

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0023.13
(195 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
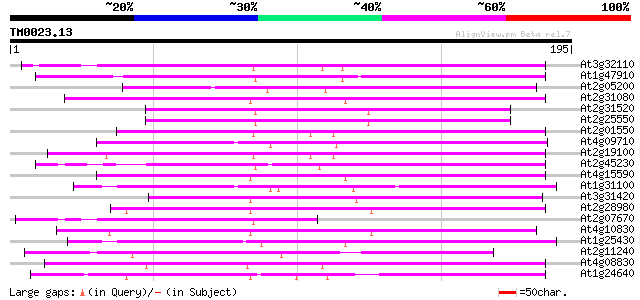
Score E
Sequences producing significant alignments: (bits) Value
At3g32110 non-LTR reverse transcriptase, putative 99 2e-21
At1g47910 reverse transcriptase, putative 87 7e-18
At2g05200 putative non-LTR retroelement reverse transcriptase 80 5e-16
At2g31080 putative non-LTR retroelement reverse transcriptase 78 3e-15
At2g31520 putative non-LTR retroelement reverse transcriptase 77 4e-15
At2g25550 putative non-LTR retroelement reverse transcriptase 77 4e-15
At2g01550 putative non-LTR retroelement reverse transcriptase 77 8e-15
At4g09710 RNA-directed DNA polymerase -like protein 76 1e-14
At2g19100 putative non-LTR retroelement reverse transcriptase 75 2e-14
At2g45230 putative non-LTR retroelement reverse transcriptase 75 2e-14
At4g15590 reverse transcriptase like protein 72 2e-13
At1g31100 hypothetical protein 72 2e-13
At3g31420 hypothetical protein 70 7e-13
At2g28980 putative non-LTR retroelement reverse transcriptase 69 1e-12
At2g07670 putative non-LTR retrolelement reverse transcriptase 69 2e-12
At4g10830 putative protein 67 6e-12
At1g25430 hypothetical protein 67 6e-12
At2g11240 pseudogene 66 1e-11
At4g08830 putative protein 66 1e-11
At1g24640 hypothetical protein 65 2e-11
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 98.6 bits (244), Expect = 2e-21
Identities = 64/185 (34%), Positives = 104/185 (55%), Gaps = 10/185 (5%)
Query: 5 VQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKY 64
VQ+L LD + ++ L +EE LL +F VL E + QKSR KW GD+NTK+
Sbjct: 841 VQEL--LDLHQSDDLLKKEEE-----LLKDFDVVLEQEEVVWMQKSREKWFVHGDRNTKF 893
Query: 65 FHSLINWRKRTNSIMGLMI-DGNWVEDPQAIKTEVQNYFEQCFT-NSLGTLV-RLDEISF 121
FH+ R+R N I L DG W+ + Q ++T +Y+++ ++ + L +V +L + F
Sbjct: 894 FHTSTIIRRRRNQIEMLQDNDGRWLSNAQELETHAIDYYKRLYSLDDLDAVVEQLPQEGF 953
Query: 122 KSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLED 181
++S+AD LT F EV+GA+ G K+P PD F F Q+ W+++ + + + D
Sbjct: 954 TALSEADFSSLTKPFSPLEVEGAIRSMGKYKAPGPDGFQPVFYQQGWEVVGESVTKFVMD 1013
Query: 182 FWNRG 186
F++ G
Sbjct: 1014 FFSSG 1018
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 86.7 bits (213), Expect = 7e-18
Identities = 61/180 (33%), Positives = 89/180 (48%), Gaps = 7/180 (3%)
Query: 10 ELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLI 69
ELD + ++ S EE E L E + R E YQKSRS W+K GD N+K+FH+L
Sbjct: 87 ELDVAQRDDDRSREEITELTLRLKEAY---RDEEQYWYQKSRSLWMKLGDNNSKFFHALT 143
Query: 70 NWRKRTNSIMGLMID-GNWVEDPQAIKTEVQNYFEQCFTNSLGTLV--RLDEISFKSISD 126
R+ N I GL + G W + I+ +YF+ FT + + L E+ I+D
Sbjct: 144 KQRRARNRITGLHDENGIWSIEDDDIQNIAVSYFQNLFTTANPQVFDEALGEVQV-LITD 202
Query: 127 ADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRG 186
N+LLT EV+ A++ +K+P PD F QK W ++K D ++ F G
Sbjct: 203 RINDLLTADATECEVRAALFMIHPEKAPGPDGMTALFFQKSWAIIKSDLLSLVNSFLQEG 262
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 80.5 bits (197), Expect = 5e-16
Identities = 51/147 (34%), Positives = 74/147 (49%), Gaps = 4/147 (2%)
Query: 40 RLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLMIDGNWV--EDPQAIKTE 97
+L E Q+SR WL GD+NT YFH++ +RT + + +M D N V + I
Sbjct: 225 KLEEQFWKQRSRVLWLHSGDRNTGYFHAVTR-NRRTQNRLTVMEDINGVAQHEEHQISQI 283
Query: 98 VQNYFEQCFTN-SLGTLVRLDEISFKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEP 156
+ YF+Q FT+ S G +DE +S DN+ LT EEVK AV+ K+P P
Sbjct: 284 ISGYFQQIFTSESDGDFSVVDEAIEPMVSQGDNDFLTRIPNDEEVKDAVFSINASKAPGP 343
Query: 157 DEFNFKFIQKFWDLLKDDFHRVLEDFW 183
D F F +W ++ D R + F+
Sbjct: 344 DGFTAGFYHSYWHIISTDVGREIRLFF 370
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 78.2 bits (191), Expect = 3e-15
Identities = 55/170 (32%), Positives = 85/170 (49%), Gaps = 3/170 (1%)
Query: 20 LSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIM 79
LS++ + + LL E VL E L +QKSR K+++ GD+NT +FH+ R+R N I
Sbjct: 173 LSDDLLAKEEVLLKEMDLVLEQEETLWFQKSREKYIELGDRNTTFFHTSTIIRRRRNRIE 232
Query: 80 GLM-IDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVR--LDEISFKSISDADNELLTGRF 136
L D WV D ++ Y+++ ++ + VR L F SIS+A+ L F
Sbjct: 233 SLKGDDDRWVTDKVELEAMALTYYKRLYSLEDVSEVRNMLPTGGFASISEAEKAALLQAF 292
Query: 137 GMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRG 186
EV AV G K+P PD + F Q+ W+ + R + +F+ G
Sbjct: 293 TKAEVVSAVKSMGRFKAPGPDGYQPVFYQQCWETVGPSVTRFVLEFFETG 342
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 77.4 bits (189), Expect = 4e-15
Identities = 44/129 (34%), Positives = 68/129 (52%), Gaps = 2/129 (1%)
Query: 48 QKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLMID-GNWVEDPQAIKTEVQNYFEQCF 106
QKSR++W+KEGDQNT YFH+ R N + +M D G + I Q++F F
Sbjct: 494 QKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFFTNIF 553
Query: 107 TNSLGTLVRLDEISFKS-ISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQ 165
+ + + +D FKS +++ N LT F E+ A+ + G DK+P PD +F +
Sbjct: 554 STNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTARFYK 613
Query: 166 KFWDLLKDD 174
WD++ D
Sbjct: 614 NCWDIVGYD 622
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 77.4 bits (189), Expect = 4e-15
Identities = 44/129 (34%), Positives = 68/129 (52%), Gaps = 2/129 (1%)
Query: 48 QKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLMID-GNWVEDPQAIKTEVQNYFEQCF 106
QKSR++W+KEGDQNT YFH+ R N + +M D G + I Q++F F
Sbjct: 720 QKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFFTNIF 779
Query: 107 TNSLGTLVRLDEISFKS-ISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQ 165
+ + + +D FKS +++ N LT F E+ A+ + G DK+P PD +F +
Sbjct: 780 STNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTARFYK 839
Query: 166 KFWDLLKDD 174
WD++ D
Sbjct: 840 NCWDIVGYD 848
>At2g01550 putative non-LTR retroelement reverse transcriptase
Length = 1449
Score = 76.6 bits (187), Expect = 8e-15
Identities = 46/155 (29%), Positives = 77/155 (49%), Gaps = 6/155 (3%)
Query: 38 VLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLMI-DGNWVEDPQAIKT 96
+ + E L Q S+ WLK GD+N K FH R NSI + DG+ IK
Sbjct: 764 IASIEEKYLKQVSKLHWLKVGDKNNKTFHRAATARAAQNSIREIQKEDGSTATTKDDIKN 823
Query: 97 EVQNYFE---QCFTNSLG--TLVRLDEISFKSISDADNELLTGRFGMEEVKGAVWECGGD 151
E + +F+ Q N T+ +L + S A+ ++LT +E++GA++ D
Sbjct: 824 ETERFFQEFLQLIPNDYEGITVEKLTSLLPYHCSPAEKDMLTASVSAKEIRGALFSMPND 883
Query: 152 KSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRG 186
KSP PD + +F ++ WD++ +F ++ F+ +G
Sbjct: 884 KSPGPDGYTSEFYKRAWDIIGAEFVLAVKSFFEKG 918
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 76.3 bits (186), Expect = 1e-14
Identities = 48/160 (30%), Positives = 78/160 (48%), Gaps = 4/160 (2%)
Query: 31 LLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLMIDGNWVE- 89
+ E R E Q SR +WL GD+N YFH+ R+ N++ ++ DG+ E
Sbjct: 262 ITQELEAAYRQEELFWKQWSRVQWLNSGDRNKGYFHATTRTRRMLNNL-SVIEDGSGQEF 320
Query: 90 -DPQAIKTEVQNYFEQCFTNSLGT-LVRLDEISFKSISDADNELLTGRFGMEEVKGAVWE 147
+ + I + + +YF+ FT S + L + E IS NE L + E+K A++
Sbjct: 321 HEEEQIASTISSYFQNIFTTSNNSDLQVVQEALSPIISSHCNEELIKISSLLEIKEALFS 380
Query: 148 CGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRGC 187
DK+P PD F+ F +WD+++ D R + F+ C
Sbjct: 381 ISADKAPGPDGFSASFFHAYWDIIEADVSRDIRSFFVDSC 420
>At2g19100 putative non-LTR retroelement reverse transcriptase
Length = 1447
Score = 75.5 bits (184), Expect = 2e-14
Identities = 54/182 (29%), Positives = 87/182 (47%), Gaps = 9/182 (4%)
Query: 14 KEEEEGLSEEENNERKWLL---DEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLIN 70
K++E L+ N K + D + V L E L +KS+ WL GD+N K FH +
Sbjct: 730 KKQESTLNNPTPNAMKEEVEAHDRWEHVAGLEEKFLKKKSKLHWLDGGDKNNKAFHRAVV 789
Query: 71 WRKRTNSIMGLMI-DGNWVEDPQAIKTEVQNYFE---QCFTNSLG--TLVRLDEISFKSI 124
R+ NSI + DG+ IK + +F Q N T+ L ++
Sbjct: 790 TREAQNSISEIQCQDGSVTAKGDEIKAYAERFFREFLQLIPNEYEGVTMADLQDLLPFRC 849
Query: 125 SDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWN 184
S+ ++ELLT EE+K ++ DKSP PD F +F + W++L ++F ++ F+
Sbjct: 850 SETEHELLTRVVTAEEIKKVLFSMPNDKSPGPDGFTSEFFKATWEILGNEFILAIQSFFA 909
Query: 185 RG 186
+G
Sbjct: 910 KG 911
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 75.1 bits (183), Expect = 2e-14
Identities = 55/180 (30%), Positives = 85/180 (46%), Gaps = 21/180 (11%)
Query: 10 ELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLI 69
EL R ++E LS+E NNE ++FW +KSR W++ GD+NTKYFH+
Sbjct: 317 ELARLKKE--LSQEYNNE-----EQFWQ----------EKSRIMWMRNGDRNTKYFHAAT 359
Query: 70 NWRKRTNSIMGLMID--GNWVEDPQAIKTEVQNYFEQCF-TNSLGTLVRLDEISFKSISD 126
R+ N I L+ + W D + + + YF++ F + +G V E +SD
Sbjct: 360 KNRRAQNRIQKLIDEEGREWTSD-EDLGRVAEAYFKKLFASEDVGYTVEELENLTPLVSD 418
Query: 127 ADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRG 186
N L EEV+ A + K P PD N Q+FW+ + D +++ F+ G
Sbjct: 419 QMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETMGDQITEMVQAFFRSG 478
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 71.6 bits (174), Expect = 2e-13
Identities = 48/159 (30%), Positives = 80/159 (50%), Gaps = 3/159 (1%)
Query: 31 LLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLM-IDGNWVE 89
LL EF +L E L +QKSR K L GD+NT +FH+ R+R N I L + WV
Sbjct: 136 LLKEFDVLLHQEETLWFQKSREKLLALGDRNTTFFHTSTVIRRRRNRIEMLKDSEDRWVT 195
Query: 90 DPQAIKTEVQNYFEQCFTNSLGTLVR--LDEISFKSISDADNELLTGRFGMEEVKGAVWE 147
+ +A++ +Y+ + ++ ++VR L F ++ + L F +EV AV
Sbjct: 196 EKEALEKLAMDYYRKLYSLEDVSVVRGTLPTEGFPRLTREEKNNLNRPFTRDEVVVAVRS 255
Query: 148 CGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRG 186
G K+P PD + F Q+ W+ + + + + +F+ G
Sbjct: 256 MGRFKAPGPDGYQPVFYQQCWETVGESVSKFVMEFFESG 294
>At1g31100 hypothetical protein
Length = 1090
Score = 71.6 bits (174), Expect = 2e-13
Identities = 50/175 (28%), Positives = 83/175 (46%), Gaps = 14/175 (8%)
Query: 23 EENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLM 82
E +RKWL+ +++ E Q+SR W+ EGD NT YFH + + RK N+I ++
Sbjct: 245 EMEAQRKWLI-----LVKAEESFFCQRSRVTWMGEGDSNTSYFHRMADSRKAVNTI-HII 298
Query: 83 IDGNWVE-DPQ-AIKTEVQNYFEQCFTNSLGTLVRLDE-----ISFKSISDADNELLTGR 135
ID N V+ D Q IK YF +G + + E + F+ D EL
Sbjct: 299 IDDNGVKIDTQLGIKEHCIEYFSNLLGGEVGPPMLIQEDFDLLLPFRCSHDQKKELAMS- 357
Query: 136 FGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRGCGLK 190
F +++K A + +K+ PD F +F ++ W ++ + + +F+ LK
Sbjct: 358 FSRQDIKSAFFSFPSNKTSGPDGFPVEFFKETWSVIGTEVTDAVSEFFTSSVLLK 412
>At3g31420 hypothetical protein
Length = 1491
Score = 70.1 bits (170), Expect = 7e-13
Identities = 42/141 (29%), Positives = 71/141 (49%), Gaps = 4/141 (2%)
Query: 49 KSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLM-IDGNWVEDPQAIKTEVQNYFEQCFT 107
KSRS+WL GD+NT+YFHS R+ N ++ + DG+ + I NYF+ +
Sbjct: 522 KSRSRWLNAGDRNTRYFHSTTKTRRCRNRLLSVQDSDGDICRGDENIAKVAINYFDDLYK 581
Query: 108 NSLGTLVRLDEI---SFKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFI 164
++ T +R ++ + I+D NE L E++ +V+ ++P+PD F F
Sbjct: 582 STPNTSLRYADVFQGFQQKITDEINEDLIRPVTELEIEESVFSVAPSRTPDPDGFTADFY 641
Query: 165 QKFWDLLKDDFHRVLEDFWNR 185
Q+FW +K + F+ R
Sbjct: 642 QQFWPDIKQKVIDEVTRFFER 662
>At2g28980 putative non-LTR retroelement reverse transcriptase
Length = 1529
Score = 69.3 bits (168), Expect = 1e-12
Identities = 44/158 (27%), Positives = 71/158 (44%), Gaps = 7/158 (4%)
Query: 36 WTVL-RLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLM-IDGNWVEDPQA 93
WT L L E L QKS+ W+ GD N YFH RK NSI + + ++ +
Sbjct: 643 WTHLSELEEGFLKQKSKLHWMNVGDGNNSYFHKAAQVRKMRNSIREIRGPNAETLQTSEE 702
Query: 94 IKTEVQNYFEQCFTNSLGTLVRLDEISFKSI-----SDADNELLTGRFGMEEVKGAVWEC 148
IK E + +F + G + +++ S D +LT EE++ ++
Sbjct: 703 IKGEAERFFNEFLNRQSGDFHGISVEDLRNLMSYRCSVTDQNILTREVTGEEIQKVLFAM 762
Query: 149 GGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRG 186
+KSP PD + +F + W L DF ++ F+ +G
Sbjct: 763 PNNKSPGPDGYTSEFFKATWSLTGPDFIAAIQSFFVKG 800
>At2g07670 putative non-LTR retrolelement reverse transcriptase
Length = 913
Score = 68.6 bits (166), Expect = 2e-12
Identities = 41/106 (38%), Positives = 64/106 (59%), Gaps = 8/106 (7%)
Query: 3 EVVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNT 62
+VVQ+L E+++ + LS+EE L+ EF VL E L +QKSR KW++ GD+NT
Sbjct: 775 KVVQELLEINQTDNL--LSKEEE-----LIKEFDVVLEQEEVLWFQKSREKWVELGDRNT 827
Query: 63 KYFHSLINWRKRTNSIMGLMI-DGNWVEDPQAIKTEVQNYFEQCFT 107
KYFH++ R+R N I L DG+WV Q ++ +Y+ + ++
Sbjct: 828 KYFHTMTVVRRRRNRIEMLKADDGSWVSQQQELEKMAVDYYSRLYS 873
>At4g10830 putative protein
Length = 1294
Score = 67.0 bits (162), Expect = 6e-12
Identities = 44/172 (25%), Positives = 83/172 (47%), Gaps = 5/172 (2%)
Query: 17 EEGLSEEENNERKWLLD---EFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRK 73
++ +S ER+ + E R E QKSR++W+KEGD+NT++FH+ R
Sbjct: 666 DKAMSSVNRTERRTISHIQRELTVAYRDEERYWQQKSRNQWMKEGDRNTEFFHACTKTRF 725
Query: 74 RTNSIMGLM-IDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDEISFKSI-SDADNEL 131
N ++ + +G + I Q +F + + ++ + +D FK I ++ N+
Sbjct: 726 SVNRLVTIKDEEGMIYRGDKEIGVHAQEFFTKVYESNGRPVSIIDFAGFKPIVTEQINDD 785
Query: 132 LTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFW 183
LT E+ A+ G DK+P PD +F + W+++ D + ++ F+
Sbjct: 786 LTKDLSDLEIYNAICHIGDDKAPGPDGLTARFYKSCWEIVGPDVIKEVKIFF 837
>At1g25430 hypothetical protein
Length = 1213
Score = 67.0 bits (162), Expect = 6e-12
Identities = 51/176 (28%), Positives = 80/176 (44%), Gaps = 12/176 (6%)
Query: 21 SEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMG 80
S E ERKW + + E QKSR W EGD NTKYFH + + R +NSI
Sbjct: 332 SFELEAERKWHI-----LTAAEESFFRQKSRISWFAEGDGNTKYFHRMADARNSSNSISA 386
Query: 81 LMIDGN--WVEDPQAIKTEVQNYFEQCFTNSLGTLVR----LDEISFKSISDADNELLTG 134
L DGN V+ + I +YF + + + ++ + S A L
Sbjct: 387 L-YDGNGKLVDSQEGILDLCASYFGSLLGDEVDPYLMEQNDMNLLLSYRCSPAQVCELES 445
Query: 135 RFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRGCGLK 190
F E+++ A++ +KS PD F +F W ++ + +++F++ GC LK
Sbjct: 446 TFSNEDIRAALFSLPRNKSCGPDGFTAEFFIDSWSIVGAEVTDAIKEFFSSGCLLK 501
>At2g11240 pseudogene
Length = 1044
Score = 66.2 bits (160), Expect = 1e-11
Identities = 50/168 (29%), Positives = 79/168 (46%), Gaps = 14/168 (8%)
Query: 6 QKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRL----HE*LLYQKSRSKWLKEGDQN 61
QKL E +R++ EE +S +++N+ LL T L L E Q+SR WL GD+N
Sbjct: 128 QKLIEENRQKLEEAMSSQDHNQE--LLSTINTNLLLAYKAEEEYWKQRSRQLWLALGDKN 185
Query: 62 TKYFHSLINWRKRTNSIMGLMIDGNWVEDPQA-IKTEVQNYFEQCFTNSLGTLVRLDEIS 120
+ YFH++ R N + + E +A I + YF++ F+ + G
Sbjct: 186 SGYFHAITRGRTVINKFSVIEKEDGVPEYEEAGILNVISEYFQKLFSANEGARAA----- 240
Query: 121 FKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFW 168
+I +A ++ EE+K A + DK+P PD F+ F Q W
Sbjct: 241 --TIKEAIKPFISPEQNPEEIKSACFSIHADKAPGPDGFSASFFQSNW 286
>At4g08830 putative protein
Length = 947
Score = 65.9 bits (159), Expect = 1e-11
Identities = 47/179 (26%), Positives = 90/179 (50%), Gaps = 5/179 (2%)
Query: 13 RKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LL--YQKSRSKWLKEGDQNTKYFHSLIN 70
+K++EE + E + + + + +LR E L+ + KSR KW+ GD+NT YFH+
Sbjct: 58 KKKKEELMREIQVVQDALDVHQSDDMLRKEEDLIKAFDKSREKWIALGDRNTNYFHTTTI 117
Query: 71 WRKRTNSIMGL-MIDGNWVEDPQAIKTEVQNYFEQCFT--NSLGTLVRLDEISFKSISDA 127
R+R N + L +G W+ D + ++ Y+++ ++ + L + L + + ++
Sbjct: 118 IRRRQNRVERLHNEEGTWIFDAKELEALAVQYYQRLYSMDDVLQVVNPLPQDGLERLTRE 177
Query: 128 DNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRG 186
+ L F EV+ A+ G K+P PD + F Q W+++ + R DF++ G
Sbjct: 178 EVLSLNKPFLATEVEVAIRSMGKYKAPGPDGYQPIFYQSCWEVVGNSVIRFALDFFSSG 236
>At1g24640 hypothetical protein
Length = 1270
Score = 65.5 bits (158), Expect = 2e-11
Identities = 56/198 (28%), Positives = 91/198 (45%), Gaps = 29/198 (14%)
Query: 8 LNELDRKEEEEGLSEEENNERKWLLDEFWTVL--------RLHE*LLYQKSRSKWLKEGD 59
LN LD+ + E E+E + W + + +VL R E QKSR KWL+ G+
Sbjct: 254 LNSLDKINQLEAALEKEQS-LVWPIFQRVSVLKKDLAKAYREEEAYWKQKSRQKWLRSGN 312
Query: 60 QNTKYFHSLINWRKRTNSIMGLM-IDGNWVEDPQAIKTEV-QNYFEQCFTNS-------- 109
+N+KYFH+ + ++ I L ++GN ++ +A K EV YF F +S
Sbjct: 313 RNSKYFHAAVKQNRQRKRIEKLKDVNGN-MQTSEAAKGEVAAAYFGNLFKSSNPSGFTDW 371
Query: 110 -LGTLVRLDEISFKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFW 168
G + R+ E+ NE L G +E+K AV+ +P PD + F Q +W
Sbjct: 372 FSGLVPRVSEVM--------NESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFFQHYW 423
Query: 169 DLLKDDFHRVLEDFWNRG 186
+ + ++ F+ G
Sbjct: 424 STVGNQVTSEVKKFFADG 441
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.139 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,068,778
Number of Sequences: 26719
Number of extensions: 230410
Number of successful extensions: 1170
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1075
Number of HSP's gapped (non-prelim): 67
length of query: 195
length of database: 11,318,596
effective HSP length: 94
effective length of query: 101
effective length of database: 8,807,010
effective search space: 889508010
effective search space used: 889508010
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0023.13


