

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0018.6
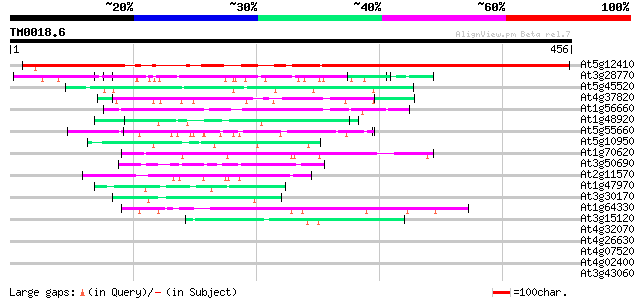
(456 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g12410 unknown protein 363 e-101
At3g28770 hypothetical protein 57 2e-08
At5g45520 putative protein 50 2e-06
At4g37820 unknown protein 48 1e-05
At1g56660 hypothetical protein 47 2e-05
At1g48920 unknown protein 47 2e-05
At5g55660 putative protein 46 4e-05
At5g10950 putative protein 46 4e-05
At1g70620 unknown protein 46 5e-05
At3g50690 putative protein 45 6e-05
At2g11570 hypothetical protein 44 1e-04
At1g47970 Unknown protein (T2J15.12) 44 1e-04
At3g30170 unknown protein 44 2e-04
At1g64330 unknown protein 44 2e-04
At3g15120 chaperone-like ATPase 42 7e-04
At4g32070 putative protein 42 0.001
At4g26630 unknown protein 42 0.001
At4g07520 hypothetical protein 42 0.001
At4g02400 41 0.001
At3g43060 putative protein 41 0.002
>At5g12410 unknown protein
Length = 376
Score = 363 bits (933), Expect = e-101
Identities = 215/450 (47%), Positives = 277/450 (60%), Gaps = 82/450 (18%)
Query: 11 AAGGRKRKR---YLPHNKPVKK-GSYPLHPGVQGVFITCEGGREHQASREALNILESFYE 66
A+G + +KR Y P N+PVKK GSYPL PGVQG FI+C+GGRE QA++EA+N+++SF+E
Sbjct: 2 ASGDQSKKRKQHYRPQNRPVKKKGSYPLKPGVQGFFISCDGGREFQAAQEAINVIDSFFE 61
Query: 67 DLVDGEHLSVKE-LLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLK 125
+L+ G V L P NKK+TF+ S+ ++DEEE DE+N
Sbjct: 62 ELIQGTDSKVNPGLFGNPINKKVTFSYSE-------DEDEEE--------DESN------ 100
Query: 126 LDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDED 185
NG E +EN G G K E D
Sbjct: 101 ------------------NGEE--------EENKGDGDKAVVSEGGNDL----------- 123
Query: 186 AGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGD 245
+ EK E D++ ++ EI + + +V A N+T E D
Sbjct: 124 --VNEKEIASEGVNDQV------------NEKEIASEGSC----EVKQLAENETVKEEED 165
Query: 246 NDNIKTKTANELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDEELRELGDKKKRRFVKLES 305
N K +E P K C + P+ + EKSID LI+ EL+ELGDK KRRF+KL+
Sbjct: 166 KGNQKNG-GDEPPRKKTCTEEANPLAKVNENAEKSIDKLIEAELKELGDKSKRRFMKLDP 224
Query: 306 GCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMSRFILRMLPIEVSCYASKEEISR 365
GCNG+VFIQM+K+DGD SPK+IV +TSAA+T+KHMSRFILR+LPIEVSCY S+EEISR
Sbjct: 225 GCNGLVFIQMKKRDGDPSPKEIVQHAMTSAAATKKHMSRFILRLLPIEVSCYPSEEEISR 284
Query: 366 AIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEIIDAVAKSVPGPHKVDLKNPDKTI 425
AIKPLVEQ FP+ T+ P KFAV+Y ARANTG+DRM+II+ +AKS+P PHKVDL NP+ TI
Sbjct: 285 AIKPLVEQYFPIETENPRKFAVLYGARANTGLDRMKIINTIAKSIPAPHKVDLSNPEMTI 344
Query: 426 VVEIARTVCLIGIIEKYKELSKYNLRELTS 455
VVEI +TVCLIG++EKYKEL+KYNLR+LTS
Sbjct: 345 VVEIIKTVCLIGVVEKYKELAKYNLRQLTS 374
>At3g28770 hypothetical protein
Length = 2081
Score = 57.0 bits (136), Expect = 2e-08
Identities = 68/269 (25%), Positives = 107/269 (39%), Gaps = 49/269 (18%)
Query: 77 KELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQV-------GEGDEANGDKKLKLDSS 129
KE N+PK+ K + E + +E + Q GE DE+ + ++ DS
Sbjct: 1450 KEEKNKPKDDKKNTTEQSGGKKESMESESKEAENQQKSQATTQGESDESKNEILMQADSQ 1509
Query: 130 VDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDS--------- 180
D HA + G + K ++ D A +T+ DE+K + + DS
Sbjct: 1510 ADTHA----NSQGDSDESKNEILMQADSQA--DSQTDSDESKNEILMQADSQADSQTDSD 1563
Query: 181 --------SVDEDAGIGE-----KAEGDEANGDKMPKMDSSVNENAG-HDNEIGGKENPH 226
D A IGE K +G E NGD++ K +S E G H+ GK N +
Sbjct: 1564 ESKNEILMQADSQAKIGESLEDNKVKGKEDNGDEVGKENSKTIEVKGRHEESKDGKTNEN 1623
Query: 227 KIDDVHAAAGNK----TDDNEGDNDNIKTKTANELPVVKQCCKTNVPVQDSSNKVEKSID 282
+V G+K + N G D+IK + + V + + + E S D
Sbjct: 1624 GGKEVSTEEGSKDSNIVERNGGKEDSIKEGSEDGKTV-------EINGGEELSTEEGSKD 1676
Query: 283 TLIDE--ELRELGDKKKRRFVKLESGCNG 309
I+E E +E K+ + K+E G G
Sbjct: 1677 GKIEEGKEGKENSTKEGSKDDKIEEGMEG 1705
Score = 49.7 bits (117), Expect = 3e-06
Identities = 67/293 (22%), Positives = 116/293 (38%), Gaps = 48/293 (16%)
Query: 84 KNKKITFADSDSSSSGDD--EDDEEEVQVQVGEGD-----EANGDKKLKLDSSVDEHAGI 136
++KK+ +S + S D +D +EE Q+ GE EA G KK +S ++
Sbjct: 707 EDKKLENKESQTDSKDDKSVDDKQEEAQIYGGESKDDKSVEAKGKKK---ESKENKKTKT 763
Query: 137 GEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDE---KPKLDSSVDEDAGIGEKAE 193
E V N E + + E G K E +AK E KL S+ + D ++
Sbjct: 764 NENRV-RNKEENVQGNKKESEKVEKGEKKESKDAKSVETKDNKKLSSTENRDEA--KERS 820
Query: 194 GDEANGDKMPKMD------SSVNENAGHDNEIGGKENPHKIDDVHA--AAGNKTDDNEGD 245
G++ DK D NEN G D +G KE+ + D + NK + +
Sbjct: 821 GEDNKEDKEESKDYQSVEAKEKNENGGVDTNVGNKEDSKDLKDDRSVEVKANKEESMKKK 880
Query: 246 NDNIKTKTANELPVVKQCCKT-NVPVQDSSNK-------------VEKSIDTLIDEELRE 291
+ ++ + V+ ++ VQ S + E++ DT+ ++
Sbjct: 881 REEVQRNDKSSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKDTINTSSKQK 940
Query: 292 LGDKKKRRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMSR 344
DKKK++ S M+KK+ DK K+ V+ + +K ++
Sbjct: 941 GKDKKKKKKESKNS--------NMKKKEEDK--KEYVNNELKKQEDNKKETTK 983
Score = 47.0 bits (110), Expect = 2e-05
Identities = 56/247 (22%), Positives = 99/247 (39%), Gaps = 28/247 (11%)
Query: 70 DGEHLSVKELLNRPKNKKITFADSDSSSSGDDED-DEEEVQVQVGEGDEANG----DKKL 124
+GE + + L N+ K++ +S + + ++ +E+ Q Q G + N +K
Sbjct: 593 NGESVKNENLENKEDKKELKDDESVGAKTNNETSLEEKREQTQKGHDNSINSKIVDNKGG 652
Query: 125 KLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDE 184
DS+ ++ +G+ N K S V+ G +G+E K + K ++
Sbjct: 653 NADSNKEKEVHVGDSTNDNNMESKEDTKSEVEVKKNDGSSEKGEEGKENNKDSMED---- 708
Query: 185 DAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEG 244
+K E E+ D K D SV++ GG+ K D A G K + E
Sbjct: 709 -----KKLENKESQTDS--KDDKSVDDKQEEAQIYGGES---KDDKSVEAKGKKKESKE- 757
Query: 245 DNDNIKTKT-----ANELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDEELRELGDKKKRR 299
N KTKT N+ V+ K + V+ K K ++ ++ ++L + R
Sbjct: 758 ---NKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDAKSVETKDNKKLSSTENRD 814
Query: 300 FVKLESG 306
K SG
Sbjct: 815 EAKERSG 821
Score = 44.3 bits (103), Expect = 1e-04
Identities = 68/291 (23%), Positives = 126/291 (42%), Gaps = 27/291 (9%)
Query: 4 EGKGDAAAAGGRKRKRYLPHNK--PVKKGSYPLHPGV--QGVFITCEGGREHQASREALN 59
+G G + GG L NK +K+G+ + G + E E + + +
Sbjct: 224 KGDGVEGSNGGDVSMENLQGNKVEDLKEGNNVVENGETKENNGENVESNNEKEVEGQGES 283
Query: 60 ILESFYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDE----DDEEEVQVQVGEG 115
I +S E ++ + VK + KN + ++ + G++ D+E+EV+ Q GE
Sbjct: 284 IGDSAIEKNLESKE-DVKSEVEAAKNDGSSMTENLGEAQGNNGVSTIDNEKEVEGQ-GES 341
Query: 116 DEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEK 175
E + D + L+S D + + E A A + KL+ + N +T E KG +
Sbjct: 342 IE-DSDIEKNLESKEDVKSEV-EAAKNAGSSMTGKLEEAQRNNGVSTNETMNSENKGSGE 399
Query: 176 PKLDSSV-----DEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDD 230
D V DED K E E NG+ + ++ AG++ + G+ +K+ +
Sbjct: 400 STNDKMVNATTNDEDHKKENKEETHENNGESV--KGENLENKAGNEESMKGENLENKVGN 457
Query: 231 VHAAAGNKTDDNEGDNDNIK------TKTANELPVVKQCCK-TNVPVQDSS 274
GN + + + +N++ K ++ +NE+ + K+ K NV +Q S
Sbjct: 458 -EELKGNASVEAKTNNESSKEEKREESQRSNEVYMNKETTKGENVNIQGES 507
Score = 41.6 bits (96), Expect = 0.001
Identities = 48/196 (24%), Positives = 82/196 (41%), Gaps = 17/196 (8%)
Query: 54 SREALNILESFYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVG 113
S+EA N +S D + S E+L + ++ ADS S S D ++ + E+ +Q
Sbjct: 1367 SKEAENQQKSQATTQADSDE-SKNEILMQADSQ----ADSHSDSQADSDESKNEILMQAD 1421
Query: 114 EG--DEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAK 171
+ N ++ K +SV E+ E N KPK D + GGK E E++
Sbjct: 1422 SQATTQRNNEEDRKKQTSVAENKKQKETKEEKN---KPK-DDKKNTTEQSGGKKESMESE 1477
Query: 172 GDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDV 231
E S G E DE+ + + + DS + +A + +N +
Sbjct: 1478 SKEAENQQKSQATTQG-----ESDESKNEILMQADSQADTHANSQGDSDESKN-EILMQA 1531
Query: 232 HAAAGNKTDDNEGDND 247
+ A ++TD +E N+
Sbjct: 1532 DSQADSQTDSDESKNE 1547
Score = 40.0 bits (92), Expect = 0.003
Identities = 40/189 (21%), Positives = 81/189 (42%), Gaps = 19/189 (10%)
Query: 46 EGGREHQASREALNILESFYEDLVDGEHLSVKELLNRPK------NKKITFA----DSDS 95
E G+E + + + E+ ++G+ S KE K +K+ T D +
Sbjct: 1680 EEGKEGKENSTKEGSKDDKIEEGMEGKENSTKESSKDGKINEIHGDKEATMEEGSKDGGT 1739
Query: 96 SSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSV 155
+S+G D D + V++ + D D K + ++E E +V N E D+S+
Sbjct: 1740 NSTGKDSKDSKSVEINGVKDDSLKDDSK---NGDINEINNGKEDSVKDNVTEIQGNDNSL 1796
Query: 156 DENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGH 215
+ + GD+ ++ +++++ G + GD NG+ +S+V+ N +
Sbjct: 1797 TNSTS--SEPNGDKLDTNKDSMKNNTMEAQGG----SNGDSTNGETEETKESNVSMNNQN 1850
Query: 216 DNEIGGKEN 224
++G EN
Sbjct: 1851 MQDVGSNEN 1859
Score = 34.3 bits (77), Expect = 0.15
Identities = 53/246 (21%), Positives = 91/246 (36%), Gaps = 41/246 (16%)
Query: 66 EDLVDGEHLSVKELLNRPKNK---KITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDK 122
E L D KE N K K+ +SD ++E+ E +++ + + DK
Sbjct: 1121 EKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDK 1180
Query: 123 KLKLDS------SVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKP 176
K K S E EK + N ++ K +SV+EN K + + K KP
Sbjct: 1181 KEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKK-QTSVEEN-----KKQKETKKEKNKP 1234
Query: 177 KLDSSVDEDAGIGEK---------------------AEGDEANGDKMPKMDSSVNENAGH 215
K D G+K A+ DE+ + + + DS + ++
Sbjct: 1235 KDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHS-- 1292
Query: 216 DNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANELPVVKQCC-KTNVPVQDSS 274
D++ E+ ++I + A T + D K + E K+ + N P D
Sbjct: 1293 DSQADSDESKNEI--LMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKK 1350
Query: 275 NKVEKS 280
N ++S
Sbjct: 1351 NTTKQS 1356
Score = 32.3 bits (72), Expect = 0.56
Identities = 49/249 (19%), Positives = 88/249 (34%), Gaps = 54/249 (21%)
Query: 77 KELLNRPKNKKITFA-------DSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSS 129
K+ N+PK+ K +S S S + E+ ++ + DE+ + ++ DS
Sbjct: 1228 KKEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQ 1287
Query: 130 VDEHA----------------GIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGD 173
D H+ + N E K +SV EN K + + +
Sbjct: 1288 ADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAEN-----KKQKETKEEK 1342
Query: 174 EKPKLDSSVDEDAGIGEK---------------------AEGDEANGDKMPKMDSSVNEN 212
KPK D G+K A+ DE+ + + + DS + +
Sbjct: 1343 NKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSH 1402
Query: 213 AGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANELPVVKQCC-KTNVPVQ 271
+ D++ E+ ++I + A T + D K + E K+ + N P
Sbjct: 1403 S--DSQADSDESKNEI--LMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKD 1458
Query: 272 DSSNKVEKS 280
D N E+S
Sbjct: 1459 DKKNTTEQS 1467
>At5g45520 putative protein
Length = 1167
Score = 50.4 bits (119), Expect = 2e-06
Identities = 75/331 (22%), Positives = 131/331 (38%), Gaps = 53/331 (16%)
Query: 46 EGGREHQASREALNILESFYEDLVDGEHLS------VKELLNRP---KNKKITFADSDSS 96
E G++H +E ++ +++V+ E + ++ N+P K K D D
Sbjct: 626 EDGKKHDEGKEERSLKS---DEVVEEEKKTSPSEEATEKFQNKPGDQKGKSNVEGDGDKG 682
Query: 97 SSG-DDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSV 155
+ ++E ++EV+ + + DE +K D S E G G+K A+ E K D
Sbjct: 683 KADLEEEKKQDEVEAEKSKSDEIVEGEKKPDDKSKVEKKGDGDKE-NADLDEGKKRDEVE 741
Query: 156 DENAGIGGKTEGDEAKGDEKPKLDS--SVDEDAGIGEKAEGDEANGDKMPKMDSSVNENA 213
+ + G EGD + + +D+ ++ +D EK EGD G P+ +E
Sbjct: 742 AKKSESGKVVEGDGKESPPQESIDTIQNMTDDQTKVEK-EGDRDKGKVDPEEGKKHDEVE 800
Query: 214 G----HDNEIGGKENPHKIDDVHAAAGNKTDDNE-GD--------------------NDN 248
G DN + G + + A NK DD++ GD +D
Sbjct: 801 GGIWKSDNGVEGVDKASPSRESTDAIENKPDDHQRGDKQEEKGDGEKEKVNLEEWKKHDE 860
Query: 249 IKTKTANELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDEELRELGD-----------KKK 297
IK +++ + V K + P + K D + +++E GD KK
Sbjct: 861 IKEESSKQDNVTGGDVKKSPPKESKDTMESKRDDQKENSKVQEKGDVDKGKAADLDEGKK 920
Query: 298 RRFVKLESGCNGVVFIQMRKKDGDKSPKDIV 328
VK ES + V +K+ + KDI+
Sbjct: 921 ENDVKAESSKSDKVIEGDEEKNPPQKSKDII 951
Score = 35.8 bits (81), Expect = 0.050
Identities = 49/213 (23%), Positives = 74/213 (34%), Gaps = 38/213 (17%)
Query: 121 DKKLK--LDSSVDEHAGIGEKAVGANG-------AEKPKLDSSVDENAGIGGKTEGDEAK 171
DKKLK ++ D+ G+ + G E ++E A + + DE K
Sbjct: 576 DKKLKDLMEREDDQVQNYGQTSKEEKGNVEETGKQEDGDQGDGINEEANLEDGKKHDEGK 635
Query: 172 GDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDV 231
+ K D V+E+ E E +K N D E K D+V
Sbjct: 636 EERSLKSDEVVEEEKKTSPSEEATEKFQNKPGDQKGKSNVEGDGDKGKADLEEEKKQDEV 695
Query: 232 HAAAGNKTDDNEGDNDNIKTKTANELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDEELRE 291
A + EG+ D +KVEK D D+E +
Sbjct: 696 EAEKSKSDEIVEGEKK-----------------------PDDKSKVEKKGDG--DKENAD 730
Query: 292 LGDKKKRRFVKLESGCNGVVFIQMRKKDGDKSP 324
L + KKR V+ + +G V + DG +SP
Sbjct: 731 LDEGKKRDEVEAKKSESGKVV----EGDGKESP 759
Score = 35.8 bits (81), Expect = 0.050
Identities = 52/195 (26%), Positives = 76/195 (38%), Gaps = 26/195 (13%)
Query: 55 REALNILESFYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGE 114
+E+ + +ES +D E+ V+E + K K AD D +D E +V E
Sbjct: 882 KESKDTMESKRDD--QKENSKVQEKGDVDKGKA---ADLDEGKKENDVKAESSKSDKVIE 936
Query: 115 GDEA-NGDKKLK--LDSSVDEHAGIG---EKAVGA-NGAEKPKLDSSVDENAGIGGKTEG 167
GDE N +K K + S D+H I EK G NG + KLD + KT
Sbjct: 937 GDEEKNPPQKSKDIIQSKPDDHREISKVQEKVDGEKNGDDLKKLDGGGEAKTQKSRKTTK 996
Query: 168 DEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNE-----IGGK 222
+ PK D+++ ED E S G N+ IG +
Sbjct: 997 FGKNVSDHPKRDNTMAEDI--------HETQSPSRKSKSSKAKWLTGRGNQTKKWGIGWQ 1048
Query: 223 ENPHKIDDVHAAAGN 237
E+P +DD + N
Sbjct: 1049 EDP-VVDDTYTLPSN 1062
>At4g37820 unknown protein
Length = 532
Score = 47.8 bits (112), Expect = 1e-05
Identities = 49/217 (22%), Positives = 90/217 (40%), Gaps = 26/217 (11%)
Query: 84 KNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGE----K 139
KN+K + D S E ++E GEG E +K+ K DSS E + E +
Sbjct: 317 KNEKDASSSQDESKEEKPERKKKEESSSQGEGKEEEPEKREKEDSSSQEESKEEEPENKE 376
Query: 140 AVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANG 199
++ E+ ++ + + EG+E K EK +S E+ +K E E+
Sbjct: 377 KEASSSQEENEIKETEIKEKEESSSQEGNENKETEKKSSESQRKENTNSEKKIEQVEST- 435
Query: 200 DKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANELPV 259
DSS + K + K D+ +GN T + E ++D+ KT++
Sbjct: 436 ------DSSNTQ----------KGDEQKTDESKRESGNDTSNKETEDDSSKTESEK---- 475
Query: 260 VKQCCKTNVPVQDSSNKVEKSIDTLIDEELRELGDKK 296
K+ N +++ N+ E++ L +++ D +
Sbjct: 476 -KEENNRNGETEETQNEQEQTKSALEISHTQDVKDAR 511
Score = 45.1 bits (105), Expect = 8e-05
Identities = 65/308 (21%), Positives = 120/308 (38%), Gaps = 52/308 (16%)
Query: 72 EHLSVKELLNRPKN--------KKITFADSDSSSS-GDDEDDEEEVQVQVGEG--DEANG 120
E++SV E + PKN K+++ +++ S G+ ++ + E+ + GE ++NG
Sbjct: 198 ENVSVHEDESGPKNEVLEGSVIKEVSLNTTENGSDDGEQQETKSELDSKTGEKGFSDSNG 257
Query: 121 D---KKLKLDSSVDEHAGIGEKAVGANGAEK--PKLDSSVDENAGIGGKTEGDEAKGDEK 175
+ L ++ + G G++G + + DE + E + K K
Sbjct: 258 ELPETNLSTSNATETTESSGSDESGSSGKSTGYQQTKNEEDEKEKVQSSEEESKVKESGK 317
Query: 176 PKLDSSVDEDAGIGEK------------AEGDEANGDKMPKMDSSVNENA------GHDN 217
+ D+S +D EK EG E +K K DSS E + +
Sbjct: 318 NEKDASSSQDESKEEKPERKKKEESSSQGEGKEEEPEKREKEDSSSQEESKEEEPENKEK 377
Query: 218 EIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANELPVVKQCCKTNVPVQ-----D 272
E + ++I + ++ EG N+N +T+ + K+ + ++ D
Sbjct: 378 EASSSQEENEIKETEIKEKEESSSQEG-NENKETEKKSSESQRKENTNSEKKIEQVESTD 436
Query: 273 SSNKVEKSIDTLIDEELRELGD-----------KKKRRFVKLESGCNGVVFIQMRKKDGD 321
SSN +K + DE RE G+ K K E+ NG +++
Sbjct: 437 SSN-TQKGDEQKTDESKRESGNDTSNKETEDDSSKTESEKKEENNRNGETEETQNEQEQT 495
Query: 322 KSPKDIVH 329
KS +I H
Sbjct: 496 KSALEISH 503
Score = 38.5 bits (88), Expect = 0.008
Identities = 35/162 (21%), Positives = 69/162 (41%), Gaps = 10/162 (6%)
Query: 101 DEDDEEEVQVQVGEGD----EANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVD 156
DE ++EE G GD + NGD+ ++ + EK G E+ DS+
Sbjct: 81 DEVEDEEGSKNEGGGDVSTDKENGDEIVEREEEEKAVEENNEKEAEGTGNEEGNEDSNNG 140
Query: 157 ENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAG-- 214
E+ + ++EG +E+ + + +DA E G E ++ +++ N+
Sbjct: 141 ESEKVVDESEGGNEISNEEAREINYKGDDAS-SEVMHGTEEKSNEKVEVEGESKSNSTEN 199
Query: 215 ---HDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKT 253
H++E G K + + + N T++ D + +TK+
Sbjct: 200 VSVHEDESGPKNEVLEGSVIKEVSLNTTENGSDDGEQQETKS 241
>At1g56660 hypothetical protein
Length = 522
Score = 47.4 bits (111), Expect = 2e-05
Identities = 57/259 (22%), Positives = 104/259 (40%), Gaps = 29/259 (11%)
Query: 77 KELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGI 136
KE ++KK+ + GD E ++EE + + E D+ +K K + ++
Sbjct: 201 KEESKSNEDKKVK-GKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKKEKDESC 259
Query: 137 GEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDE 196
E+ EK + D S ++ K +G + KG++ K D G+K + +
Sbjct: 260 AEEKKKKPDKEKKEKDESTEKE---DKKLKGKKGKGEKPEKEDE--------GKKTKEHD 308
Query: 197 ANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANE 256
A +M + E N+ K+ ID+V DD+EG+ K K
Sbjct: 309 ATEQEMDDEAADHKEGKKKKNKDKAKKKETVIDEVCEKETKDKDDDEGETKQKKNKKKE- 367
Query: 257 LPVVKQCCKTNVPVQDSSNKVEKSIDTLI----------DEELRELGDKKKRRFVKLESG 306
K+ K V++ K E ++T + + E +E D ++++ K+E G
Sbjct: 368 ----KKSEKGEKDVKEDKKK-ENPLETEVMSRDIKLEEPEAEKKEEDDTEEKKKSKVEGG 422
Query: 307 CNGVVFIQMRKKDGDKSPK 325
+ +KKD K+ K
Sbjct: 423 -ESEEGKKKKKKDKKKNKK 440
>At1g48920 unknown protein
Length = 557
Score = 47.4 bits (111), Expect = 2e-05
Identities = 40/183 (21%), Positives = 73/183 (39%), Gaps = 19/183 (10%)
Query: 94 DSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDS 153
+SS D E +EEE +V A+ + DSS D+ +KAV A
Sbjct: 62 ESSDDSDSESEEEEKAKKVPAKKAASSSDESSDDSSSDDEPA-PKKAVAAT--------- 111
Query: 154 SVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENA 213
N + K++ D + D D S DE+ + +K NG K +SS +++
Sbjct: 112 ----NGTVAKKSKDDSSSSD-----DDSSDEEVAVTKKPAAAAKNGSVKAKKESSSEDDS 162
Query: 214 GHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANELPVVKQCCKTNVPVQDS 273
++E K + + DD++ D+++ K T P + ++ +
Sbjct: 163 SSEDEPAKKPAAKIAKPAAKDSSSSDDDSDEDSEDEKPATKKAAPAAAKAASSSDSSDED 222
Query: 274 SNK 276
S++
Sbjct: 223 SDE 225
Score = 43.9 bits (102), Expect = 2e-04
Identities = 49/223 (21%), Positives = 82/223 (35%), Gaps = 9/223 (4%)
Query: 70 DGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEAN---GDKKLKL 126
D E ++ P K + +D S S D++ + V G A D
Sbjct: 68 DSESEEEEKAKKVPAKKAASSSDESSDDSSSDDEPAPKKAVAATNGTVAKKSKDDSSSSD 127
Query: 127 DSSVDEHAGIGEKAVGA--NGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDE 184
D S DE + +K A NG+ K K +SS ++++ + A KP S
Sbjct: 128 DDSSDEEVAVTKKPAAAAKNGSVKAKKESSSEDDSSSEDEPAKKPAAKIAKPAAKDSSSS 187
Query: 185 DAGIGEKAEGDEANGDKMP----KMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTD 240
D E +E ++ K K SS + + +E E P + A+ +
Sbjct: 188 DDDSDEDSEDEKPATKKAAPAAAKAASSSDSSDEDSDEESEDEKPAQKKADTKASKKSSS 247
Query: 241 DNEGDNDNIKTKTANELPVVKQCCKTNVPVQDSSNKVEKSIDT 283
D +++ +++ E P K V + SS K K+ T
Sbjct: 248 DESSESEEDESEDEEETPKKKSSDVEMVDAEKSSAKQPKTPST 290
Score = 36.6 bits (83), Expect = 0.029
Identities = 46/174 (26%), Positives = 66/174 (37%), Gaps = 30/174 (17%)
Query: 66 EDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLK 125
E + + S E +P K A DSSSS DD D++ E DE KK
Sbjct: 155 ESSSEDDSSSEDEPAKKPAAKIAKPAAKDSSSSDDDSDEDSE--------DEKPATKKAA 206
Query: 126 LDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDED 185
+ A ++ +++ + S DE + K D K SS DE
Sbjct: 207 --------PAAAKAASSSDSSDEDSDEESEDEKPA--------QKKADTKASKKSSSDES 250
Query: 186 AGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKT 239
+ E E + + ++ PK SS E D E + P K AA G+KT
Sbjct: 251 S---ESEEDESEDEEETPKKKSSDVEMV--DAEKSSAKQP-KTPSTPAAGGSKT 298
>At5g55660 putative protein
Length = 759
Score = 46.2 bits (108), Expect = 4e-05
Identities = 64/278 (23%), Positives = 117/278 (42%), Gaps = 31/278 (11%)
Query: 48 GREHQASREALNILESFYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDD--E 105
G+E Q + + ES E+ E + + + + + + T D + + +++ D E
Sbjct: 29 GKEVQENASGKEVQESKKEEDTGLEKMEIDDEGKQHEGESET-GDKEVEVTEEEKKDVGE 87
Query: 106 EEVQVQVGEGDEANGDKKLKLDSSV-----DEHAGIGEKAVGANG--AEKPKLDSSVD-- 156
++ Q + + DE DK LK D V +E A + E A+ AE P+ + +
Sbjct: 88 DKEQPEADKMDEDTDDKNLKADDGVSGVATEEDAVMKESVESADNKDAENPEGEQEKESK 147
Query: 157 ENAGIGGKTEGDEA-----------KGDEKPKLDS--SVDED---AGIGEKAEGDEANGD 200
E GGK G+E KGD+ + + +VDED + EK E + A +
Sbjct: 148 EEKLEGGKANGNEEGDTEEKLVGGDKGDDVDEAEKVENVDEDDKEEALKEKNEAELAEEE 207
Query: 201 KMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANELPVV 260
+ K + N D E K +++D + ++ +D E + ++ K E
Sbjct: 208 ETNKGEEVKEANKEDDVEADTKVAEPEVEDKKTESKDENEDKEEEKEDEKEDEKEESNDD 267
Query: 261 KQCCKTNVPVQD--SSNKVEKSI-DTLIDEELRELGDK 295
K+ K ++ + K EK+ T DEE +++ K
Sbjct: 268 KEDKKEDIKKSNKRGKGKTEKTRGKTKSDEEKKDIEPK 305
Score = 46.2 bits (108), Expect = 4e-05
Identities = 56/228 (24%), Positives = 100/228 (43%), Gaps = 36/228 (15%)
Query: 93 SDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAG---IGEKAVGANGAEK- 148
SD+ S + +++ +VQ + +E G +K+++D +H G G+K V EK
Sbjct: 24 SDAISGKEVQENASGKEVQESKKEEDTGLEKMEIDDEGKQHEGESETGDKEVEVTEEEKK 83
Query: 149 -----------PKLDSSVDE-----NAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKA 192
K+D D+ + G+ G ++A K ++S+ ++DA E
Sbjct: 84 DVGEDKEQPEADKMDEDTDDKNLKADDGVSGVATEEDAV--MKESVESADNKDA---ENP 138
Query: 193 EGDEANGDKMPKMD-SSVNENAGHDNE---IGGKENPHKIDDVHAAAGNKTDDNEGDNDN 248
EG++ K K++ N N D E +GG K DDV A + D + +
Sbjct: 139 EGEQEKESKEEKLEGGKANGNEEGDTEEKLVGG----DKGDDVDEAEKVENVDEDDKEEA 194
Query: 249 IKTKTANELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDEELRELGDKK 296
+K K EL ++ K V++++ + + DT + E E+ DKK
Sbjct: 195 LKEKNEAELAEEEETNK-GEEVKEANKEDDVEADTKVAEP--EVEDKK 239
Score = 40.4 bits (93), Expect = 0.002
Identities = 57/252 (22%), Positives = 94/252 (36%), Gaps = 44/252 (17%)
Query: 75 SVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHA 134
S K+ + K + A SD S + EDDEEE + Q E +E + + D S DE
Sbjct: 488 SQKKTEEATRTNKKSVAHSDDESEEEKEDDEEEEKEQEVEEEEEENENGIP-DKSEDEAP 546
Query: 135 GIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEK-------------PKLDSS 181
+ E E+ + ++ + G +T D+ + K P ++
Sbjct: 547 QLSESEENVESEEESEEETKKKKR---GSRTSSDKKESAGKSRSKKTAVPTKSSPPKKAT 603
Query: 182 VDEDAGIGEKAEGDEAN----GDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGN 237
AG +K++ D K K + E A + KE P V G
Sbjct: 604 QKRSAGKRKKSDDDSDTSPKASSKRKKTEKPAKEQAAAPLKSVSKEKP-----VIGKRGG 658
Query: 238 KTDD--NEGDNDNIKT-----------KTANELPVVKQC-CKTNVPVQDSSNKVEKSIDT 283
K D E ++ +KT TA ++K+ K N+ + + SI
Sbjct: 659 KGKDKNKEPSDEELKTAIIDILKGVDFNTATFTDILKRLDAKFNISLASK----KSSIKR 714
Query: 284 LIDEELRELGDK 295
+I +EL +L D+
Sbjct: 715 MIQDELTKLADE 726
Score = 33.5 bits (75), Expect = 0.25
Identities = 33/137 (24%), Positives = 53/137 (38%), Gaps = 11/137 (8%)
Query: 84 KNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGA 143
K +++ A+ + D + E EV+ + E + N DK+ + + ++ EK
Sbjct: 211 KGEEVKEANKEDDVEADTKVAEPEVEDKKTESKDENEDKEEEKEDEKED-----EKEESN 265
Query: 144 NGAEKPKLDSSVDENAGIG------GKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEA 197
+ E K D G G GKT+ DE K D +PK D + E A
Sbjct: 266 DDKEDKKEDIKKSNKRGKGKTEKTRGKTKSDEEKKDIEPKTPFFSDRPVRERKSVERLVA 325
Query: 198 NGDKMPKMDSSVNENAG 214
DK + V + G
Sbjct: 326 VVDKDSSREFHVEKGKG 342
>At5g10950 putative protein
Length = 395
Score = 46.2 bits (108), Expect = 4e-05
Identities = 58/234 (24%), Positives = 89/234 (37%), Gaps = 58/234 (24%)
Query: 64 FYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEG-------- 115
FYED KE L+ K + + D+ S D+ +EE + EG
Sbjct: 70 FYED-------GDKETLDLKKERWELIEEDDAESESDEISLQEESAGESSEGTPEPPKGK 122
Query: 116 ---------DEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKT- 165
DEA KK K+DSS A EK A KP+ + + + IGGK
Sbjct: 123 GKASLKGRTDEAMPKKKQKIDSSSKSKAKEVEKK-----ASKPETEKN-GKMGDIGGKIV 176
Query: 166 -----------------EGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGD-------K 201
+ + +K +KPK+ S + + ++ E +EA D
Sbjct: 177 AAASRMSERFRSKGNVDQKETSKASKKPKMSSKLTKRKHTDDQDEDEEAGDDIDTSSEEA 236
Query: 202 MPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDD---NEGDNDNIKTK 252
PK+ S N NA E E+ K+ + + +K +D NE +D + K
Sbjct: 237 KPKVLKSCNSNADEVAENSSDEDEPKVLKTNNSKADKDEDEEENETSDDEAEPK 290
Score = 36.6 bits (83), Expect = 0.029
Identities = 32/146 (21%), Positives = 57/146 (38%), Gaps = 9/146 (6%)
Query: 112 VGEGDEANGDKKLKLDSSV---------DEHAGIGEKAVGANGAEKPKLDSSVDENAGIG 162
V + + + KK K+ S + DE G+ ++ KPK+ S + NA
Sbjct: 192 VDQKETSKASKKPKMSSKLTKRKHTDDQDEDEEAGDDIDTSSEEAKPKVLKSCNSNADEV 251
Query: 163 GKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGK 222
+ DE + +S D+D E D+ K K+ +S ++N +++ K
Sbjct: 252 AENSSDEDEPKVLKTNNSKADKDEDEEENETSDDEAEPKALKLSNSNSDNGENNSSDDEK 311
Query: 223 ENPHKIDDVHAAAGNKTDDNEGDNDN 248
E N D+ GDN++
Sbjct: 312 EITISKITSKKIKSNTADEENGDNED 337
Score = 34.7 bits (78), Expect = 0.11
Identities = 36/147 (24%), Positives = 59/147 (39%), Gaps = 15/147 (10%)
Query: 84 KNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGA 143
K K D D + D + EE + +V + +N D+ + S DE K +
Sbjct: 212 KRKHTDDQDEDEEAGDDIDTSSEEAKPKVLKSCNSNADEVAENSSDEDE-----PKVLKT 266
Query: 144 NGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMP 203
N ++ K D +EN +T DEA +PK + ++ GE D+ +
Sbjct: 267 NNSKADK-DEDEEEN-----ETSDDEA----EPKALKLSNSNSDNGENNSSDDEKEITIS 316
Query: 204 KMDSSVNENAGHDNEIGGKENPHKIDD 230
K+ S ++ D E G E+ K D
Sbjct: 317 KITSKKIKSNTADEENGDNEDGEKAVD 343
Score = 33.9 bits (76), Expect = 0.19
Identities = 27/109 (24%), Positives = 52/109 (46%), Gaps = 12/109 (11%)
Query: 83 PKNKKITFADSDSSSSGDDEDDEE-------EVQVQVGEGDEANGDKKLKLDSSVDEHAG 135
PK K++ ++SD+ + +D++E +++ DE NGD + + +VDE +
Sbjct: 289 PKALKLSNSNSDNGENNSSDDEKEITISKITSKKIKSNTADEENGDNE-DGEKAVDEMSD 347
Query: 136 IGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDE 184
GE V +D+ + + GK E +E +G+E D+ +E
Sbjct: 348 -GEPLVSFLKKSPEGIDA---KRKKMKGKKEEEEEEGEENAGKDTKAEE 392
>At1g70620 unknown protein
Length = 897
Score = 45.8 bits (107), Expect = 5e-05
Identities = 65/282 (23%), Positives = 113/282 (40%), Gaps = 43/282 (15%)
Query: 92 DSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEK---AVGANGAEK 148
D+D+ ++ D DE V+ +G G N ++ + D A K AVG N
Sbjct: 539 DADTDAASDANADENGVE-SLGVGSRHNVSQQPSTEKLPDPEAMASAKLDPAVGVNANSG 597
Query: 149 PKLDSSVDENAGIGGKTEGDEAKGDEK---PKLDSSVDEDAGIGEKAEGDEANGDKMPKM 205
S +++ + + G T D+ G K S +D+D K D + DK +
Sbjct: 598 KNSKSGLEDYSQMPGSTRKDDEAGSTKISDVSASSGLDDDTSGSRKEHPDRTDSDKDAIL 657
Query: 206 DSSVNENAGHDNEIGGKENPHK-----IDD---------VHAAAGNKTDDNEGDN-DNIK 250
D +N+G ++ +++ +K + D V G + D++ D+ D +K
Sbjct: 658 DEPHVKNSGVKSDCNLRQDSNKPYGKDLSDEVSTDRSRIVETKGGKEKGDSQNDSKDRMK 717
Query: 251 ---TKTANELPVVKQCCKTNVP-VQDSSNKVEKSIDTLIDEELRELGDKKKRRFVKLESG 306
K+A ++ V+ K+ P V+ S VE+ T E+ + +K+K
Sbjct: 718 ENDLKSAEKVKGVESNKKSTDPHVKKDSRDVERPHRTNSKEDRGKRKEKEKEE------- 770
Query: 307 CNGVVFIQMRKKDGDKSPKDIVHRIVTSAAST----RKHMSR 344
+ R + + S KD R TS S+ RK SR
Sbjct: 771 ------ERSRHRRAENSSKDKRRRSPTSNESSDDSKRKSRSR 806
>At3g50690 putative protein
Length = 447
Score = 45.4 bits (106), Expect = 6e-05
Identities = 46/169 (27%), Positives = 74/169 (43%), Gaps = 23/169 (13%)
Query: 89 TFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEK 148
T A+ + DDEDDEE+ + E +E GD +E G GE G AE
Sbjct: 154 TDAEGNERPESDDEDDEED---EEDEEEEEEGD---------EEDPGSGE-IDGDERAEA 200
Query: 149 PKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMD-S 207
P++ + E + G + DE DE+ + E A G ANG ++ ++
Sbjct: 201 PRMSNGHSER--VDGVVDVDE---DEESDAEDDESEQA-TGVNGTSYRANGFRLEAVNGE 254
Query: 208 SVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANE 256
V E+ G D+E G +E D V ++ +D+E + D + + +E
Sbjct: 255 EVREDDGDDSESGEEEVGEDNDVVEV---HEIEDSENEEDGVDDEEDDE 300
Score = 40.0 bits (92), Expect = 0.003
Identities = 32/156 (20%), Positives = 70/156 (44%), Gaps = 4/156 (2%)
Query: 85 NKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGAN 144
N++ D D +DE++EEE + E +GD++ + + H+ + V +
Sbjct: 159 NERPESDDEDDEEDEEDEEEEEEGDEEDPGSGEIDGDERAEAPRMSNGHSERVDGVVDVD 218
Query: 145 GAEKPKLDSSVDENA-GIGG---KTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGD 200
E+ + E A G+ G + G + ++ +D+ GE+ G++ +
Sbjct: 219 EDEESDAEDDESEQATGVNGTSYRANGFRLEAVNGEEVREDDGDDSESGEEEVGEDNDVV 278
Query: 201 KMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAG 236
++ +++ S NE G D+E +E+ + + +A G
Sbjct: 279 EVHEIEDSENEEDGVDDEEDDEEDEEEEEVDNADRG 314
Score = 34.7 bits (78), Expect = 0.11
Identities = 51/225 (22%), Positives = 90/225 (39%), Gaps = 35/225 (15%)
Query: 46 EGGREHQASREALNILESFYE------DLVDGEHLSVKELLNRPKNKKITFADSDSSSSG 99
E E S +A + + Y + V+GE + + + ++ D+D
Sbjct: 222 ESDAEDDESEQATGVNGTSYRANGFRLEAVNGEEVREDDGDDSESGEEEVGEDNDVVEVH 281
Query: 100 DDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENA 159
+ ED E E E D+ +++ ++D++ ++ +G +G+ + NA
Sbjct: 282 EIEDSENEEDGVDDEEDDEEDEEEEEVDNA--------DRGLGGSGSTSRLM------NA 327
Query: 160 G-IGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNE 218
G I G +GD+ DE D D GE +G E +G+ D V E E
Sbjct: 328 GEIDGHEQGDD---DE--------DGDGETGEDDQGVEDDGE-FADEDDDVEEEDEESGE 375
Query: 219 IGGKENPHKIDDVHAAAGNKTDDNEGDND-NIKTKTANELPVVKQ 262
G P + H A GN + DND + + + ++LP+ Q
Sbjct: 376 -GYLVQPVSQVEDHDAVGNDIEPINEDNDPDEEEEVEDDLPIPDQ 419
Score = 33.5 bits (75), Expect = 0.25
Identities = 34/159 (21%), Positives = 66/159 (41%), Gaps = 20/159 (12%)
Query: 143 ANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDA---------GIGEKAE 193
A G E+P+ D DE + E +E +GDE+ +D D G E+ +
Sbjct: 156 AEGNERPESDDEDDEE---DEEDEEEEEEGDEEDPGSGEIDGDERAEAPRMSNGHSERVD 212
Query: 194 G-DEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTK 252
G + + D+ + +E A N + N +++ V+ + D ++ ++ +
Sbjct: 213 GVVDVDEDEESDAEDDESEQATGVNGTSYRANGFRLEAVNGEEVREDDGDDSESGEEEVG 272
Query: 253 TANELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDEELRE 291
N++ V + ++DS N+ + D DEE E
Sbjct: 273 EDNDVVEVHE-------IEDSENEEDGVDDEEDDEEDEE 304
>At2g11570 hypothetical protein
Length = 305
Score = 44.3 bits (103), Expect = 1e-04
Identities = 51/227 (22%), Positives = 94/227 (40%), Gaps = 52/227 (22%)
Query: 60 ILESFYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEAN 119
I+++ D + +L + +L + K D + ++G+ D G+
Sbjct: 28 IVKTAVADAMQDVYLRLDKLERDAETSKTKEKDREEEATGEKTD---------GKTGFGG 78
Query: 120 GDKKLKLDSSVD--EHAGI----------GEKAVGANGAEKPKLDSSVD-----ENAGIG 162
GD + D+ D ++ G+ GEKAVG +G E K D + + +G
Sbjct: 79 GDFRGDCDTGFDGGDYRGVAETGLVEVTSGEKAVGESGHEALKKDGMLKLVADVKEIDVG 138
Query: 163 GKTEGDEAKGDE----KP-----------------KLDSSVDED--AGIGEKAEGDEANG 199
K DE GD+ KP ++ SVD++ +G+ + +G+ +G
Sbjct: 139 DKAVDDEMVGDDVVETKPGFGNTVNEILRTDDVLGVVEKSVDDELRSGVKKSVDGELRSG 198
Query: 200 DKMPKMDSSV-NENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGD 245
D+ ++ SV N +NE GG++ H V + K+ D++ D
Sbjct: 199 DEPSCLEKSVINNEPTSENETGGEKRVHFF--VEGSGFEKSSDDKSD 243
>At1g47970 Unknown protein (T2J15.12)
Length = 198
Score = 44.3 bits (103), Expect = 1e-04
Identities = 43/181 (23%), Positives = 72/181 (39%), Gaps = 40/181 (22%)
Query: 70 DGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQ------------------VQ 111
D + VK++ ++++ D D S GDD+DDEEE + VQ
Sbjct: 25 DSDEEQVKDV-----GEEVSDEDDDDGSEGDDDDDEEEEEDDDDDDDVQVLQSLGGPPVQ 79
Query: 112 VGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIG--------G 163
E ++ GD+ D + D+ G+ + E D V++ +G G
Sbjct: 80 SAEDEDEEGDE----DGNGDDDDDDGDDDDDDDDDE----DEDVEDEGDLGTEYLVRPVG 131
Query: 164 KTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKE 223
+ E +E D +P+ ++ V+ED GE E D + G + D E G E
Sbjct: 132 RAEDEEDASDFEPE-ENGVEEDIDEGEDDENDNSGGAGKSEAPPKRKRAPEEDEEDSGDE 190
Query: 224 N 224
+
Sbjct: 191 D 191
>At3g30170 unknown protein
Length = 995
Score = 43.5 bits (101), Expect = 2e-04
Identities = 39/154 (25%), Positives = 57/154 (36%), Gaps = 21/154 (13%)
Query: 84 KNKKITFADSDSSSSGDDEDDEEEVQVQ-----------VGEGDEANGDKKLKLDSSVDE 132
K +KI D D + D D E++ V VG D+A G + + DE
Sbjct: 67 KRRKIRVGDEDEAEIDPDPDYEDDCAVHGDEDCDAYTDAVGGDDDATGG-----EGNDDE 121
Query: 133 HAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKA 192
G G+ A G G + L D + G + + D L E AG+ E
Sbjct: 122 AVGGGDDATGCGGNDDEALGYDDDATSDGGNDDDDGGVEEDANLNLAEEFPEFAGVEEDC 181
Query: 193 EGDEA-----NGDKMPKMDSSVNENAGHDNEIGG 221
D+ DK+P SS +E+ E+ G
Sbjct: 182 SDDDPPDDVWEEDKIPDPFSSDDEDESRTREVHG 215
Score = 30.8 bits (68), Expect = 1.6
Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 8/81 (9%)
Query: 168 DEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKE---N 224
D+A G E + DE G G+ A G N D+ D + G+D++ GG E N
Sbjct: 110 DDATGGE-----GNDDEAVGGGDDATGCGGNDDEALGYDDDATSDGGNDDDDGGVEEDAN 164
Query: 225 PHKIDDVHAAAGNKTDDNEGD 245
+ ++ AG + D ++ D
Sbjct: 165 LNLAEEFPEFAGVEEDCSDDD 185
>At1g64330 unknown protein
Length = 555
Score = 43.5 bits (101), Expect = 2e-04
Identities = 68/312 (21%), Positives = 129/312 (40%), Gaps = 47/312 (15%)
Query: 92 DSDSSSSGDDEDD---------EEEVQVQVGEGDEAN---GDKKLKLDSSVDEHAGIGEK 139
DS SSSS D + D E E+++ + ++AN D K+KL ++ DEH ++
Sbjct: 95 DSSSSSSSDSDSDKKSKRNGRGENEIELLKKQMEDANLEIADLKMKL-ATTDEH----KE 149
Query: 140 AVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAE-GDEAN 198
AV + E K E DE G+ + + + E+ + EK E E
Sbjct: 150 AVESEHQEILK------------KLKESDEICGNLRVETEKLTSENKELNEKLEVAGETE 197
Query: 199 GDKMPKMDSSVNENAGHDNEIGGKENPHK--IDDVHAAAG--NKTDDNEGDNDNIKTKTA 254
D K++ E G + E+ K H+ +++V+ G N+T+ K
Sbjct: 198 SDLNQKLEDVKKERDGLEAELASKAKDHESTLEEVNRLQGQKNETEAELEREKQEKPALL 257
Query: 255 NELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDEE---LRELGDKKKRRFVKLESGCNGVV 311
N++ V++ ++ ++ K I+ L +E +++L D K+ LE + +
Sbjct: 258 NQINDVQKALLEQEAAYNTLSQEHKQINGLFEEREATIKKLTDDYKQAREMLEEYMSKME 317
Query: 312 FIQMRKKDGDK---SPKDIVHRIVTSAASTRKHMSR-------FILRMLPIEVSCYASKE 361
+ R ++ K S + + + + S R + R + +M IEV S +
Sbjct: 318 ETERRMQETGKDVASRESAIVDLEETVESLRNEVERKGDEIESLMEKMSNIEVKLRLSNQ 377
Query: 362 EISRAIKPLVEQ 373
++ + L E+
Sbjct: 378 KLRVTEQVLTEK 389
>At3g15120 chaperone-like ATPase
Length = 1954
Score = 42.0 bits (97), Expect = 7e-04
Identities = 45/191 (23%), Positives = 76/191 (39%), Gaps = 17/191 (8%)
Query: 144 NGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMP 203
N A+KP +D E++ GGK D + + D+S ED E E +A+
Sbjct: 165 NKAKKP-VDVKESESSEDGGKESDTSNSEDVQKESDTSNSEDESASESEESMQADSAARE 223
Query: 204 KMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTD---------DNEGDNDNI----K 250
K + A + EN ++D + + TD D EG+++ K
Sbjct: 224 KYQE---KKATKRSVFLESENEAEVDRTETESEDGTDSTDNEIDDSDEEGESETQCSAEK 280
Query: 251 TKTANELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDEELRELGDKKKRRFVKLESGCNGV 310
T + E V + TNV ++ N+ ++ L +E + D+KK V + NG
Sbjct: 281 TGSETEANVEEMRADTNVTMEAVQNESRNQMEELENEIEMGVEDEKKEMSVIVSESGNGT 340
Query: 311 VFIQMRKKDGD 321
+ K+ D
Sbjct: 341 GIREDENKEMD 351
Score = 35.8 bits (81), Expect = 0.050
Identities = 51/229 (22%), Positives = 91/229 (39%), Gaps = 45/229 (19%)
Query: 80 LNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEK 139
LN+ K K + +S+SS G E D + E D +N S DE A E+
Sbjct: 164 LNKAK-KPVDVKESESSEDGGKESDTSNSEDVQKESDTSN---------SEDESASESEE 213
Query: 140 AVGANGAEKPKLDSS---------VDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGE 190
++ A+ A + K EN +TE + G + D+ +D+ GE
Sbjct: 214 SMQADSAAREKYQEKKATKRSVFLESENEAEVDRTETESEDGTD--STDNEIDDSDEEGE 271
Query: 191 --------------KAEGDEANGDKMPKMDSSVNENAG------HDNEIGGKENPHKIDD 230
+A +E D M++ NE+ ++ E+G ++ ++
Sbjct: 272 SETQCSAEKTGSETEANVEEMRADTNVTMEAVQNESRNQMEELENEIEMGVEDEKKEMSV 331
Query: 231 VHAAAGNKTDDNEGDN---DNIKTKTANELPVVK-QCCKTNVPVQDSSN 275
+ + +GN T E +N D I +++ N +++ + K V V S N
Sbjct: 332 IVSESGNGTGIREDENKEMDVIVSESGNGTGILEGENKKMEVMVSGSGN 380
>At4g32070 putative protein
Length = 811
Score = 41.6 bits (96), Expect = 0.001
Identities = 39/124 (31%), Positives = 54/124 (43%), Gaps = 16/124 (12%)
Query: 76 VKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAG 135
VKE L + K KK S G DE+ + V V +GDEA G K K + S + +
Sbjct: 201 VKERLRKSKKKK--------KSGGKDEELKSPKVVVVDKGDEAEGRNKPKEEKS--DKSD 250
Query: 136 IGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEK-----PKLDSSVDEDAGIGE 190
I K G +K S + GG G+E K ++K ++ +S D G G
Sbjct: 251 IDGKIGGKREEKKTSFKSDKGQKKKSGGNKAGEERKVEDKVVVMDKEVIASEIVDGG-GS 309
Query: 191 KAEG 194
K EG
Sbjct: 310 KKEG 313
>At4g26630 unknown protein
Length = 763
Score = 41.6 bits (96), Expect = 0.001
Identities = 56/243 (23%), Positives = 94/243 (38%), Gaps = 41/243 (16%)
Query: 48 GREHQASREALNILESFYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDD--- 104
G+E+ +E + + ED+ + +++ + + + K T +D +
Sbjct: 28 GKENAGGKETQELAKD--EDMAEPDNMEIDAQIKKDDEKAETEDKESEVKKNEDNAETQK 85
Query: 105 -EEEVQVQVGEG-------DEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVD 156
EE+V+V EG DE KK + D V + ++ V + K D
Sbjct: 86 MEEKVEVTKDEGQAEATNMDEDADGKKEQTDDGVSVEDTVMKENVESKDNNYAKDDEKET 145
Query: 157 ENAGIGGKTEGDEAKGD------EKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVN 210
+ I TE D K E K + + D + G K EG + DK MD V
Sbjct: 146 KETDI---TEADHKKAGKEDIQHEADKANGTKDGNTG-DIKEEGTLVDEDKGTDMDEKV- 200
Query: 211 ENAGHDNEIGGKENPHKID-------DVHAAAG----NKTDD------NEGDNDNIKTKT 253
EN + ++ E K D +V AA +K +D +E DN+ +++K
Sbjct: 201 ENGDENKQVENVEGKEKEDKEENKTKEVEAAKAEVDESKVEDEKEGSEDENDNEKVESKD 260
Query: 254 ANE 256
A E
Sbjct: 261 AKE 263
Score = 35.8 bits (81), Expect = 0.050
Identities = 39/164 (23%), Positives = 67/164 (40%), Gaps = 11/164 (6%)
Query: 86 KKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAV---- 141
+K + A + ++G E E + E D D ++K D DE A +K
Sbjct: 20 EKPSEAMAGKENAGGKETQELAKDEDMAEPDNMEIDAQIKKD---DEKAETEDKESEVKK 76
Query: 142 GANGAEKPKLDSSVD--ENAGIGGKTEGDEAKGDEKPKLDSSVD-EDAGIGEKAEGDEAN 198
+ AE K++ V+ ++ G T DE +K + D V ED + E E + N
Sbjct: 77 NEDNAETQKMEEKVEVTKDEGQAEATNMDEDADGKKEQTDDGVSVEDTVMKENVESKDNN 136
Query: 199 GDKMPKMDSSVNENAGHDNEIGGKEN-PHKIDDVHAAAGNKTDD 241
K + ++ + D++ GKE+ H+ D + T D
Sbjct: 137 YAKDDEKETKETDITEADHKKAGKEDIQHEADKANGTKDGNTGD 180
Score = 32.3 bits (72), Expect = 0.56
Identities = 46/234 (19%), Positives = 90/234 (37%), Gaps = 47/234 (20%)
Query: 71 GEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSV 130
G S + ++ K+++ T S + DDE +EE+ + + E ++A
Sbjct: 491 GSSSSKRSAKSQKKSEEATKVVKKSLAHSDDESEEEKEEEEKQEEEKAE----------- 539
Query: 131 DEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGE 190
EK + +EN GI K+E DE P+ S ++D E
Sbjct: 540 ----------------EKEEKKEEENEN-GIPDKSE------DEAPQPSESEEKDES-EE 575
Query: 191 KAEGDEANGDKMPKMDSSVNENAGHDNE----IGGKEN-PHKIDDVHAAAGNKTDDNEGD 245
+E + + ++ + E+AG + K + P KI ++A K D++ D
Sbjct: 576 HSEEETTKKKRGSRLSAGKKESAGRARNKKAVVAAKSSPPEKITQKRSSAKRKKTDDDSD 635
Query: 246 N----DNIKTKTANEL---PVVKQCCKTNVPVQDSSNKVEKSIDTLIDEELREL 292
+ + K+ N + P + PV+ + +K D ++ + E+
Sbjct: 636 TSPKASSKRKKSENPIKASPAPSKSASKEKPVKRAGKGKDKPSDKVLKNAIVEI 689
>At4g07520 hypothetical protein
Length = 734
Score = 41.6 bits (96), Expect = 0.001
Identities = 30/131 (22%), Positives = 60/131 (44%), Gaps = 13/131 (9%)
Query: 100 DDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENA 159
+DE+ E+E + E +E K+ VD+ + + + N E+ K +E+
Sbjct: 40 EDENCEQEPPKNLHEPEEE------KISEEVDDEEPMQSQGMEENPEEEEKEGEEEEESE 93
Query: 160 GIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEI 219
EGD+ + + ++ + E+ GE+ E +E + D+ P + EN + +
Sbjct: 94 ------EGDDVEPMQSQGMEENPKEEEKEGEEEESEEIDDDE-PMPSHGMEENPQEEEKE 146
Query: 220 GGKENPHKIDD 230
+ENP ++DD
Sbjct: 147 REEENPEELDD 157
Score = 33.5 bits (75), Expect = 0.25
Identities = 24/121 (19%), Positives = 49/121 (39%), Gaps = 2/121 (1%)
Query: 127 DSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDA 186
D + +E A K N ++P + E I + + +E + ++ + +E+
Sbjct: 26 DMNREEVAAEENKFEDENCEQEPPKNLHEPEEEKISEEVDDEEPM--QSQGMEENPEEEE 83
Query: 187 GIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDN 246
GE+ E E D P + EN + + G +E +IDD + ++N +
Sbjct: 84 KEGEEEEESEEGDDVEPMQSQGMEENPKEEEKEGEEEESEEIDDDEPMPSHGMEENPQEE 143
Query: 247 D 247
+
Sbjct: 144 E 144
>At4g02400
Length = 870
Score = 41.2 bits (95), Expect = 0.001
Identities = 53/246 (21%), Positives = 95/246 (38%), Gaps = 37/246 (15%)
Query: 72 EHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEAN-----GDKKLKL 126
E L + L+R N ++ SS D+ DDEEE+ + + +K LK
Sbjct: 404 EQLQINATLSRKMNS------TNDGSSSDESDDEEELSCGSDQDTPSKLIAKAREKTLKT 457
Query: 127 DSSVD-EHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDED 185
D ++G+ A +K +++ + G E + G++ PK + V
Sbjct: 458 MEDDDVPNSGLLSLPFMARAMKKKNEEANEEAKRAFGEYKELENFGGEDNPKKSADVSGR 517
Query: 186 AGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPH--KIDDVHAAAGN-----K 238
G ++ + K K + +N+ DN++ G EN + D + A N +
Sbjct: 518 RVFGATSKVEAPKESK--KDSDNFYDNSDSDNDMEGIENNDLGAVGDTASPARNTGAITE 575
Query: 239 TDDNEGDNDNIKTKTANELPVV----------------KQCCKTNVPVQDSSNKVEKSID 282
T+ GD +N +KT ++ + K+ KT VP+ +K E +
Sbjct: 576 TEKCCGDVENPASKTTFDVALFASGSWKKMKGSQNAESKKAPKTRVPISKGQDKKESRDE 635
Query: 283 TLIDEE 288
D E
Sbjct: 636 ESEDSE 641
>At3g43060 putative protein
Length = 585
Score = 40.8 bits (94), Expect = 0.002
Identities = 37/143 (25%), Positives = 60/143 (41%), Gaps = 31/143 (21%)
Query: 131 DEHAGIGEKAVGANGAEKPKLDSSVDENAGIG-GKTEGDEAKGDEKPKLDSSVDEDAGIG 189
DE AG+G A+ D+ G G G E +E DE+ + + DE AG+
Sbjct: 422 DEMAGVGATGKKAS-----------DDIGGSGFGNEEQEEDASDEEGAKEENDDEMAGVE 470
Query: 190 EK---------AEGDEANGDKMPKMDSSVNENA------GHDNEIGGKENPHKIDDVHAA 234
E+ EG E + D+M ++ +E G + E +E P + DD
Sbjct: 471 EEDAEQVDATDEEGAEGDNDEMAGVEEETSEKVDATDEEGAEEEASNREGPEEGDD---- 526
Query: 235 AGNKTDDNEGDNDNIKTKTANEL 257
A N+ EGD++ + + E+
Sbjct: 527 ASNREGVEEGDDEMAEKEEVAEI 549
Score = 33.1 bits (74), Expect = 0.33
Identities = 23/96 (23%), Positives = 39/96 (39%), Gaps = 4/96 (4%)
Query: 92 DSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKL 151
++D +G +E+D E+V EG E + D+ + V+E A GAE+
Sbjct: 461 ENDDEMAGVEEEDAEQVDATDEEGAEGDNDEM----AGVEEETSEKVDATDEEGAEEEAS 516
Query: 152 DSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAG 187
+ E EG E DE + + + +G
Sbjct: 517 NREGPEEGDDASNREGVEEGDDEMAEKEEVAEISSG 552
Score = 32.0 bits (71), Expect = 0.73
Identities = 22/110 (20%), Positives = 45/110 (40%), Gaps = 7/110 (6%)
Query: 92 DSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKL 151
D+ +E+D+E V+ + ++ + + + DE AG+ E+ K+
Sbjct: 451 DASDEEGAKEENDDEMAGVEEEDAEQVDATDEEGAEGDNDEMAGVEEET-------SEKV 503
Query: 152 DSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDK 201
D++ +E A +GD+ + + D + EK E E + K
Sbjct: 504 DATDEEGAEEEASNREGPEEGDDASNREGVEEGDDEMAEKEEVAEISSGK 553
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.308 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,180,373
Number of Sequences: 26719
Number of extensions: 558690
Number of successful extensions: 3699
Number of sequences better than 10.0: 280
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 235
Number of HSP's that attempted gapping in prelim test: 2800
Number of HSP's gapped (non-prelim): 697
length of query: 456
length of database: 11,318,596
effective HSP length: 103
effective length of query: 353
effective length of database: 8,566,539
effective search space: 3023988267
effective search space used: 3023988267
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0018.6


