

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0017.9
(387 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
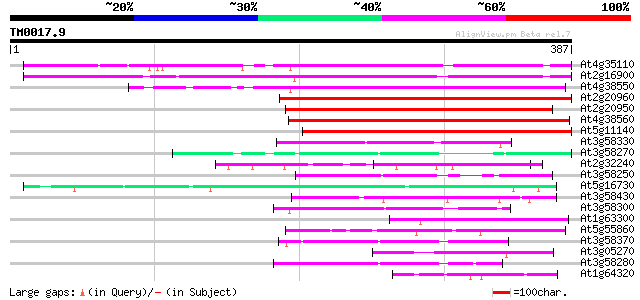
Score E
Sequences producing significant alignments: (bits) Value
At4g35110 unknown protein 172 4e-43
At2g16900 unknown protein 164 8e-41
At4g38550 Phospholipase like protein 139 2e-33
At2g20960 pEARLI 4 protein 135 4e-32
At2g20950 unknown protein 134 8e-32
At4g38560 Phospholipase like protein 132 3e-31
At5g11140 putative protein 128 6e-30
At3g58330 putative protein 59 6e-09
At3g58270 unknown protein 49 4e-06
At2g32240 putative myosin heavy chain 49 6e-06
At3g58250 putative protein 48 1e-05
At5g16730 putative protein 45 9e-05
At3g58430 putative protein 44 1e-04
At3g58300 putative protein 44 2e-04
At1g63300 hypothetical protein 44 2e-04
At5g55860 myosin heavy chain-like 43 3e-04
At3g58370 putative protein 43 3e-04
At3g05270 unknown protein 42 4e-04
At3g58280 putative protein 42 6e-04
At1g64320 bZIP transcription factor, putative 42 6e-04
>At4g35110 unknown protein
Length = 386
Score = 172 bits (435), Expect = 4e-43
Identities = 126/404 (31%), Positives = 205/404 (50%), Gaps = 50/404 (12%)
Query: 10 SNPNCPHASNPYHQCTQACSKKTSHRTKPHAKNTSSSGAAPPPSNFDRRKKPGSRPEPPV 69
++P+C +A NP+H+C AC +K + SS +F R+KK S +PP
Sbjct: 7 AHPDCVYADNPFHECASACLEKIAQGHVKKNTKKQSSRLLSFSGSFGRKKKE-SNSQPPT 65
Query: 70 VAGAVPAAKVEAIQKSNASSPISNHS-----EIKKV---ESRN-----------DDHNHS 110
A P +N +SP +HS +KK E++N D NH
Sbjct: 66 PLSARPYQNGRG-GFANGNSPKVHHSVAPPASVKKKIVSETKNSFTSSSSGDPDDFFNHK 124
Query: 111 VPVSGQIHVPLPVPDVKHVHEEDQQKNGVEHLANPIQTKQEDKTASDVH----PGEGLAT 166
+PL ++ + K G++ I E + S + PG
Sbjct: 125 PEKKPSQTIPLSSNNLVDQSKVVSPKPGIQEHNGKIGEGGETRLFSFLSLPRSPG----- 179
Query: 167 TFKGGSIDFSFSGIIPQGNEDNDDKG---DTESVVSESRVPVGKYHVKESFASILESILD 223
K + DFS +E+N++ G D ESV+S++ V VGKY V+ ++IL ++++
Sbjct: 180 --KESNDDFS-----DDDDENNNEIGVELDLESVMSDTFVSVGKYRVRSGSSTILSAVIE 232
Query: 224 KYGDIGASCHLESLVLRSYYIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLR 283
K+GDI +C LES +RS Y+EC+C ++QEL+S + L+K K+ E++A+LKD+ES +
Sbjct: 233 KHGDIAQNCKLESDSMRSRYLECLCSLMQELRSTPVGQLSKVKVKEMLAVLKDLESVNIE 292
Query: 284 VAWLRSALDEIAENVEVINQHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIK 343
VAWLRS L+E A++ E +V+ K D +++ RE+LE++ L E+EV ++K
Sbjct: 293 VAWLRSVLEEFAQSQE------DVENEKERHDGLVKAKREELEAQETDLVRMEKEVVEVK 346
Query: 344 KRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRKSLIDELL 387
+RI E ++ E+E + +E+ ++E KS IDELL
Sbjct: 347 RRIEETRAQMVEIEAERLRMEKMGF----KMEKFKGKSFIDELL 386
>At2g16900 unknown protein
Length = 382
Score = 164 bits (415), Expect = 8e-41
Identities = 118/392 (30%), Positives = 193/392 (49%), Gaps = 30/392 (7%)
Query: 10 SNPNCPHASNPYHQCTQACSKKTSHRTKPHAKNTSSSGAAPPPSNFDRRKKPGSRPEPPV 69
++P+C ++SNP+H+C C +K S S P +F ++K P P
Sbjct: 7 AHPDCRYSSNPFHECASDCLEKISQGRGNKNSKKQGSKILSLPGSFGKKKTESQPPSPLS 66
Query: 70 VAGAVPAAKVEAIQKSNASSPISNHSEIKKVESRNDDHN-HSVPVSGQIHVPLPVPDVKH 128
A + + SP++ +KK + + HS+ G I + L +
Sbjct: 67 TRNYQNGAANTPKVRQSRPSPVA----MKKTPVPEANKSLHSLSSDG-ISIDLNGQNDSF 121
Query: 129 VHEEDQQKNGVEHLANPIQTKQEDKTASDV-HPGEGLATTFKGGSIDFSFSGIIPQGNED 187
H++++ V N + + + + H G T S+ S N+D
Sbjct: 122 NHKQEKPSRTVPLSPNSMADRGKPLSPRPQGHEHSGKNDTASEISLFNVVSPPRSCANDD 181
Query: 188 NDDKGDTE-----------SVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLES 236
+DD + SV+S+S V VGKY V S ++IL+SI+DK+GDI A+C LES
Sbjct: 182 DDDDDENNGYEEGVELDLISVMSDSCVSVGKYRVNSSVSTILQSIIDKHGDIAANCKLES 241
Query: 237 LVLRSYYIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAE 296
+RS Y+EC+C ++QEL S + LT+ K+ E+VA+LKD+ES + V W+RS L+E A
Sbjct: 242 ASMRSRYLECLCSLMQELGSTPVGQLTELKVKEMVAVLKDLESVNIDVGWMRSVLEEFA- 300
Query: 297 NVEVINQHQE-VDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKE 355
Q+QE D+ K + + S ++++E + LA E+EVA+ + R+ E+ L E
Sbjct: 301 ------QYQENTDSEKERQEGLVRSKKQEMEIQEADLARIEKEVAEARLRVEEMKAELAE 354
Query: 356 LELKSAELEQSMLSSKSEVENLHRKSLIDELL 387
LE + +E+ +VE K+ +DELL
Sbjct: 355 LETERLRMEEMGF----KVEKYKGKTFLDELL 382
>At4g38550 Phospholipase like protein
Length = 612
Score = 139 bits (351), Expect = 2e-33
Identities = 85/303 (28%), Positives = 163/303 (53%), Gaps = 19/303 (6%)
Query: 83 QKSNASSPISNHSEIKKVESRNDDHNHSVPVSGQIHVPLPVPDVKHVHEEDQQKNGVEHL 142
QKS SP+ S +DD NHS + +I PL + E+ +
Sbjct: 324 QKSAPPSPVHG-----PYYSSSDDDNHSTYLYPEIRSPLR----SRIVSENSTPVHHNYQ 374
Query: 143 ANPIQTKQEDKTASDVHPGEGLATTFKGGSIDFSFSGIIPQGNEDNDDKG--DTESVVSE 200
+T ++DK P E S F+ I N + G +++S++SE
Sbjct: 375 IVAAETYEQDK---QFEPPE-----LPDESQSFTMQEITKMRGLKNYESGKEESQSMISE 426
Query: 201 SRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQELQSDSIM 260
+ V V Y V++S + L++I+DK+GDI AS L+++ RSYY+E + VV EL+ +
Sbjct: 427 AYVSVANYRVRQSVSETLQAIIDKHGDIAASSKLQAMATRSYYLESLAAVVMELKKTVLR 486
Query: 261 HLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREMES 320
LTK+++ E+ A++KD+ES ++ V+WL++A+ E+AE VE Q+ K +R++ +
Sbjct: 487 DLTKTRVAEIAAVVKDMESVKINVSWLKTAVTELAEAVEYFGQYDTAKVEKEVCERDLTA 546
Query: 321 LREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRK 380
+ ++E L ++E+E+ + ++++ ++ RL +LE+K ++L +++ +S+V +
Sbjct: 547 KKGEMEEMTAELVKREKEIKECREKVTVVAGRLGQLEMKGSKLNKNLDLFQSKVHKFQGE 606
Query: 381 SLI 383
+++
Sbjct: 607 AVL 609
>At2g20960 pEARLI 4 protein
Length = 748
Score = 135 bits (340), Expect = 4e-32
Identities = 67/201 (33%), Positives = 129/201 (63%)
Query: 187 DNDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIEC 246
+N+ + +S++SES V VG+Y V+ S ++ L+ IL K+GDI + L+SL RSYY++
Sbjct: 548 NNEIRDVMQSILSESYVSVGQYKVRASVSTTLQHILQKHGDIASGSKLQSLATRSYYLDM 607
Query: 247 VCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQE 306
+ VV ELQ+ + +L +S++ E+VAI+KD+ES +++ WL+ L+EI E V+ ++H+
Sbjct: 608 LASVVFELQTTPLKYLKESRVVEMVAIVKDIESVKIKAGWLKPVLEEIVEAVKHYDEHKM 667
Query: 307 VDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQS 366
K +R++ +++ E +++ L EKE+++ + + ++ E++ +L +L++K A L +S
Sbjct: 668 SVVEKEVWERDVLLAKQETEKQVKELGEKEKKIKEWRAKMTEMAAKLGDLDMKRARLHKS 727
Query: 367 MLSSKSEVENLHRKSLIDELL 387
S+V+ K L+ +L
Sbjct: 728 FTFLSSKVDKFQGKPLLQGIL 748
Score = 35.0 bits (79), Expect = 0.070
Identities = 24/86 (27%), Positives = 37/86 (42%), Gaps = 16/86 (18%)
Query: 12 PNCPHASNPYHQCTQACSKKTS----HRTKP----HAKNTSSSGAAPPPSNFDRRKKPGS 63
P C +A NP+H+CT C ++ + H+ + K T S PP S + P
Sbjct: 10 PGCSNAGNPFHECTAICFERVNSPDVHKKEKKLFGFGKRTPSRDQTPPASPARGSRSP-- 67
Query: 64 RPEPPVVAGAVPAAKVEAIQKSNASS 89
+A KVE+ +S+ SS
Sbjct: 68 ------LASYFAKKKVESSAESSPSS 87
>At2g20950 unknown protein
Length = 520
Score = 134 bits (337), Expect = 8e-32
Identities = 64/184 (34%), Positives = 120/184 (64%)
Query: 191 KGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFV 250
K + + +SE+ V V Y V+ S A+ LE+I+DK+GDI AS L+S RS+Y+E +
Sbjct: 326 KEEIQLAISETYVSVANYKVRMSIAATLEAIIDKHGDIAASSKLQSTSTRSFYLESLAAA 385
Query: 251 VQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAA 310
V EL+S ++ LTK+++ E+ A++KD++S ++ V+WL++A+ E+AE VE ++
Sbjct: 386 VMELKSTALRDLTKTRVAEIAAVVKDMDSVKIDVSWLKTAVTELAEAVEYYGKYDTAKIV 445
Query: 311 KANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSS 370
+ DREM + ++++E E L +E+E + ++R+ E++ RL +LE+K +++++
Sbjct: 446 REECDREMTAGKKEMEEMREELRRREKETKECRERVTELAGRLGQLEMKDVRVKKNLELY 505
Query: 371 KSEV 374
+S+V
Sbjct: 506 ESKV 509
>At4g38560 Phospholipase like protein
Length = 521
Score = 132 bits (332), Expect = 3e-31
Identities = 70/194 (36%), Positives = 118/194 (60%)
Query: 193 DTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQ 252
+ +S++SES V VG Y V+ S +S L+ ILDK+GDI + L+SL +SY +E + VV
Sbjct: 327 EMKSMISESYVSVGSYKVRASVSSTLQKILDKHGDIASGSKLQSLRTKSYSLETLAAVVL 386
Query: 253 ELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKA 312
ELQS + L ++++ E+++++ D ES ++R WLR L+EI E + H+ K
Sbjct: 387 ELQSTPLKKLKQARVLEMLSVVIDAESVKIRAGWLREILNEILEAAHHYDGHETTVVEKE 446
Query: 313 NTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKS 372
+R+M RE+++ E + KE+E D +K + E++ RL EL++K A LE+ + S
Sbjct: 447 GRERDMLLEREEMKKIQEEVRLKEKEAKDFRKGVMEMAGRLGELKMKRARLEKRLAFLSS 506
Query: 373 EVENLHRKSLIDEL 386
+VE +SL++ +
Sbjct: 507 KVEKFEGESLLENV 520
Score = 32.7 bits (73), Expect = 0.35
Identities = 11/26 (42%), Positives = 16/26 (61%)
Query: 13 NCPHASNPYHQCTQACSKKTSHRTKP 38
+CP++ NPYH+C C K+ S P
Sbjct: 8 DCPNSGNPYHECHDQCFKRISSGDVP 33
>At5g11140 putative protein
Length = 241
Score = 128 bits (321), Expect = 6e-30
Identities = 64/185 (34%), Positives = 119/185 (63%)
Query: 203 VPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQELQSDSIMHL 262
V VG+Y VK S +S L+SI DKYGDI ++ L+SL R+Y++E + VV ELQS + L
Sbjct: 57 VSVGEYKVKASLSSTLQSIFDKYGDITSNSKLQSLSTRTYHLETLAEVVIELQSTPLRRL 116
Query: 263 TKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREMESLR 322
++S+ E++AI+ D+E+A++RV WLRS L+E+ E ++ + K + + +
Sbjct: 117 SESRATEILAIVDDIETAKIRVGWLRSVLEEVLEATRYFDRCEMAVMEKKAGEHRLLLAK 176
Query: 323 EQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRKSL 382
+++E ++ LAEKE+E+ + ++++ + + +L LE+K L++ ++ +S+VE +S+
Sbjct: 177 QEMELSLKKLAEKEKEMKEFREKLMKTTGKLGSLEMKRTCLDKRLVFLRSKVEKFQGQSV 236
Query: 383 IDELL 387
++L
Sbjct: 237 FQDIL 241
>At3g58330 putative protein
Length = 173
Score = 58.5 bits (140), Expect = 6e-09
Identities = 43/166 (25%), Positives = 82/166 (48%), Gaps = 9/166 (5%)
Query: 185 NEDNDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYI 244
++D D +S E V V +HV S +++ +L K+ D+ ++ L++ L++ Y+
Sbjct: 12 SDDGDFSPSQDSSEDEETVEVNGFHVLPSQENLVSQMLKKHPDLTSNFDLKNQQLKNAYM 71
Query: 245 ECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQH 304
+ V + E S S L+ +++ + L D+ A L+V WLR LDE + +
Sbjct: 72 D-VLVDISETLSQSTKALSMEDLDKAESTLFDLTKAGLKVGWLRQKLDE----AYLKKEK 126
Query: 305 QEVDAAKANTDREMESLREQLESEMESLAEKEQ----EVADIKKRI 346
Q + + E + R+ S++ES +KE+ +A +KKR+
Sbjct: 127 QRISGVRIRELEEQVNKRKLTLSDLESDLKKEKANQLSMAQLKKRM 172
>At3g58270 unknown protein
Length = 343
Score = 49.3 bits (116), Expect = 4e-06
Identities = 57/275 (20%), Positives = 109/275 (38%), Gaps = 52/275 (18%)
Query: 113 VSGQIHVPLPVPDVKHVHEEDQQKNGVEHLANPIQTKQEDKTASDVHPGEGLATTFKGGS 172
V+G++ + + V ++ + + D E + +E+ P E TT
Sbjct: 121 VNGELKIVVDVSVLEVIGKLDVPVESEETTTKALSELEENDV-----PEESEETTKALSK 175
Query: 173 IDFSFSGIIPQGNEDNDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASC 232
+D + G E ND + SV + + V + V S + I +++ DI +
Sbjct: 176 VDEN------DGAESNDSLKEASSV--KESMDVNGFRVLPSQVETVSCIFERHPDIASEF 227
Query: 233 HLESLVLRSYYIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALD 292
++ LRS Y+ + +++ + S L+K +++ A L + A L + WL L+
Sbjct: 228 GPKNQHLRSAYMNVLLSLIKTM-CQSTQELSKDDLSDADAALAYLTDAGLNLNWLEEKLE 286
Query: 293 EIAENVEVINQHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDR 352
E++E +KE E A + R+ EI +
Sbjct: 287 EVSE-------------------------------------KKENEEAG-ETRVHEIEEE 308
Query: 353 LKELELKSAELEQSMLSSKSEVENLHRKSLIDELL 387
LKEL+LK + LE + K++V D+++
Sbjct: 309 LKELKLKCSNLEAQLEKEKADVSVARAPFSFDDIV 343
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 48.5 bits (114), Expect = 6e-06
Identities = 61/242 (25%), Positives = 113/242 (46%), Gaps = 46/242 (19%)
Query: 143 ANPIQTK------QEDKTASDVHPGEGLAT------TFKGGSIDFSFSGIIPQGNEDN-- 188
AN +QTK ++++TA+++ + T +G + S + N+ N
Sbjct: 1025 ANELQTKLSALEAEKEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSHTEENNQVNAM 1084
Query: 189 --DDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIEC 246
K + +SV+++ + V+ S A L S ++K + A E VL S++ E
Sbjct: 1085 FQSTKEELQSVIAKLEEQLT---VESSKADTLVSEIEKLRAVAA----EKSVLESHFEEL 1137
Query: 247 VCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDE---IAENVEVINQ 303
+ + K+++ E V ++ +A ++VA L S L E IA +V+N+
Sbjct: 1138 ----------EKTLSEVKAQLKENV---ENAATASVKVAELTSKLQEHEHIAGERDVLNE 1184
Query: 304 H-----QEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKR-IPEISDRLKELE 357
+E+ AA+++ D E + Q +SE+ES +K QE + KK+ + E +K+LE
Sbjct: 1185 QVLQLQKELQAAQSSID-EQKQAHSQKQSELESALKKSQEEIEAKKKAVTEFESMVKDLE 1243
Query: 358 LK 359
K
Sbjct: 1244 QK 1245
Score = 42.4 bits (98), Expect = 4e-04
Identities = 30/119 (25%), Positives = 60/119 (50%), Gaps = 3/119 (2%)
Query: 252 QELQSDSIMHLTKS--KINELVAILKDV-ESAQLRVAWLRSALDEIAENVEVINQHQEVD 308
+EL S H K E +LK ESA+ + S EI E E ++++++V+
Sbjct: 218 EELHKQSASHADSESQKALEFSELLKSTKESAKEMEEKMASLQQEIKELNEKMSENEKVE 277
Query: 309 AAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSM 367
AA ++ E+ +++E+L L E EQ+V+ + I E++ L++ + + ++ +
Sbjct: 278 AALKSSAGELAAVQEELALSKSRLLETEQKVSSTEALIDELTQELEQKKASESRFKEEL 336
Score = 35.0 bits (79), Expect = 0.070
Identities = 29/107 (27%), Positives = 51/107 (47%), Gaps = 13/107 (12%)
Query: 293 EIAENVEVINQHQEVDAAKANTDREMESLRE---QLESEMESLAEK----EQEVADIKKR 345
E A++V V Q + ++ + + + RE+ +E +LE E+E +A + E E +K
Sbjct: 63 EKADHVPVEEQKEVIERSSSGSQRELHESQEKAKELELELERVAGELKRYESENTHLKDE 122
Query: 346 IPEISDRLKELELKSAELE------QSMLSSKSEVENLHRKSLIDEL 386
+ ++L+E E K +LE Q + E + KSL D L
Sbjct: 123 LLSAKEKLEETEKKHGDLEVVQKKQQEKIVEGEERHSSQLKSLEDAL 169
Score = 33.5 bits (75), Expect = 0.20
Identities = 28/107 (26%), Positives = 55/107 (51%), Gaps = 8/107 (7%)
Query: 276 DVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREME-----SLREQLESEME 330
++E A ++ L S ++E+ + + + + D A+ N +E S +L++++
Sbjct: 975 ELEDALSKLKNLESTIEELGAKCQGLEK-ESGDLAEVNLKLNLELANHGSEANELQTKLS 1033
Query: 331 SL-AEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVEN 376
+L AEKEQ +++ I D K+L + +L QS +SS +E N
Sbjct: 1034 ALEAEKEQTANELEASKTTIEDLTKQLTSEGEKL-QSQISSHTEENN 1079
Score = 32.7 bits (73), Expect = 0.35
Identities = 25/119 (21%), Positives = 57/119 (47%), Gaps = 2/119 (1%)
Query: 255 QSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANT 314
++D+++ S +EL LK +E +A + +N+E+ + + A
Sbjct: 435 KTDALLSQALSNNSELEQKLKSLEELHSEAGSAAAAATQ--KNLELEDVVRSSSQAAEEA 492
Query: 315 DREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSE 373
+++ L + + + AE EQ++ ++ + + LKEL KS+EL+ ++ ++ E
Sbjct: 493 KSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKELSEKSSELQTAIEVAEEE 551
Score = 32.0 bits (71), Expect = 0.59
Identities = 34/119 (28%), Positives = 55/119 (45%), Gaps = 10/119 (8%)
Query: 269 ELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREMESLREQLESE 328
+L +LK+ E+ + VA + S +A EV N+ +E K T E S + L S+
Sbjct: 390 KLAEVLKEKEALEANVAEVTS---NVATVTEVCNELEE----KLKTSDENFSKTDALLSQ 442
Query: 329 -MESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRKSLIDEL 386
+ + +E EQ++ +++ E K+ ELE + SS E KS I EL
Sbjct: 443 ALSNNSELEQKLKSLEELHSEAGSAAAAATQKNLELEDVVRSSSQAAE--EAKSQIKEL 499
Score = 30.8 bits (68), Expect = 1.3
Identities = 29/125 (23%), Positives = 60/125 (47%), Gaps = 10/125 (8%)
Query: 250 VVQELQSDSIMHLTK---SKINELVAILKDVESAQLRVAWLRSALD----EIAENVEVIN 302
VVQ+ Q + I+ + S++ L L+ ++ + ++ A D E+ + + +
Sbjct: 142 VVQKKQQEKIVEGEERHSSQLKSLEDALQSHDAKDKELTEVKEAFDALGIELESSRKKLI 201
Query: 303 QHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAE 362
+ +E A ++ E L +Q S +S ++K E +++ K E + KE+E K A
Sbjct: 202 ELEEGLKRSAEEAQKFEELHKQSASHADSESQKALEFSELLKSTKESA---KEMEEKMAS 258
Query: 363 LEQSM 367
L+Q +
Sbjct: 259 LQQEI 263
Score = 29.3 bits (64), Expect = 3.8
Identities = 27/118 (22%), Positives = 59/118 (49%), Gaps = 6/118 (5%)
Query: 275 KDVESAQLRVAWLRSALDEIAE-NVEVINQHQEVDAAKANTDREMESLREQLESEMESLA 333
++ + AQ + S + +AE N ++ + QE++ + E E+ ++LE +E
Sbjct: 876 QEFDQAQEKSLQSSSESELLAETNNQLKIKIQELEGLIGSGSVEKETALKRLEEAIERFN 935
Query: 334 EKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRK-----SLIDEL 386
+KE E +D+ +++ ++++E + + E + K E+E+ K S I+EL
Sbjct: 936 QKETESSDLVEKLKTHENQIEEYKKLAHEASGVADTRKVELEDALSKLKNLESTIEEL 993
Score = 29.3 bits (64), Expect = 3.8
Identities = 21/82 (25%), Positives = 46/82 (55%), Gaps = 7/82 (8%)
Query: 296 ENVEVINQHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKE 355
+N E+ Q + ++ +RE++ L E+ SE+++ EVA+ +K+ + + +++E
Sbjct: 509 KNAELEQQLNLLQLKSSDAERELKELSEK-SSELQTAI----EVAEEEKK--QATTQMQE 561
Query: 356 LELKSAELEQSMLSSKSEVENL 377
+ K++ELE S+ S + L
Sbjct: 562 YKQKASELELSLTQSSARNSEL 583
Score = 28.1 bits (61), Expect = 8.5
Identities = 25/128 (19%), Positives = 61/128 (47%), Gaps = 11/128 (8%)
Query: 254 LQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVE----VINQHQEVDA 309
L+++ + T++ I+EL L+ ++++ R S L ++ + +++ + +++
Sbjct: 302 LETEQKVSSTEALIDELTQELEQKKASESRFKEELSVLQDLDAQTKGLQAKLSEQEGINS 361
Query: 310 AKANTDREMESLR-------EQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAE 362
A +E E L E+L + E LAE +E ++ + E++ + + E
Sbjct: 362 KLAEELKEKELLESLSKDQEEKLRTANEKLAEVLKEKEALEANVAEVTSNVATVTEVCNE 421
Query: 363 LEQSMLSS 370
LE+ + +S
Sbjct: 422 LEEKLKTS 429
>At3g58250 putative protein
Length = 317
Score = 47.8 bits (112), Expect = 1e-05
Identities = 37/178 (20%), Positives = 87/178 (48%), Gaps = 26/178 (14%)
Query: 198 VSESR-VPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQELQS 256
VSES + V +HV S A ++S+ + + DI +++ L++ Y+ + +++ ++
Sbjct: 146 VSESESMDVNGFHVLPSQAKYVKSLFEIHPDIATKFRIKNQYLKTGYMNVLLCLIETVRR 205
Query: 257 DSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDR 316
S ++K+ + + L+ + ++ WL+ LD++ Q +E +AA
Sbjct: 206 -SPKEISKNDLADAYVALESLTDHGFKLDWLKKKLDQVT-------QKKEKEAAG----- 252
Query: 317 EMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEV 374
E+ M + E E+ D+K + ++ +L +++ K ++LE ++ K++V
Sbjct: 253 ---------ETRMHEIGE---ELKDLKLKCSDLEAQLDKVKWKCSDLEAQLVKEKADV 298
>At5g16730 putative protein
Length = 853
Score = 44.7 bits (104), Expect = 9e-05
Identities = 74/386 (19%), Positives = 152/386 (39%), Gaps = 30/386 (7%)
Query: 10 SNPNCPHASNPYHQCTQACSKKTSHRTKPHAKNT---SSSGAAPPPSNFDRRKKPGSRPE 66
SN N P + P+ S+ + R+ P++K++ S PP R E
Sbjct: 40 SNNNSPSTTTPH-------SRLSLDRSSPNSKSSVERRSPKLPTPPEKSQARVAAVKGTE 92
Query: 67 PPVVAGAVPAAKVEAIQKSNASSPISNHSEIKKVESRNDDHNHSVPVSGQIHVPLPVPDV 126
P + K E ++K+N + K ++ + V+ ++ L
Sbjct: 93 SPQTTTRLSQIK-EDLKKANERISSLEKDKAKALDELKQAKKEAEQVTLKLDDALKAQ-- 149
Query: 127 KHVHEEDQQKN------GVEHLAN-PIQTKQEDKTASDVHPGEGLATTFKGGSIDFSFSG 179
KHV E + + G+E + N + K+E +T + H + A ++
Sbjct: 150 KHVEENSEIEKFQAVEAGIEAVQNNEEELKKELETVKNQHASDSAALVAVRQELEKINEE 209
Query: 180 IIPQGNEDNDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVL 239
+ + + E + + K + S + L+++LD + A E +
Sbjct: 210 LAAAFDAKSKALSQAEDASKTAEIHAEKVDILSSELTRLKALLDSTREKTAISDNEMVAK 269
Query: 240 RSYYIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENV- 298
I + ++ + K I E + + D+E+A++ + S +E
Sbjct: 270 LEDEIVVLKRDLESARGFEAEVKEKEMIVEKLNV--DLEAAKMAESNAHSLSNEWQSKAK 327
Query: 299 EVINQHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRI----PEISDRLK 354
E+ Q +E + + + +ES+ +QLE + L + E E+ D+K+RI ++ + +
Sbjct: 328 ELEEQLEEANKLERSASVSLESVMKQLEGSNDKLHDTETEITDLKERIVTLETTVAKQKE 387
Query: 355 ELELKSAEL---EQSMLSSKSEVENL 377
+LE+ L E+ + ++ EVE L
Sbjct: 388 DLEVSEQRLGSVEEEVSKNEKEVEKL 413
Score = 38.1 bits (87), Expect = 0.008
Identities = 31/130 (23%), Positives = 62/130 (46%), Gaps = 5/130 (3%)
Query: 257 DSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQ-EVDAAKANTD 315
+S+M + ++L ++ + R+ L + + + E++EV Q V+ + +
Sbjct: 348 ESVMKQLEGSNDKLHDTETEITDLKERIVTLETTVAKQKEDLEVSEQRLGSVEEEVSKNE 407
Query: 316 REMESLREQLES---EMESLAEKEQEVADIKKRIPEISDR-LKELELKSAELEQSMLSSK 371
+E+E L+ +LE+ E +KEQ+ +R+ E + L +LE E E+S + +
Sbjct: 408 KEVEKLKSELETVKEEKNRALKKEQDATSRVQRLSEEKSKLLSDLESSKEEEEKSKKAME 467
Query: 372 SEVENLHRKS 381
S LH S
Sbjct: 468 SLASALHEVS 477
Score = 32.7 bits (73), Expect = 0.35
Identities = 31/138 (22%), Positives = 70/138 (50%), Gaps = 14/138 (10%)
Query: 261 HLTKSKINELVAILKDV-ESAQLRVAWLRSALDEIAENVEVINQHQEV-----DAAKANT 314
H +++I++L ++K E + + R +D + VE +H E + +AN
Sbjct: 493 HEYETQIDDLKLVIKATNEKYENMLDEARHEIDVLVSAVEQTKKHFESSKKDWEMKEANL 552
Query: 315 DREMESLREQLES------EMESLAEKEQEVADIK-KRIPEISDRLKELELKSAELEQSM 367
++ + E + S +++L ++ +E AD K+ + D LKE+E + L++++
Sbjct: 553 VNYVKKMEEDVASMGKEMNRLDNLLKRTEEEADAAWKKEAQTKDSLKEVEEEIVYLQETL 612
Query: 368 LSSKSEVENLHRKSLIDE 385
+K+E L +++L+D+
Sbjct: 613 GEAKAESMKL-KENLLDK 629
Score = 29.6 bits (65), Expect = 2.9
Identities = 24/101 (23%), Positives = 46/101 (44%), Gaps = 8/101 (7%)
Query: 270 LVAILKDVESAQLRVAWLRSALDEIAENVE-VINQHQEVDAAKANTDREMESLREQLESE 328
L ++ ++V + V L+S L+ + E + + Q+ + E L LES
Sbjct: 396 LGSVEEEVSKNEKEVEKLKSELETVKEEKNRALKKEQDATSRVQRLSEEKSKLLSDLESS 455
Query: 329 MESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLS 369
E E+E+ KK + ++ L E+ + EL++ +LS
Sbjct: 456 KE---EEEKS----KKAMESLASALHEVSSEGRELKEKLLS 489
>At3g58430 putative protein
Length = 552
Score = 44.3 bits (103), Expect = 1e-04
Identities = 44/198 (22%), Positives = 89/198 (44%), Gaps = 19/198 (9%)
Query: 195 ESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQEL 254
E ++ +V V + V S + SI +++ DI ++ LR+ C+ F++ +
Sbjct: 335 EDSSTQGKVDVNGFQVLPSQVEYVRSIFERHPDIAVEFRAKNQHLRT---SCMIFLLSLI 391
Query: 255 QS--DSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEV----INQHQEVD 308
++ S+ L+ + E L V+ A +V WL LD++ E E + Q QE++
Sbjct: 392 ETLCQSLEELSNEDLVEADIALTYVKDAGFKVDWLEKKLDQVKEKKEKELSDMVQLQEME 451
Query: 309 AAKANTDREMESLREQLESEMESLAEKE-----QEVADIKKRIPEISDRLKELE----LK 359
+ L + + + + L+ +V ++K++ ++ D+ KE E +
Sbjct: 452 DHLLKLKQSCSDLDDLVVKKKDELSVTRSPLSFDDVDWLEKKLDQVKDK-KEREQSGLAR 510
Query: 360 SAELEQSMLSSKSEVENL 377
ELE+ +L K + NL
Sbjct: 511 LHELEEYLLKLKQKCSNL 528
>At3g58300 putative protein
Length = 181
Score = 43.9 bits (102), Expect = 2e-04
Identities = 39/173 (22%), Positives = 78/173 (44%), Gaps = 24/173 (13%)
Query: 183 QGNEDNDDK----------GDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASC 232
+G+E +DD+ + S + V V + + +S S + +I +K+ DI +
Sbjct: 6 EGSESSDDEQRYSSSSEEWSEESSEDAHEIVEVNGFQILDSQVSQVNAIFEKHPDIASDF 65
Query: 233 HLESLVLRSYYIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALD 292
+L++ L++ Y+ + ++ L LT +N+ + D+ A L++ WLR LD
Sbjct: 66 NLKNQQLKNAYMGVLLDLIMTLCRPP-KELTMDDLNKADRTVLDLTKAGLKLQWLRQKLD 124
Query: 293 EIAENVEVINQHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKR 345
E E K + + L EQ++ +L++ E +D+KK+
Sbjct: 125 EAFLKKE----------KKRDIGARIRELEEQVKKRKLNLSDLE---SDLKKQ 164
>At1g63300 hypothetical protein
Length = 1029
Score = 43.9 bits (102), Expect = 2e-04
Identities = 29/128 (22%), Positives = 64/128 (49%), Gaps = 5/128 (3%)
Query: 263 TKSKINELVAILKDVESAQL----RVAWLRSALDEIAENVEVIN-QHQEVDAAKANTDRE 317
TK + E A L+ ++ +++ +R + +A ++VI E + A + E
Sbjct: 764 TKKSVMEAEASLQRENMKKIELESKISLMRKESESLAAELQVIKLAKDEKETAISLLQTE 823
Query: 318 MESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENL 377
+E++R Q + SL+E + E+ KK++ + LK+ E A LE+ + S++ +
Sbjct: 824 LETVRSQCDDLKHSLSENDLEMEKHKKQVAHVKSELKKKEETMANLEKKLKESRTAITKT 883
Query: 378 HRKSLIDE 385
+++ I++
Sbjct: 884 AQRNNINK 891
Score = 37.0 bits (84), Expect = 0.018
Identities = 31/116 (26%), Positives = 57/116 (48%), Gaps = 12/116 (10%)
Query: 261 HLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREMES 320
H+ + KI +L ++E + L ++++A + E++ Q + K + E
Sbjct: 465 HILEQKITDLY---NEIEIYKRDKDELEIQMEQLALDYEILKQQNHDISYKLEQSQLQEQ 521
Query: 321 LREQLE--SEMESLAEKEQEV----ADIKKRIPEISD---RLKELELKSAELEQSM 367
L+ Q E S + + E E +V A++KK+ E S+ R+KELE + LE+ M
Sbjct: 522 LKIQYECSSSLVDVTELENQVESLEAELKKQSEEFSESLCRIKELESQMETLEEEM 577
Score = 32.3 bits (72), Expect = 0.45
Identities = 37/152 (24%), Positives = 67/152 (43%), Gaps = 13/152 (8%)
Query: 185 NEDNDDKGDTESVVSESRVPVGKYHVKESFASILES-ILDKYGDIGA-SCHLESLVLRSY 242
+ED+ D+ E +V K HV ILE I D Y +I + L ++
Sbjct: 442 DEDDHDQKALEDLV--------KKHVDAKDTHILEQKITDLYNEIEIYKRDKDELEIQME 493
Query: 243 YIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVIN 302
+ ++++ D L +S++ E + I + S+ + V L + ++ + E+
Sbjct: 494 QLALDYEILKQQNHDISYKLEQSQLQEQLKIQYECSSSLVDVTELENQVESL--EAELKK 551
Query: 303 QHQEVDAAKANTDREMESLREQLESEMESLAE 334
Q +E + +E+ES E LE EME A+
Sbjct: 552 QSEEFSESLCRI-KELESQMETLEEEMEKQAQ 582
Score = 28.5 bits (62), Expect = 6.5
Identities = 61/300 (20%), Positives = 109/300 (36%), Gaps = 76/300 (25%)
Query: 64 RPEPPVVAGAVPAAKVEAIQKSNASSPISNHSEIKKVESRNDDHNHSVPVSGQIHVPLPV 123
R E +A + K+ +K A S + +E++ V S+ DD HS+ + +
Sbjct: 793 RKESESLAAELQVIKLAKDEKETAISLLQ--TELETVRSQCDDLKHSLS-ENDLEMEKHK 849
Query: 124 PDVKHVHEEDQQKNGVEHLANPIQTKQEDKTASDVHPGEGLATTFKGGSIDFSFSGIIPQ 183
V HV E ++K E +AN + +E +TA I
Sbjct: 850 KQVAHVKSELKKKE--ETMANLEKKLKESRTA------------------------ITKT 883
Query: 184 GNEDNDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYY 243
+N +KG PVG + + A + + I G I
Sbjct: 884 AQRNNINKGS----------PVGAHGGSKEVAVMKDKIKLLEGQIKLK------------ 921
Query: 244 IECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQ 303
L+S S M + K K + + R+ L + LD+ ++ +
Sbjct: 922 -------ETALESSSNMFIEKEK------------NLKNRIEELETKLDQNSQEMS---- 958
Query: 304 HQEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAEL 363
E + + ++ L ++ES E E E+ ++++R EIS R E+E + +L
Sbjct: 959 --ENELLNGQENEDIGVLVAEIESLRECNGSMEMELKEMRERYSEISLRFAEVEGERQQL 1016
>At5g55860 myosin heavy chain-like
Length = 649
Score = 42.7 bits (99), Expect = 3e-04
Identities = 43/203 (21%), Positives = 89/203 (43%), Gaps = 18/203 (8%)
Query: 191 KGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFV 250
KG +S S V VG+ F S+ +++ + +G+ S E V R + V
Sbjct: 5 KGRRDSSDSSPIVEVGEIDTSAPFQSVKDAV-NLFGEAAFSA--EKPVFRKPNPQSAEKV 61
Query: 251 VQELQSDSIMHLTKSKINELVAILKDVES----AQLRVAWLRSALDEIAENVEVINQHQE 306
L + +HL + ++N+L LK+ E+ A + W + +DE+ +E +N+ +
Sbjct: 62 ---LVKQTELHLAQKELNKLKEQLKNAETIREQALSELEWSKRTVDELTRKLEAVNESR- 117
Query: 307 VDAAKANTDREMESLRE------QLESEMESLAEKEQEVADIKKRIPEISDRLKELELKS 360
D+A T+ + E + S ++ +E ++ K + L+++ S
Sbjct: 118 -DSANKATEAAKSLIEEAKPGNVSVASSSDAQTRDMEEYGEVCKELDTAKQELRKIRQVS 176
Query: 361 AELEQSMLSSKSEVENLHRKSLI 383
E+ ++ + S+VE + S +
Sbjct: 177 NEILETKTVALSKVEEAKKVSKV 199
Score = 28.5 bits (62), Expect = 6.5
Identities = 16/57 (28%), Positives = 30/57 (52%)
Query: 319 ESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVE 375
E L E+ +S E + + E+ ++K E+ + E+E + +L + SKSE+E
Sbjct: 316 EKLVEEEKSLQELVESLKAELKNVKMEHDEVEAKEAEIESVAGDLHLKLSRSKSELE 372
>At3g58370 putative protein
Length = 219
Score = 42.7 bits (99), Expect = 3e-04
Identities = 40/163 (24%), Positives = 75/163 (45%), Gaps = 12/163 (7%)
Query: 186 EDND----DKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRS 241
+DND D + S V ES + V + V S ++ I +++ D + ++ L+S
Sbjct: 54 DDNDAVSFDSLNETSPVKES-IDVNGFQVLPSQVESVKCIFERHPDFASKFRPKNRHLKS 112
Query: 242 YYIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVI 301
Y+ + +++ L LT ++E + VE+ LR+ WL L E+
Sbjct: 113 TYMTVLLGLIKTL-CQLPEELTDDDLDEASVAVSYVENGGLRLDWLEKKLAEVKA----- 166
Query: 302 NQHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIKK 344
+ ++V+ KA R E L++ + +E A E+E AD+ +
Sbjct: 167 -KKKKVETGKARLQRAEEELQKLNQKCLELKAFLEKENADVSE 208
>At3g05270 unknown protein
Length = 603
Score = 42.4 bits (98), Expect = 4e-04
Identities = 33/135 (24%), Positives = 65/135 (47%), Gaps = 23/135 (17%)
Query: 251 VQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAA 310
V++LQ + ++ +K +I L + LK++E +++E ++ ++QE++
Sbjct: 355 VEKLQLEMALNGSKEQIEALQSRLKEIEG-------------KLSEMKKLEAENQELELL 401
Query: 311 KANTDREMESLREQLESEMESLAEKEQEVAD----------IKKRIPEISDRLKELELKS 360
+ ++ME L+ QL +L+E E A+ KK++ +RLKE E K
Sbjct: 402 LGESGKQMEDLQRQLNKAQVNLSELETRRAEKLELTMCLNGTKKQLETSQNRLKETERKL 461
Query: 361 AELEQSMLSSKSEVE 375
EL+ + +K E
Sbjct: 462 TELQTLLHLTKDAKE 476
Score = 35.0 bits (79), Expect = 0.070
Identities = 30/109 (27%), Positives = 56/109 (50%), Gaps = 4/109 (3%)
Query: 269 ELVAILKDVESAQLRVAWLRSALDEIAENVEV--INQHQEVDAAKANTDREMESLREQLE 326
+L A V + + R + L SAL E + Q+Q+++ A N +E E+ + QLE
Sbjct: 95 QLDASTSKVSALEDRNSHLDSALKECVRQLWQGREEQNQKIEEAINNKCKEWETTKSQLE 154
Query: 327 SEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVE 375
+ +E L + Q+V ++ +L+ LE +++ L+ +LS EV+
Sbjct: 155 ARIEEL-QARQDVTTSSVH-EDLYPKLEALEKENSALKLQLLSKSEEVK 201
Score = 33.5 bits (75), Expect = 0.20
Identities = 25/107 (23%), Positives = 50/107 (46%), Gaps = 12/107 (11%)
Query: 275 KDVESAQLRVAWLRSALDEIAENVEVINQHQE-VDAAKANTDREMESLREQLESEMESLA 333
K++E + V L+ L + + + E V+ K + + +EQ+E+ L
Sbjct: 320 KELEKSNAHVNQLKHELKTSLRRISELEEKVEMVEVEKLQLEMALNGSKEQIEALQSRLK 379
Query: 334 EKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRK 380
E E +++++KK LE ++ ELE + S ++E+L R+
Sbjct: 380 EIEGKLSEMKK-----------LEAENQELELLLGESGKQMEDLQRQ 415
Score = 32.0 bits (71), Expect = 0.59
Identities = 14/65 (21%), Positives = 36/65 (54%)
Query: 316 REMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVE 375
R+ ++LE + + + E+ +RI E+ ++++ +E++ +LE ++ SK ++E
Sbjct: 313 RKHSESNKELEKSNAHVNQLKHELKTSLRRISELEEKVEMVEVEKLQLEMALNGSKEQIE 372
Query: 376 NLHRK 380
L +
Sbjct: 373 ALQSR 377
Score = 31.2 bits (69), Expect = 1.0
Identities = 22/97 (22%), Positives = 46/97 (46%), Gaps = 20/97 (20%)
Query: 303 QHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVADI--------------KKRIPE 348
+H E + ++ + L+ +L++ + ++E E++V + K++I
Sbjct: 314 KHSESNKELEKSNAHVNQLKHELKTSLRRISELEEKVEMVEVEKLQLEMALNGSKEQIEA 373
Query: 349 ISDRLKELELKSAELEQSMLSSKSEVENLHRKSLIDE 385
+ RLKE+E K +E++ K E EN + L+ E
Sbjct: 374 LQSRLKEIEGKLSEMK------KLEAENQELELLLGE 404
>At3g58280 putative protein
Length = 196
Score = 42.0 bits (97), Expect = 6e-04
Identities = 36/158 (22%), Positives = 72/158 (44%), Gaps = 8/158 (5%)
Query: 183 QGNEDNDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSY 242
Q E +DD + + V + V +S + +I +K+ D+ ++ L++ +R+
Sbjct: 39 QVEELSDDSSEDLQNKDNVTIEVNGFQVLDSQVDQVNAIFEKHPDLISNFSLKNQHIRNA 98
Query: 243 YIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVIN 302
Y+ + + + L S S LT +NE + D+ A L + WLR D+ E
Sbjct: 99 YMHALLDLTKTL-SKSTKELTVEDMNEADNTITDLIKAGLNLDWLRQKFDQALE------ 151
Query: 303 QHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVA 340
+ D +++++ R+ +E+E+ EKE+ A
Sbjct: 152 KQIAYDTRIGELEKQVKK-RKLAVTELEADLEKEKAAA 188
>At1g64320 bZIP transcription factor, putative
Length = 476
Score = 42.0 bits (97), Expect = 6e-04
Identities = 38/128 (29%), Positives = 67/128 (51%), Gaps = 19/128 (14%)
Query: 265 SKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDR------EM 318
S+I +L LK++E + +LR+ +A N+EV + +E + K D+ E+
Sbjct: 50 SRIEDLKCQLKNLEQ---EIGFLRARNAGLAGNLEV-TKVEEKERVKGLMDQVNGMKHEL 105
Query: 319 ESLREQ-------LESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSK 371
ESLR Q LE ++E + E + ++ +K+ E +RL E E+ + E ML +
Sbjct: 106 ESLRSQKDESEAKLEKKVEEVTETKMQLKSLKEETEEERNRLSE-EIDQLKGENQMLHRR 164
Query: 372 -SEVENLH 378
SE+++LH
Sbjct: 165 ISELDSLH 172
Score = 37.4 bits (85), Expect = 0.014
Identities = 30/116 (25%), Positives = 53/116 (44%), Gaps = 13/116 (11%)
Query: 268 NELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREMESLREQLES 327
N + I K++ESA+L V+ +S ++ + +E + + L+E+L
Sbjct: 350 NRVARIGKEMESAKLWVSEKKSEVETLTAKLECSEAQETL-------------LKEKLSK 396
Query: 328 EMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRKSLI 383
+ LAE+ E + K + + R+KELE+K E +LS E R+ I
Sbjct: 397 LEKKLAEEGTEKLKLAKVLSKFETRIKELEVKVKGREVELLSLGEEKREAIRQLCI 452
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.308 0.126 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,778,992
Number of Sequences: 26719
Number of extensions: 401325
Number of successful extensions: 2995
Number of sequences better than 10.0: 344
Number of HSP's better than 10.0 without gapping: 101
Number of HSP's successfully gapped in prelim test: 245
Number of HSP's that attempted gapping in prelim test: 2393
Number of HSP's gapped (non-prelim): 738
length of query: 387
length of database: 11,318,596
effective HSP length: 101
effective length of query: 286
effective length of database: 8,619,977
effective search space: 2465313422
effective search space used: 2465313422
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0017.9


