

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0018.1
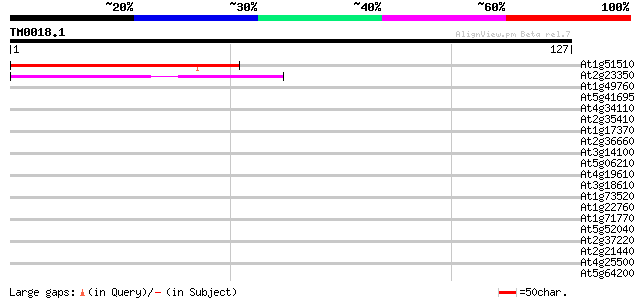
(127 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g51510 unknown protein 76 5e-15
At2g23350 putative poly(A) binding protein 40 4e-04
At1g49760 Putative Poly-A Binding Protein (F14J22.3) 37 0.002
At5g41695 putative protein 37 0.003
At4g34110 poly(A)-binding protein 36 0.006
At2g35410 putative chloroplast RNA binding protein precursor 34 0.017
At1g17370 oligouridylate binding protein, putative 32 0.066
At2g36660 putative poly(A) binding protein 32 0.086
At3g14100 oligouridylate binding protein, putative 32 0.11
At5g06210 unknown protein 31 0.15
At4g19610 putative protein 31 0.15
At3g18610 unknown protein 31 0.19
At1g73520 31 0.19
At1g22760 putative polyA-binding protein, PAB3 31 0.19
At1g71770 polyadenylate-binding protein 5 30 0.25
At5g52040 arginine/serine-rich splicing factor RSP41 homolog 30 0.33
At2g37220 putative RNA-binding protein 30 0.33
At2g21440 unknown protein (At2g21440) 30 0.33
At4g25500 splicing factor At-SRp40 30 0.43
At5g64200 unknown protein 29 0.56
>At1g51510 unknown protein
Length = 202
Score = 75.9 bits (185), Expect = 5e-15
Identities = 37/53 (69%), Positives = 45/53 (84%), Gaps = 1/53 (1%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPI-NESIRRKNARH 52
GYALIEYE+ EEA++AI +NG+ELLTQN+ VDWAFSSGP ES RRKN+R+
Sbjct: 137 GYALIEYEKKEEAQSAISAMNGAELLTQNVSVDWAFSSGPSGGESYRRKNSRY 189
>At2g23350 putative poly(A) binding protein
Length = 662
Score = 39.7 bits (91), Expect = 4e-04
Identities = 21/62 (33%), Positives = 32/62 (50%), Gaps = 6/62 (9%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRKNARHPVMNLNIY 60
GY ++++ + AKNAIE LNG L + I+V GP R++A + N+Y
Sbjct: 175 GYGFVQFDTEDSAKNAIEKLNGKVLNDKQIFV------GPFLRKEERESAADKMKFTNVY 228
Query: 61 FK 62
K
Sbjct: 229 VK 230
Score = 25.4 bits (54), Expect = 8.1
Identities = 10/31 (32%), Positives = 17/31 (54%)
Query: 2 YALIEYERAEEAKNAIENLNGSELLTQNIYV 32
+ + +E E+A A+E LNG + + YV
Sbjct: 267 FGFVNFENPEDAARAVEALNGKKFDDKEWYV 297
>At1g49760 Putative Poly-A Binding Protein (F14J22.3)
Length = 671
Score = 37.4 bits (85), Expect = 0.002
Identities = 20/62 (32%), Positives = 31/62 (49%), Gaps = 6/62 (9%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRKNARHPVMNLNIY 60
GY ++Y+ E A+ AI+ LNG L + +YV GP ++R + V N+Y
Sbjct: 174 GYGFVQYDTDEAAQGAIDKLNGMLLNDKQVYV------GPFVHKLQRDPSGEKVKFTNVY 227
Query: 61 FK 62
K
Sbjct: 228 VK 229
Score = 27.3 bits (59), Expect = 2.1
Identities = 11/32 (34%), Positives = 18/32 (55%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYV 32
G + + EEA AI +NG ++T+ +YV
Sbjct: 368 GSGFVAFSTPEEATRAITEMNGKMIVTKPLYV 399
>At5g41695 putative protein
Length = 548
Score = 37.0 bits (84), Expect = 0.003
Identities = 19/54 (35%), Positives = 28/54 (51%), Gaps = 4/54 (7%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWA----FSSGPINESIRRKNA 50
GY +E+ A+E K A+EN NG L I++D A + GP E R+ +
Sbjct: 213 GYGFVEFASADETKKALENKNGEYLHDHKIFIDVAKTAPYPPGPNYEDYLRRES 266
Score = 28.9 bits (63), Expect = 0.73
Identities = 10/33 (30%), Positives = 20/33 (60%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVD 33
G +E+ EA+ A++ +NG L ++ I++D
Sbjct: 458 GCGFVEFASVNEAQKALQKMNGENLRSREIFLD 490
>At4g34110 poly(A)-binding protein
Length = 629
Score = 35.8 bits (81), Expect = 0.006
Identities = 20/62 (32%), Positives = 28/62 (44%), Gaps = 6/62 (9%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRKNARHPVMNLNIY 60
GY ++Y E A+ AIE LNG L + +YV GP R + + N+Y
Sbjct: 165 GYGFVQYANEESAQKAIEKLNGMLLNDKQVYV------GPFLRRQERDSTANKTKFTNVY 218
Query: 61 FK 62
K
Sbjct: 219 VK 220
Score = 30.4 bits (67), Expect = 0.25
Identities = 18/68 (26%), Positives = 30/68 (43%), Gaps = 6/68 (8%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIR------RKNARHPV 54
G+ + +E A++A A+E+LNG + + YV A +R K A
Sbjct: 256 GFGFVNFENADDAARAVESLNGHKFDDKEWYVGRAQKKSERETELRVRYEQNLKEAADKF 315
Query: 55 MNLNIYFK 62
+ N+Y K
Sbjct: 316 QSSNLYVK 323
>At2g35410 putative chloroplast RNA binding protein precursor
Length = 308
Score = 34.3 bits (77), Expect = 0.017
Identities = 18/53 (33%), Positives = 29/53 (53%), Gaps = 3/53 (5%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINE---SIRRKNA 50
GY + + EEA+NAI LNG E++ + I + ++ S +E S+ NA
Sbjct: 237 GYGFVSFATREEAENAITKLNGKEIMGRPITLKFSLRSASESEDGDSVEANNA 289
>At1g17370 oligouridylate binding protein, putative
Length = 419
Score = 32.3 bits (72), Expect = 0.066
Identities = 16/38 (42%), Positives = 21/38 (55%)
Query: 2 YALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSG 39
Y + Y A AI +LNG L Q I V+WA++SG
Sbjct: 93 YGFVHYFDRRSAGLAILSLNGRHLFGQPIKVNWAYASG 130
>At2g36660 putative poly(A) binding protein
Length = 609
Score = 32.0 bits (71), Expect = 0.086
Identities = 18/74 (24%), Positives = 36/74 (48%), Gaps = 6/74 (8%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRK-NARHPVMNL-- 57
GYA + ++ E+A+ A E +NG++ ++ +YV A + +R + +H +
Sbjct: 242 GYAFVNFDNPEDARRAAETVNGTKFGSKCLYVGRAQKKAEREQLLREQFKEKHEEQKMIA 301
Query: 58 ---NIYFKTGQLVI 68
NIY K + +
Sbjct: 302 KVSNIYVKNVNVAV 315
Score = 28.1 bits (61), Expect = 1.2
Identities = 11/32 (34%), Positives = 20/32 (62%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYV 32
GY +++E+ + A AI+ LN + + + IYV
Sbjct: 153 GYGFVQFEQEDAAHAAIQTLNSTIVADKEIYV 184
>At3g14100 oligouridylate binding protein, putative
Length = 427
Score = 31.6 bits (70), Expect = 0.11
Identities = 15/38 (39%), Positives = 21/38 (54%)
Query: 2 YALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSG 39
Y + Y A AI +LNG L Q I V+WA+++G
Sbjct: 98 YGFVHYFDRRSAALAILSLNGRHLFGQPIKVNWAYATG 135
Score = 28.9 bits (63), Expect = 0.73
Identities = 10/35 (28%), Positives = 20/35 (56%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWA 35
G+ + + ++A+ AI +NG L ++ I +WA
Sbjct: 186 GFGFVSFRNQQDAQTAINEMNGKWLSSRQIRCNWA 220
>At5g06210 unknown protein
Length = 146
Score = 31.2 bits (69), Expect = 0.15
Identities = 12/35 (34%), Positives = 22/35 (62%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWA 35
G+ + + A+EA+ A+ NG +L + I+VD+A
Sbjct: 76 GFGFVTFASADEAQKALMEFNGQQLNGRTIFVDYA 110
>At4g19610 putative protein
Length = 772
Score = 31.2 bits (69), Expect = 0.15
Identities = 15/50 (30%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRKNA 50
GYA +E+ +EA NA + L + +++ ++WA + E+IR+++A
Sbjct: 699 GYAFVEFVTKQEALNAKKALASTHFYGRHLVLEWANDDNSM-EAIRKRSA 747
>At3g18610 unknown protein
Length = 636
Score = 30.8 bits (68), Expect = 0.19
Identities = 20/49 (40%), Positives = 29/49 (58%), Gaps = 2/49 (4%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSG-PINESIRRK 48
GY IE+ EEA+ A+E +NG LL +++ +D A G P N + RK
Sbjct: 425 GYGHIEFASPEEAQKALE-MNGKLLLGRDVRLDLANERGTPRNSNPGRK 472
>At1g73520
Length = 74
Score = 30.8 bits (68), Expect = 0.19
Identities = 16/53 (30%), Positives = 30/53 (56%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRKNARHP 53
G+A + YE EEA AI+ ++G L + I+V+ A + ++ + R++ P
Sbjct: 12 GFAFLRYETEEEAMKAIQGMHGKFLDGRVIFVEEAKTRSDMSRAKPRRDFPKP 64
>At1g22760 putative polyA-binding protein, PAB3
Length = 660
Score = 30.8 bits (68), Expect = 0.19
Identities = 19/67 (28%), Positives = 28/67 (41%), Gaps = 6/67 (8%)
Query: 2 YALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRK------NARHPVM 55
+ + +E E A +A+E +NG L +YV A E +RRK N
Sbjct: 271 FGFVNFECTEAAASAVEKMNGISLGDDVLYVGRAQKKSEREEELRRKFEQERINRFEKSQ 330
Query: 56 NLNIYFK 62
N+Y K
Sbjct: 331 GANLYLK 337
Score = 30.4 bits (67), Expect = 0.25
Identities = 20/69 (28%), Positives = 32/69 (45%), Gaps = 4/69 (5%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRKNARHPVMNLNIY 60
GY +++E+ E A+ AI+ LNG + + ++V E R +N P N+Y
Sbjct: 177 GYGFVQFEKEESAQAAIDKLNGMLMNDKQVFVGHFIRR---QERARDENTPTPRFT-NVY 232
Query: 61 FKTGQLVIG 69
K IG
Sbjct: 233 VKNLPKEIG 241
Score = 25.8 bits (55), Expect = 6.2
Identities = 9/32 (28%), Positives = 17/32 (53%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYV 32
G+ + Y EEA A+ +NG + + +Y+
Sbjct: 373 GFGFVAYSNPEEALRALSEMNGKMIGRKPLYI 404
>At1g71770 polyadenylate-binding protein 5
Length = 668
Score = 30.4 bits (67), Expect = 0.25
Identities = 12/32 (37%), Positives = 21/32 (65%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYV 32
GY +++E+ E A+ AI+ LNG L + ++V
Sbjct: 173 GYGFVQFEKEETAQAAIDKLNGMLLNDKQVFV 204
Score = 26.6 bits (57), Expect = 3.6
Identities = 14/47 (29%), Positives = 21/47 (43%)
Query: 2 YALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRK 48
+ + + E A A+E +NG L +YV A E +RRK
Sbjct: 267 FGFVNFVSPEAAAVAVEKMNGISLGEDVLYVGRAQKKSDREEELRRK 313
>At5g52040 arginine/serine-rich splicing factor RSP41 homolog
Length = 356
Score = 30.0 bits (66), Expect = 0.33
Identities = 13/34 (38%), Positives = 23/34 (67%)
Query: 2 YALIEYERAEEAKNAIENLNGSELLTQNIYVDWA 35
+A I+YE E+A A++ N S+L+ + I V++A
Sbjct: 132 FAFIQYEAQEDATRALDATNSSKLMDKVISVEYA 165
>At2g37220 putative RNA-binding protein
Length = 289
Score = 30.0 bits (66), Expect = 0.33
Identities = 13/40 (32%), Positives = 26/40 (64%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGP 40
G+ + Y+ ++E +NAI++L+G++L + I V A + P
Sbjct: 246 GFGFVTYDSSQEVQNAIKSLDGADLDGRQIRVSEAEARPP 285
>At2g21440 unknown protein (At2g21440)
Length = 1003
Score = 30.0 bits (66), Expect = 0.33
Identities = 13/35 (37%), Positives = 21/35 (59%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWA 35
G+A +++ ++A NAI+ NG + I VDWA
Sbjct: 373 GFAFVKFTCKKDAANAIKKFNGHMFGKRPIAVDWA 407
>At4g25500 splicing factor At-SRp40
Length = 350
Score = 29.6 bits (65), Expect = 0.43
Identities = 13/34 (38%), Positives = 23/34 (67%)
Query: 2 YALIEYERAEEAKNAIENLNGSELLTQNIYVDWA 35
+A I+YE E+A A++ N S+L+ + I V++A
Sbjct: 133 FAFIQYEAQEDATRALDASNNSKLMDKVISVEYA 166
>At5g64200 unknown protein
Length = 303
Score = 29.3 bits (64), Expect = 0.56
Identities = 17/53 (32%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRKNARHP 53
G+A + Y+ +EA A+E L+G + + I V +A GP E I + P
Sbjct: 58 GFAFVRYKYKDEAHKAVERLDGRVVDGREITVQFA-KYGPNAEKISKGRVVEP 109
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.141 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,828,716
Number of Sequences: 26719
Number of extensions: 108291
Number of successful extensions: 350
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 267
Number of HSP's gapped (non-prelim): 92
length of query: 127
length of database: 11,318,596
effective HSP length: 87
effective length of query: 40
effective length of database: 8,994,043
effective search space: 359761720
effective search space used: 359761720
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0018.1


