 138 TPVVMGDEP----DKEILKMVARTTLRTKLYEGLADQLTDIVVNSVL--CIRKPQEPIDL 191
Query: 202 DNVRVAKLLGGGLHNSTVVRGMVLK--SDAVGSIKRIEKAKVAVFVSGVDTSATDTKGTV 259
V + + ++ +V G+VL S +R E + ++ ++
Sbjct: 192 FMVEIMHMRHKFDVDTRLVEGLVLDHGSRHPDMKRRAENCHILTCNVSLEYEKSEINAGF 251
Query: 260 LIHSAEQLENYSKTEEAKVEELIKAVADSGAK---------VIVSGGAVGEMALHFCERY 310
+AEQ E E V+E ++ + + K VI++ + +L R
Sbjct: 252 FYSNAEQREAMVTAERRSVDERVQKIIELKNKVCAGNDNSFVILNQKGIDPPSLDLLARE 311
Query: 311 KLMVLKISSKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEG 370
++ L+ + + + R G AV + PD LG+ V +G T V+ +
Sbjct: 312 GIIALRRAKRRNMERLVLACGGEAVNSVDDLTPDCLGWAGLVYEHVLGEEKYTFVEQVKN 371
Query: 371 GNSVSTVVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKR-VKDFS 429
+S T++++G D + ++ AV DG+ + K +D V GA A E+ + + +
Sbjct: 372 PHS-CTILIKGPNDHTIAQIKDAVRDGLRSVKNTLEDECVVLGAGAFEVAARQHLINEVK 430
Query: 430 FKETGLDQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVC 489
G Q + FA + ++P+TLAENAGL+ ++I SL +EH GN VG+DL++G
Sbjct: 431 KTVQGRAQLGVEAFANALLVVPKTLAENAGLDTQDVIISLTSEHDKGNI-VGLDLQDGEP 489
Query: 490 KDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
D ++D + K + +L VD+VI
Sbjct: 490 VDPQLAGIFDNYSVKRQLINSGPVIASQLLLVDEVI 525
>At4g08640 hypothetical protein
Length = 214
Score = 97.4 bits (241), Expect = 2e-20
Identities = 61/159 (38%), Positives = 88/159 (54%), Gaps = 29/159 (18%)
Query: 200 NVDNVRVAKLLGGGLHNSTVVRGMVLKSDAVGSIKR-IEKAKVAVFVSGVDTSATDTKGT 258
N+D+V VAK+LG S VV G+V KSD VG+I+ ++ AKV
Sbjct: 4 NIDDVHVAKILGVDSRKSCVVCGIVFKSDDVGNIRHPLKNAKV----------------- 46
Query: 259 VLIHSAEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKIS 318
N+ K++EA +E L+K +A S KVIVS ++ EM L FC+ YK+MVL+IS
Sbjct: 47 ---------HNFGKSKEAMLETLVKDIAFSNVKVIVSRSSICEMTLRFCKIYKIMVLQIS 97
Query: 319 SKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEV 357
S ++ C GA+ + QP+ LGY+DS S+ E+
Sbjct: 98 SDIDINSLCSIVGAIDSSQHFQPH--HLGYMDSTSLSEI 134
>At5g56500 RuBisCO subunit binding-protein beta subunit precursor;
chaperonin, 60 kDa
Length = 596
Score = 60.8 bits (146), Expect = 2e-09
Identities = 124/560 (22%), Positives = 213/560 (37%), Gaps = 104/560 (18%)
Query: 25 DEAVIKNIDA-CKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHP--- 80
D IK + A +L+ + +LGP G N ++ + + ND T+ E+E++ P
Sbjct: 60 DGTAIKKLQAGVNKLADLVGVTLGPKGRNVVLESKYGSPRIVNDGVTVAREVELEDPVEN 119
Query: 81 -AAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTI 139
AK++ A + GDG ++ A L+ +++ G +P I T+ I KT
Sbjct: 120 IGAKLVRQAASKTNDLAGDGTTTSVVLAQGLIAEGVKVVAAGANPVLI----TRGIEKTT 175
Query: 140 EILDELVEKGSESMDVRDKVQVISRMRAAVASKQFGQEDTICSLVADACIQVCPKNPANF 199
+ L ++K S+ ++ + A VA+ G + +++A+A +V K
Sbjct: 176 KALVAELKKMSKEVEDSE--------LADVAAVSAGNNYEVGNMIAEAMAKVGRK----- 222
Query: 200 NVDNVRVAKLLGGGLHNSTVVRGMVLKSDAVGSIKRIEKAKVAVFVSGVDTSATDTKGTV 259
V + K L+ VV GM + + K+ D K T
Sbjct: 223 GVVTLEEGKSAENSLY---VVEGMQFDRGYISPYFVTDSEKMCAEYENCKLFLVDKKITN 279
Query: 260 LIHSAEQLENYSK--------TEEAKVEELIKAVADS--GAKVIVSGGAVGEMALHFCER 309
LE+ K E+ + E L V + G + + A G F ER
Sbjct: 280 ARDIISILEDAIKGGYPLLIIAEDIEQEPLATLVVNKLRGTIKVAALKAPG-----FGER 334
Query: 310 YKLMVLKISSKFELRRFCRTTGAVAV-----LKLSQPNPDDLGYVDSV------------ 352
+ I++ TGA + L+L + P+ LG V
Sbjct: 335 KSQYLDDIAA---------LTGATVIREEVGLQLEKVGPEVLGNAGKVVLTKDTTTIVGD 385
Query: 353 -SVEEVGGVTVTIVKN------------------EEGGNSVSTVVLRGSTDSILDDLERA 393
S EEV V +KN + V+ + + T++ L + +
Sbjct: 386 GSTEEVVKKRVEQIKNLIEAAEQDYEKEKLNERIAKLSGGVAVIQVGAQTETELKEKKLR 445
Query: 394 VDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKET--------GLDQYAIAKFAE 445
V+D +N KA ++ V G T + LA +V + KET G D I K A
Sbjct: 446 VEDALNATKAAVEEG-IVVGGGCTLLRLASKVD--AIKETLANDEEKVGAD---IVKKAL 499
Query: 446 SFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKDVTTTHVWDLHVTKF 505
S+ + + +A+NAG+N + + +S N K G + G +D+ + D
Sbjct: 500 SYPL--KLIAKNAGVNGSVVSEKVL---SSDNPKHGYNAATGKYEDLMAAGIIDPTKVVR 554
Query: 506 FALKYAADAVCTVLRVDQVI 525
L++A+ T L D V+
Sbjct: 555 CCLEHASSVAKTFLMSDCVV 574
>At5g18820 chaperonin 60 alpha chain - like protein
Length = 575
Score = 60.8 bits (146), Expect = 2e-09
Identities = 120/566 (21%), Positives = 217/566 (38%), Gaps = 91/566 (16%)
Query: 12 SMLKEGHKH-LSGLD--EAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDA 68
S+++ G K L G D E + ID +IT LGP G N +V+ D + V ND
Sbjct: 29 SVVRAGAKRILYGKDSREKLQAGIDKLADAVSIT---LGPRGRN-VVLAEKDTIKVINDG 84
Query: 69 ATIVNELE----VQHPAAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHP 124
TI +E +++ A ++ E GDG I A E+++ I G +
Sbjct: 85 VTIAKSIELPDTIENAGATLIQEVAIKMNESAGDGTTTAIILAREMIKAGSLAIAFGANA 144
Query: 125 SEIISGYTKAIAKTIEILDELVEKGSESMDVRDKVQVISRMRAAVASKQFGQEDTICSLV 184
+ +G K + + + +L +S+ V+ K + AVAS G ++ + +L+
Sbjct: 145 VSVKNGMNKTVKELVRVLQ------MKSIPVQGKNDI-----KAVASISAGNDEFVGNLI 193
Query: 185 ADACIQVCPKNPANFNVDNV-RVAKLLGGGLHNSTVVRGMVLKSDAVGSIKRIEKAKVAV 243
A+ ++ P + + + ++ G+ ++ S +KAK+ V
Sbjct: 194 AETVEKIGPDGVISIESSSTSETSVIVEEGMKFDKGYMSPHFITNQEKSTVEFDKAKILV 253
Query: 244 FVSGVDTSATDTKGTV-LIHSAEQLEN----YSKTEEAKVEELIKAVADSG----AKVIV 294
D T K V L+ QL ++ A+V E++ G A V
Sbjct: 254 ----TDQKITSAKELVPLLEKTSQLSVPLLIIAEDISAEVLEILVVNKKQGLINVAVVKC 309
Query: 295 SGGAVGEMAL----------HFCERYKLMVLKISSKFEL---RRFCRTTGAVAVLKLSQP 341
G G+ AL + M L ++ +L RR T + ++ +
Sbjct: 310 PGMLDGKKALLQDIALMTGADYLSGDLGMSLMGATSDQLGVSRRVVITANSTTIVADAST 369
Query: 342 NPD---------------DLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTVVLRGSTDSI 386
P+ D Y+ E + +T GG V+ + + G T++
Sbjct: 370 KPEIQARIAQMKKDLAETDNSYLSKKIAERIAKLT--------GG--VAVIKVGGHTETE 419
Query: 387 LDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIEL-------AKRVKDFSFKETGLDQYA 439
L+D + ++D N A ++ VPG AT I L K + + S+++ G D A
Sbjct: 420 LEDRKLRIEDAKNATFAAMREG-IVPGGGATYIHLLDEIPRIKKNLMEDSYEQIGADIVA 478
Query: 440 IAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKDVTTTHVWD 499
+A A + +A NAG++ ++ + + G + G +D+ + D
Sbjct: 479 MALTAPAM-----AIATNAGVDGSVVVQ----KTRELEWRSGYNAMSGKYEDLLNAGIAD 529
Query: 500 LHVTKFFALKYAADAVCTVLRVDQVI 525
FAL+ A +L V+
Sbjct: 530 PCRVSRFALQNAVSVAGIILTTQAVL 555
>At1g26230 chaperonin precursor, putative
Length = 611
Score = 60.5 bits (145), Expect = 2e-09
Identities = 118/559 (21%), Positives = 219/559 (39%), Gaps = 75/559 (13%)
Query: 10 IQSMLKEGHKHLSGLDEAVIKNIDA-CKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDA 68
+++ KE H + G +V K + A ++ + +LGP G N ++ N + ND
Sbjct: 36 VRAAAKEVHFNRDG---SVTKKLQAGADMVAKLLGVTLGPKGRNVVLQNKYGPPRIVNDG 92
Query: 69 ATIVNELEVQHP----AAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHP 124
T++ E+E++ P K++ AG + GDG+ +I A L+ ++I G +P
Sbjct: 93 ETVLKEIELEDPLENVGVKLVRQAGAKTNDLAGDGSTTSIILAHGLITEGIKVISAGTNP 152
Query: 125 SEIISGYTKAIAKTIEILDELVEKGSESMDVRDKVQVISRMRAAVASKQFGQEDTICSLV 184
++ G I KT + L ++E S S ++ D A VA+ G + + +++
Sbjct: 153 IQVARG----IEKTTKAL--VLELKSMSREIED------HELAHVAAVSAGNDYEVGNMI 200
Query: 185 ADACIQVCPKNPANFNVDNVRVAKLLGGGLHNSTVVRGMVLKSDAVGSIKRIEKAK-VAV 243
++A QV V + K L ++N +V GM + ++ K A
Sbjct: 201 SNAFQQV-----GRTGVVTIEKGKYL---VNNLEIVEGMQFNRGYLSPYFVTDRRKREAE 252
Query: 244 F----VSGVDTSATDTKGTVLIHSAEQLENY------SKTEEAKVEELIKAVADSGAKV- 292
F + VD T+ K I + E + E+ + +I+ KV
Sbjct: 253 FHDCKLLLVDKKITNPKDMFKILDSAVKEEFPVLIVAEDIEQDALAPVIRNKLKGNLKVA 312
Query: 293 IVSGGAVGEMALHFCERYKLMV--------LKISSKFELRRFCRTTGAVAVLKLSQPNPD 344
+ A GE H + + + +S + + T V V K S
Sbjct: 313 AIKAPAFGERKSHCLDDLAIFTGATVIRDEMGLSLEKAGKEVLGTAKRVLVTKDSTLIVT 372
Query: 345 DLGYVDSVSVEEVGGVTVTIVKNEEG-------------GNSVSTVVLRGSTDSILDDLE 391
+ G+ E V + I EE ++ + + T L D +
Sbjct: 373 N-GFTQKAVDERVSQIKNLIENTEENFQKKILNERVARLSGGIAIIQVGALTQVELKDKQ 431
Query: 392 RAVDDGVNTYKAMCKDSRTVPGAA-----ATEIELAKRVKDFSFKETGLDQYAIAKFAES 446
V+D +N K+ ++ V G AT+++ K D + ++ G + I K A S
Sbjct: 432 LKVEDALNATKSAIEEGIVVGGGCALLRLATKVDRIKETLDNTEQKIGAE---IFKKALS 488
Query: 447 FEMIPRTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKDVTTTHVWDLHVTKFF 506
+ + R +A+NA N +I + ++ NT G + + +D+ + D
Sbjct: 489 YPI--RLIAKNADTNGNIVIEKVL---SNKNTMYGYNAAKNQYEDLMLAGIIDPTKVVRC 543
Query: 507 ALKYAADAVCTVLRVDQVI 525
L++A+ T L D V+
Sbjct: 544 CLEHASSVAQTFLTSDCVV 562
>At2g28000 putative rubisco subunit binding-protein alpha subunit
Length = 586
Score = 52.0 bits (123), Expect = 8e-07
Identities = 47/162 (29%), Positives = 76/162 (46%), Gaps = 18/162 (11%)
Query: 37 QLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHP-----AAKILVLAGKA 91
+L+ +LGP G N +V++ V ND TI +E+ + AA I +A K
Sbjct: 67 KLADCVGLTLGPRGRN-VVLDEFGSPKVVNDGVTIARAIELPNAMENAGAALIREVASKT 125
Query: 92 QQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDELVEKGSE 151
+ GDG A E+++ + G +P + G I KT++ L E ++K +
Sbjct: 126 N-DSAGDGTTTASILAREIIKHGLLSVTSGANPVSLKRG----IDKTVQGLIEELQKKAR 180
Query: 152 SMDVRDKVQVISRMRAAVASKQFGQEDTICSLVADACIQVCP 193
+ RD ++ AVAS G +D I S++ADA +V P
Sbjct: 181 PVKGRDDIR-------AVASISAGNDDLIGSMIADAIDKVGP 215
>At1g55490 Rubisco subunit binding-protein beta subunit
Length = 600
Score = 50.1 bits (118), Expect = 3e-06
Identities = 41/172 (23%), Positives = 79/172 (45%), Gaps = 17/172 (9%)
Query: 25 DEAVIKNIDA-CKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHP--- 80
D I+ + A +L+ + +LGP G N ++ + + ND T+ E+E++ P
Sbjct: 64 DGTTIRRLQAGVNKLADLVGVTLGPKGRNVVLESKYGSPRIVNDGVTVAREVELEDPVEN 123
Query: 81 -AAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTI 139
AK++ A + GDG ++ A + +++ G +P I T+ I KT
Sbjct: 124 IGAKLVRQAAAKTNDLAGDGTTTSVVLAQGFIAEGVKVVAAGANPVLI----TRGIEKTA 179
Query: 140 EILDELVEKGSESMDVRDKVQVISRMRAAVASKQFGQEDTICSLVADACIQV 191
+ L ++K S+ ++ + A VA+ G D I +++A+A +V
Sbjct: 180 KALVTELKKMSKEVEDSE--------LADVAAVSAGNNDEIGNMIAEAMSKV 223
>At3g13470 chaperonin 60 beta - like protein
Length = 596
Score = 47.0 bits (110), Expect = 3e-05
Identities = 36/159 (22%), Positives = 69/159 (42%), Gaps = 16/159 (10%)
Query: 37 QLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHP----AAKILVLAGKAQ 92
+L+ + +LGP G N ++ + + ND T+ E+E++ P AK++ A
Sbjct: 73 KLADLVGVTLGPKGRNVVLESKYGSPRIVNDGVTVAREVELEDPVENIGAKLVRQAAAKT 132
Query: 93 QEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDELVEKGSES 152
+ GDG ++ A + +++ G +P I G K + L +L+ K E
Sbjct: 133 NDLAGDGTTTSVVLAQGFIAEGVKVVAAGANPVLITRGIEKTAKALVNEL-KLMSKEVED 191
Query: 153 MDVRDKVQVISRMRAAVASKQFGQEDTICSLVADACIQV 191
++ D VA+ G + S++A+A +V
Sbjct: 192 SELAD-----------VAAVSAGNNHEVGSMIAEAMSKV 219
>At3g14270 unknown protein
Length = 1791
Score = 42.0 bits (97), Expect = 8e-04
Identities = 50/212 (23%), Positives = 91/212 (42%), Gaps = 27/212 (12%)
Query: 204 VRVAKLLGGGLHNSTVVRGMVLKSDAVG--SIKRIEKAKVAVFVSGVDTSATDTKGTVLI 261
V+V L G H+S VV+G+V K + V +IEKA++ + G++
Sbjct: 440 VKVKCLASGFRHDSMVVKGVVCKKNVVNRRMSTKIEKARLLILGGGLEYQRV-------- 491
Query: 262 HSAEQLENYSKTEEAKVEELIKAVADSGAK---VIVSGGAVGEMALHFCERYKLMVLKIS 318
+ QL ++ + + + L AVA A+ +++ +V A + + ++
Sbjct: 492 --SNQLSSFDTLLQQEKDHLKMAVAKIHAERPNILLVEKSVSRFAQEYLLAKDISLVLNI 549
Query: 319 SKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSV----EEVGG-------VTVTIVKN 367
+ L R R TGA + + + LGY ++ V EE G V T++
Sbjct: 550 KRPLLDRIARCTGAQIIPSVDHLSSQKLGYCENFRVDRYPEEHGSTGQVGKKVVKTLMYF 609
Query: 368 EEGGNSVS-TVVLRGSTDSILDDLERAVDDGV 398
E + T++LRG+ + L ++ V GV
Sbjct: 610 EHCPKPLGFTILLRGANEDELKKVKHVVQYGV 641
>At4g33240 putative protein
Length = 1757
Score = 34.3 bits (77), Expect = 0.17
Identities = 50/194 (25%), Positives = 78/194 (39%), Gaps = 26/194 (13%)
Query: 175 GQEDTICSLVADACIQVCPKNPANFNVDN---VRVAKLLGGGLHNSTVVRGMVLKSDAVG 231
G D I SL +A + P + +D V+V + G S VV+G+V K +
Sbjct: 396 GWLDIITSLSWEAATLLKPDTSKSGGMDPGGYVKVKCIPCGRRSESMVVKGVVCKKNVAH 455
Query: 232 S--IKRIEKAKVAVFVSGVDTSATDTKGTVLIHSAEQLENYSKTEEAKVEELIKAVA--D 287
+IEK ++ + ++ + QL ++ + +++ L AVA D
Sbjct: 456 RRMTSKIEKPRLLILGGALEYQRI----------SNQLSSFDTLLQQEMDHLKMAVAKID 505
Query: 288 SGAKVIVSGGAVGEMALHFCERYKL-----MVLKISSKFELRRFCRTTGAVAVLKLSQPN 342
S I+ V + F + Y L +VL I L R R TGA V + Q
Sbjct: 506 SHNPDIL---LVEKSVSRFAQEYLLAKDISLVLNIKRSL-LERISRCTGAQIVPSIDQLT 561
Query: 343 PDDLGYVDSVSVEE 356
LGY D VE+
Sbjct: 562 SPKLGYCDLFHVEK 575
>At3g23990 mitochondrial chaperonin hsp60
Length = 577
Score = 33.1 bits (74), Expect = 0.39
Identities = 30/169 (17%), Positives = 67/169 (38%), Gaps = 18/169 (10%)
Query: 26 EAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELE----VQHPA 81
EA + + L+ + ++GP G N ++ VT D T+ +E +++
Sbjct: 41 EARALMLKGVEDLADAVKVTMGPKGRNVVIEQSWGAPKVTKDGVTVAKSIEFKDKIKNVG 100
Query: 82 AKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAI------ 135
A ++ A + GDG + + + G++ ++ G + A+
Sbjct: 101 ASLVKQVANATNDVAGDGTTCATVLTRAIFAEGCKSVAAGMNAMDLRRGISMAVDAVVTN 160
Query: 136 ----AKTIEILDELVEKGSESMDVRDKVQVISRMRAAVASKQFGQEDTI 180
A+ I +E+ + G+ S + ++ + A A ++ G+E I
Sbjct: 161 LKSKARMISTSEEIAQVGTISANGEREIGEL----IAKAMEKVGKEGVI 205
>At2g38060 putative Na+-dependent inorganic phosphate cotransporter
Length = 561
Score = 33.1 bits (74), Expect = 0.39
Identities = 31/123 (25%), Positives = 52/123 (42%), Gaps = 8/123 (6%)
Query: 128 ISGYTKAIAKTIEILDELVEKGSESMDVRDKVQVISRMRAAVA--SKQFGQEDTICSLVA 185
ISGY A D L+ G VR +Q I M ++ F + + ++
Sbjct: 372 ISGYYAGAAS-----DFLIRTGHSVTSVRKIMQSIGFMGPGLSLLCLNFAKSPSCAAVFM 426
Query: 186 DACIQVCPKNPANFNVDNVRVAKLLGGGLHNSTVVRGMVLKSDAVGSIKRIEKAKVAVFV 245
+ + + A F ++ +A G LH + V+R + S + SI + + KV V +
Sbjct: 427 TIALSLSSFSQAGFLLNMQDIAPQYAGFLHGNNVIRWWIFSSSSFASILFLAE-KVRVCI 485
Query: 246 SGV 248
SGV
Sbjct: 486 SGV 488
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,524,419
Number of Sequences: 26719
Number of extensions: 417438
Number of successful extensions: 1331
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 1271
Number of HSP's gapped (non-prelim): 49
length of query: 528
length of database: 11,318,596
effective HSP length: 104
effective length of query: 424
effective length of database: 8,539,820
effective search space: 3620883680
effective search space used: 3620883680
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0023.15


