

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0016.9
(219 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
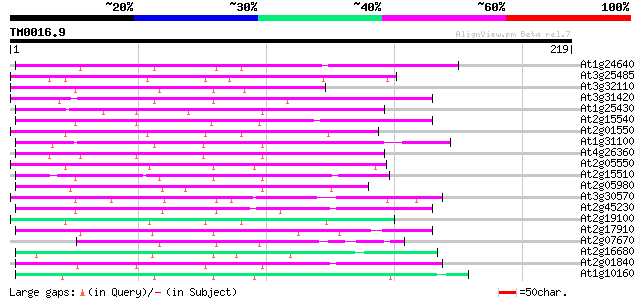
Score E
Sequences producing significant alignments: (bits) Value
At1g24640 hypothetical protein 69 1e-12
At3g25485 unknown protein 61 4e-10
At3g32110 non-LTR reverse transcriptase, putative 59 2e-09
At3g31420 hypothetical protein 59 3e-09
At1g25430 hypothetical protein 59 3e-09
At2g15540 putative non-LTR retroelement reverse transcriptase 58 4e-09
At2g01550 putative non-LTR retroelement reverse transcriptase 55 3e-08
At1g31100 hypothetical protein 55 3e-08
At4g26360 putative protein 52 2e-07
At2g05550 putative non-LTR retroelement reverse transcriptase 51 5e-07
At2g15510 putative non-LTR retroelement reverse transcriptase 49 2e-06
At2g05980 putative non-LTR retroelement reverse transcriptase 49 2e-06
At3g30570 putative reverse transcriptase 48 4e-06
At2g45230 putative non-LTR retroelement reverse transcriptase 46 1e-05
At2g19100 putative non-LTR retroelement reverse transcriptase 45 2e-05
At2g17910 putative non-LTR retroelement reverse transcriptase 45 4e-05
At2g07670 putative non-LTR retrolelement reverse transcriptase 45 4e-05
At2g16680 putative non-LTR retroelement reverse transcriptase 44 5e-05
At2g01840 putative non-LTR retroelement reverse transcriptase 44 9e-05
At1g10160 putative reverse transcriptase 42 2e-04
>At1g24640 hypothetical protein
Length = 1270
Score = 69.3 bits (168), Expect = 1e-12
Identities = 51/182 (28%), Positives = 86/182 (47%), Gaps = 11/182 (6%)
Query: 3 GDFNAILNDGERKGLNSNTYGDVH-FKHFVETYALIDLPLGNDEFTWGSTRGDG-LWSKI 60
GDFN IL++GE+ G + D F ++ L+++P + FTW RGD + ++
Sbjct: 98 GDFNDILHNGEKNGGPRRSDLDCKAFNEMIKGCDLVEMPAHGNGFTWAGRRGDHWIQCRL 157
Query: 61 DRWLVNDEAILQFDGATQS------ADHRPVSLAL-GSHDFGPKPFCFFNYWLMEDGFKK 113
DR N E F + Q+ +DHRPV + L S D F F +L ++ K+
Sbjct: 158 DRAFGNKEWFCFFPVSNQTFLDFRGSDHRPVLIKLMSSQDSYRGQFRFDKRFLFKEDVKE 217
Query: 114 MVEEWWSSAVVEGWSNFALMQKLKGLKVKIREWKENRGIRGSEKIKGLEDNLHEGECLVT 173
+ WS + +N ++ +L+ + + WK+ + +KI LE L + + LV
Sbjct: 218 AIIRTWSRG--KHGTNISVADRLRACRKSLSSWKKQNNLNSLDKINQLEAALEKEQSLVW 275
Query: 174 KI 175
I
Sbjct: 276 PI 277
>At3g25485 unknown protein
Length = 979
Score = 61.2 bits (147), Expect = 4e-10
Identities = 56/168 (33%), Positives = 79/168 (46%), Gaps = 17/168 (10%)
Query: 1 IGGDFNAILNDGER-KGLNSN-TYGDVHFKHFVETYALIDLPLGNDEFTWGSTR-GDGLW 57
I GDFN ILN E KG ++ TYG F+ V A DL +TW + + D +
Sbjct: 96 IFGDFNEILNMAEHSKGDDTRVTYGMREFQEMVNYCAFTDLSSHGSLYTWSNRQENDLIL 155
Query: 58 SKIDRWLVNDEAILQFDG------ATQSADHRP----VSLALGSHDFGPKPFCFFNYWLM 107
K+DR LVN++ + F A +DH +++ +G G KPF F N
Sbjct: 156 KKLDRVLVNNKWKMSFPQSYNVFEAGGCSDHLRCRINLNIDVGERVKGKKPFKFINAIAE 215
Query: 108 EDGFKKMVEEWWSS---AVVEGWSNFALMQKLKGLKVKIREW-KENRG 151
F+ MVE +W + + + F +KLK LK KIR+ KE G
Sbjct: 216 MQEFRPMVEAYWKTTEPVYMSTSTMFRFTKKLKALKPKIRQLAKEKMG 263
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 58.9 bits (141), Expect = 2e-09
Identities = 42/135 (31%), Positives = 68/135 (50%), Gaps = 12/135 (8%)
Query: 1 IGGDFNAILNDGERKGLNSNTYGD-VHFKHFVETYALIDLPLGNDEFTWGSTRGDGLW-- 57
IGGDFN I+ ER G N D + F ++ ++LIDL ++FTW R + +
Sbjct: 699 IGGDFNTIVRLDERSGGNGRLSSDSLAFGEWINDHSLIDLGFKGNKFTWKRGREERFFVA 758
Query: 58 SKIDRWLVNDEAILQFDGATQ------SADHRPVSLALG---SHDFGPKPFCFFNYWLME 108
++DR L A L++ A+ ++DH P+ + L S + G +PF F WL
Sbjct: 759 KRLDRVLCCAHARLKWQEASVLHLPFLASDHAPLYVQLTPEVSGNRGRRPFRFEAAWLSH 818
Query: 109 DGFKKMVEEWWSSAV 123
GFK+++ W+ +
Sbjct: 819 PGFKELLLTSWNKDI 833
>At3g31420 hypothetical protein
Length = 1491
Score = 58.5 bits (140), Expect = 3e-09
Identities = 48/176 (27%), Positives = 80/176 (45%), Gaps = 13/176 (7%)
Query: 1 IGGDFNAILNDGERKGLN---SNTYGDVHFKHFVETYALIDLPLGNDEFTWGSTRGDG-L 56
I GDFN IL++ E++G T+ D F++ V DL D F+W RGD +
Sbjct: 314 IMGDFNEILSNREKRGGRLRPERTFQD--FRNMVRGCNFTDLKSVGDRFSWAGKRGDHHV 371
Query: 57 WSKIDRWLVNDEAILQFDGATQ------SADHRPVSLALGSHDFGPKPFCFFNYWLM-ED 109
+DR + N+E F + +DHRP+ + + + F ++ L ++
Sbjct: 372 TCSLDRTMANNEWHTLFPESETVFMEYGESDHRPLVTNISAQKEERRGFFSYDSRLTHKE 431
Query: 110 GFKKMVEEWWSSAVVEGWSNFALMQKLKGLKVKIREWKENRGIRGSEKIKGLEDNL 165
GFK++V W + +L +KL + I WK+N + E+IK + L
Sbjct: 432 GFKQVVLNQWHRRNGSFEGDSSLNRKLVECRQAISRWKKNNRVNAEERIKIIRHKL 487
>At1g25430 hypothetical protein
Length = 1213
Score = 58.5 bits (140), Expect = 3e-09
Identities = 50/155 (32%), Positives = 70/155 (44%), Gaps = 12/155 (7%)
Query: 3 GDFNAILNDGERKGLNSNTYGDVHFKHFVETYA---LIDLPLGNDEFTW-GSTRGDGLWS 58
GDFN +LN E S D++ + F + L DL + FTW + +
Sbjct: 143 GDFNQVLNPQEHSNPVSLNV-DINMRDFRDCLLAAELSDLRYKGNTFTWWNKSHTTPVAK 201
Query: 59 KIDRWLVNDE------AILQFDGATQSADHRPVSLALGSHDFGPK-PFCFFNYWLMEDGF 111
KIDR LVND + L G+ +DH + L K PF FFNY L F
Sbjct: 202 KIDRILVNDSWNALFPSSLGIFGSLDFSDHVSCGVVLEETSIKAKRPFKFFNYLLKNLDF 261
Query: 112 KKMVEEWWSSAVVEGWSNFALMQKLKGLKVKIREW 146
+V + W + V G S F + +KLK LK I+++
Sbjct: 262 LNLVRDNWFTLNVVGSSMFRVSKKLKALKKPIKDF 296
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 57.8 bits (138), Expect = 4e-09
Identities = 45/180 (25%), Positives = 77/180 (42%), Gaps = 19/180 (10%)
Query: 3 GDFNAILNDGERKGLNSNTYGD-VHFKHFVETYALIDLPLGNDEFTW---------GSTR 52
GDFN I+ + E++G + + F +E +ID P F+W G R
Sbjct: 135 GDFNEIIGNHEKRGGKKRSESSFLPFCCMIENCGMIDFPSTGSLFSWVGKRSCGVAGRKR 194
Query: 53 GDGLWSKIDRWLVNDEAILQFDGAT------QSADHRPVSLALGSHDFGP-KPFCFFNYW 105
D + ++DR + N+E + + +DH+P+ ++ + + P K F F W
Sbjct: 195 RDLIKCRLDRAMGNEEWHSIYSHTNVEYLQHRGSDHKPLLASIQNKPYRPYKHFIFDKRW 254
Query: 106 LMEDGFKKMVEEWWSSAVVEGWSNFALMQKLKGLKVKIREWKENRGIRGSEKIKGLEDNL 165
+ + GFK+ V+E W A QK+K + I WK++ + I L L
Sbjct: 255 INKPGFKESVQEGW--AFPSRGEGVPFFQKIKNCRQTISIWKKSNKTNTEKLILELHSQL 312
>At2g01550 putative non-LTR retroelement reverse transcriptase
Length = 1449
Score = 55.1 bits (131), Expect = 3e-08
Identities = 50/161 (31%), Positives = 71/161 (44%), Gaps = 17/161 (10%)
Query: 1 IGGDFNAILNDGERKGLNSN---TYGDVHFKHFVETYALIDLPLGNDEFTWGSTR-GDGL 56
I GDFN IL+ E + + T G F+ V + DL FTW + R D +
Sbjct: 554 IFGDFNEILDMDEHSRMEDHPAVTSGMRDFQSLVNYCSFSDLASHGPLFTWCNKRDNDPI 613
Query: 57 WSKIDRWLVNDEAILQFDG------ATQSADHRPVSLAL----GSHDFGPKPFCFFNYWL 106
W K+DR +VN+ + + A +DH + L G+ G KPF F N
Sbjct: 614 WKKLDRVMVNEAWKMVYPQSYNVFEAGGCSDHLRCRINLNMNSGAQVRGNKPFKFVNAVA 673
Query: 107 MEDGFKKMVEEWWSSAV---VEGWSNFALMQKLKGLKVKIR 144
+ FK +VE +W + S F +KLK LK K+R
Sbjct: 674 DMEEFKPLVENFWRETEPIHMSTSSLFRFTKKLKALKPKLR 714
>At1g31100 hypothetical protein
Length = 1090
Score = 55.1 bits (131), Expect = 3e-08
Identities = 55/181 (30%), Positives = 74/181 (40%), Gaps = 19/181 (10%)
Query: 3 GDFNAILNDGERK---GLNSNTYGDVHFKHFVETYALIDLPLGNDEFTWGSTRGDG-LWS 58
GDFN +L E LN N V F+ + L DL + FTW + +
Sbjct: 54 GDFNQVLCPAEHSQATSLNVNRRMKV-FRDCLFEAELCDLVFKGNTFTWWNKSATRPVAK 112
Query: 59 KIDRWLVNDEAILQFD------GATQSADHRPVSLALGSHDFGPK-PFCFFNYWLMEDGF 111
K+DR LVN+ +F G +DH + + K PF F+N+ L F
Sbjct: 113 KLDRILVNESWCSRFPSAYAVFGEPDFSDHASCGVIINPLMHREKRPFRFYNFLLQNPDF 172
Query: 112 KKMVEEWWSSAVVEGWSNFALMQKLKGLKVKIREWKENRGIRGSEKIKGLEDNLHEGECL 171
+V E W S V G S F + +KLK LK IR + E LE + E L
Sbjct: 173 ISLVGELWYSINVVGSSMFKMSKKLKALKNPIRTF-------SMENFSNLEKRVKEAHNL 225
Query: 172 V 172
V
Sbjct: 226 V 226
>At4g26360 putative protein
Length = 1141
Score = 52.4 bits (124), Expect = 2e-07
Identities = 47/154 (30%), Positives = 69/154 (44%), Gaps = 10/154 (6%)
Query: 3 GDFNAILNDGER-KGLNSNTYGDVH-FKHFVETYALIDLPLGNDEFTW-GSTRGDGLWSK 59
GDFN +L+ E + ++ N + F+ + L DL FTW ++ + K
Sbjct: 135 GDFNQVLHPHEHSRHVSLNVDRRIRDFRECLLDAELSDLVYKGSSFTWWNKSKTRPVAKK 194
Query: 60 IDRWLVNDEAILQFD------GATQSADHRPVSLALGSHDFGPK-PFCFFNYWLMEDGFK 112
IDR LVN+ F G +DH + L K PF FFN+ L F
Sbjct: 195 IDRILVNESWSNLFPSSFGLFGPPDFSDHASCGVVLELDPIKAKRPFKFFNFLLKNPEFL 254
Query: 113 KMVEEWWSSAVVEGWSNFALMQKLKGLKVKIREW 146
+V + W S V G S F + +KLK LK I+++
Sbjct: 255 NLVWDVWYSTNVVGSSMFRVSKKLKALKKPIKDF 288
>At2g05550 putative non-LTR retroelement reverse transcriptase
Length = 977
Score = 50.8 bits (120), Expect = 5e-07
Identities = 49/162 (30%), Positives = 69/162 (42%), Gaps = 15/162 (9%)
Query: 1 IGGDFNAILNDGERKGLNSN---TYGDVHFKHFVETYALIDLPLGNDEFTWGSTRG-DGL 56
I GDFN L+ E +SN T G F+ + + L D+ +TW + + D +
Sbjct: 346 IFGDFNVTLDMKEHSNCDSNPVVTSGMRDFQQMIRSCDLSDMASHGPLYTWCNKKEQDLI 405
Query: 57 WSKIDRWLVNDEAILQFDGATQ------SADHRPVSLALGSHDF----GPKPFCFFNYWL 106
K+DR +VND F A +DH + L GPKPF F N +
Sbjct: 406 LKKLDRVMVNDIWAQSFSQAYNVFEAGGCSDHLRCRIMLNDAAERSVKGPKPFKFVNAII 465
Query: 107 MEDGFKKMVEEWWSSAVVEGWSNFALMQKLKGLKV-KIREWK 147
+ FK MV+E W+ S L + K LK+ K WK
Sbjct: 466 EMEDFKPMVKEHWNKTEPLFMSTSTLFRFSKKLKILKPGIWK 507
>At2g15510 putative non-LTR retroelement reverse transcriptase
Length = 1138
Score = 49.3 bits (116), Expect = 2e-06
Identities = 42/158 (26%), Positives = 72/158 (44%), Gaps = 17/158 (10%)
Query: 3 GDFNAILNDGERKGLNSNTYGD---VHFKHFVETYALIDLPLGNDEFTWGSTRGDGLW-- 57
GDFN IL++GE++G GD + FK +E +++LP + F+WG R + LW
Sbjct: 101 GDFNVILHNGEKRG--GPRRGDSSFISFKDMLECCDMLELPSKGNPFSWGGKR-NSLWIQ 157
Query: 58 SKIDRWLVNDEAILQFDGATQ------SADHRPVSLALGSHDFGPK-PFCFFNYWLMEDG 110
++DR N F + Q +D RPV + L K F F +
Sbjct: 158 CRLDRCFGNKNWFRHFPVSNQEFLDKRGSDRRPVLVRLSKTKEEYKGNFRFDKRLFNQPL 217
Query: 111 FKKMVEEWWSSAVVEGWSNFALMQKLKGLKVKIREWKE 148
K+ +++ W+ G ++++LK + + WK+
Sbjct: 218 VKEAIQQAWNGNQRVG--GMQVLERLKKCRSALSRWKQ 253
>At2g05980 putative non-LTR retroelement reverse transcriptase
Length = 1352
Score = 48.9 bits (115), Expect = 2e-06
Identities = 44/152 (28%), Positives = 69/152 (44%), Gaps = 14/152 (9%)
Query: 3 GDFNAILNDGERKGLNSNTY---GDVHFKHFVETYALIDLPLGNDEFTWGSTRGDGLWS- 58
GDFN IL+ + ++ + G F+ V +L D+ FTW + R +GL +
Sbjct: 378 GDFNEILDGEDHSNFVNSPFIPQGMRDFQDVVRYCSLADMSYHGPVFTWCNKREEGLVNK 437
Query: 59 KIDRWLVND------EAILQFDGATQSADHRPVSLALGSHDFGPK-PFCFFNYWLMEDGF 111
K+DR LVN+ A+ A +DH + S + PK PF F N F
Sbjct: 438 KLDRVLVNEVWTRKFPAVYSIFEAGGISDHLRCRIKFASGNTQPKGPFKFNNVLTTLPEF 497
Query: 112 KKMVEEWWSSAVV---EGWSNFALMQKLKGLK 140
++V+++W + + F +KLKGLK
Sbjct: 498 LQIVKDFWDGTLALFHPTSAMFRFSKKLKGLK 529
>At3g30570 putative reverse transcriptase
Length = 1099
Score = 48.1 bits (113), Expect = 4e-06
Identities = 53/189 (28%), Positives = 87/189 (45%), Gaps = 28/189 (14%)
Query: 1 IGGDFNAILNDGERKGLNSNTYGD-VHFKHFVETYALID-LPLGNDEFTWGSTRGDGLWS 58
IG DFN I+ ER G + D + F ++ +L+ + + GS R D +
Sbjct: 183 IGEDFNMIVRVDERSGGSGPLSPDSIAFGEWINDSSLLTWVSRVTNSVGSGSVRRDFFVA 242
Query: 59 K-IDRWLVNDEAILQFDGATQS------ADHRPV----SLALGSHDFGPKPFCFFNYWLM 107
K +DR L A L++ AT + +DH P+ +L +G + F +PF F WL
Sbjct: 243 KRLDRVLCCSHARLKWQEATLTHLPFLASDHAPLYLQLALGVGKNRFR-RPFRFEAAWLS 301
Query: 108 EDGFKKMVEEWWSSAVVEGWSNFALMQKLKGLKVKIREW-KENRGIRGSEK------IKG 160
GFK+++ W N + LKG++ +R+W KE G +K IK
Sbjct: 302 HPGFKELLLASWK-------GNIPTPEALKGIQTTLRKWNKEVFGDIQEKKEKLMVEIKT 354
Query: 161 LEDNLHEGE 169
++D+L+ +
Sbjct: 355 VQDSLYSNQ 363
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 46.2 bits (108), Expect = 1e-05
Identities = 39/174 (22%), Positives = 80/174 (45%), Gaps = 15/174 (8%)
Query: 3 GDFNAILNDGERKGLNSNTYGD-VHFKHFVETYALIDLPLGNDEFTWGSTRGDGLWS-KI 60
GDFN +++ E+ G + + F+ + + L ++ +F+W R D L ++
Sbjct: 135 GDFNELVDPSEKIGGPARKESSCLEFRQMLNSCGLWEVNHSGYQFSWYGNRNDELVQCRL 194
Query: 61 DRWLVNDEAILQFDGATQS------ADHRPVSLALGSHDFGPKPFCFFNY---WLMEDGF 111
DR + N + F A + +DH P+ L ++ + + F Y W+ +GF
Sbjct: 195 DRTVANQAWMELFPQAKATYLQKICSDHSPLINNLVGDNW--RKWAGFKYDKRWVQREGF 252
Query: 112 KKMVEEWWSSAVVEGWSNFALMQKLKGLKVKIREWKENRGIRGSEKIKGLEDNL 165
K ++ +WS + +N +M+K+ + +I +WK + +I+ L+ L
Sbjct: 253 KDLLCNFWSQQSTK--TNALMMEKIASCRREISKWKRVSKPSSAVRIQELQFKL 304
>At2g19100 putative non-LTR retroelement reverse transcriptase
Length = 1447
Score = 45.4 bits (106), Expect = 2e-05
Identities = 48/167 (28%), Positives = 67/167 (39%), Gaps = 17/167 (10%)
Query: 1 IGGDFNAILNDGERKGLNSN---TYGDVHFKHFVETYALIDLPLGNDEFTWGSTRG-DGL 56
I GDFN L E + N + G F+ V +L D+ +TW + R D +
Sbjct: 547 IFGDFNETLELEEHSKVEDNPVVSMGMRDFRSMVNYCSLTDMAHHGPLYTWSNKREHDLI 606
Query: 57 WSKIDRWLVNDEAILQFDG------ATQSADHRPVSLAL----GSHDFGPKPFCFFNYWL 106
K+DR +VND F A DH + L GS G +PF F N
Sbjct: 607 AKKLDRVMVNDVWTQSFPQSYSVFEAGGCLDHLRGRINLNDGPGSIVRGKRPFKFVNVLT 666
Query: 107 MEDGFKKMVEEWWSSA---VVEGWSNFALMQKLKGLKVKIREWKENR 150
+ FK V+ +W + S F +KLK LK +R + R
Sbjct: 667 EMEDFKPTVDSYWKETEPIFLSTSSLFRFSKKLKSLKPLLRNLAKER 713
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 44.7 bits (104), Expect = 4e-05
Identities = 44/174 (25%), Positives = 79/174 (45%), Gaps = 15/174 (8%)
Query: 3 GDFNAILNDGERKGLNSNTYGDVH-FKHFVETYALIDLPLGNDEFTWGSTRGD-GLWSKI 60
GDFN I ++ E+ G + F+H + ++ +L + FTWG R D + K+
Sbjct: 113 GDFNDIRSNDEKLGGPRRSPSSFQCFEHMLLNCSMHELGSTGNSFTWGGNRNDQWVQCKL 172
Query: 61 DRWLVNDEAILQFDGATQ------SADHRPVSLALGSHDFGPKPFCFFNYWLMEDGF-KK 113
DR N F A Q +DHRPV + + + + ++ L +D + +
Sbjct: 173 DRCFGNPAWFSIFPNAHQWFLEKFGSDHRPVLVKFTNDNELFRGQFRYDKRLDDDPYCIE 232
Query: 114 MVEEWWSSAVVEGW--SNFALMQKLKGLKVKIREWKENRGIRGSEKIKGLEDNL 165
++ W+SA+ +G S F+L++ + + V WK + +IK L +L
Sbjct: 233 VIHRSWNSAMSQGTHSSFFSLIECRRAISV----WKHSSDTNAQSRIKRLRKDL 282
>At2g07670 putative non-LTR retrolelement reverse transcriptase
Length = 913
Score = 44.7 bits (104), Expect = 4e-05
Identities = 37/139 (26%), Positives = 66/139 (46%), Gaps = 20/139 (14%)
Query: 27 FKHFVETYALIDLPLGNDEFTWGSTRGDGLW--SKIDRWLVNDEAILQFDGATQS----- 79
F ++ +LID+ ++FTW R + + ++DR L + L++ A+ +
Sbjct: 632 FGEWINELSLIDMGFKGNKFTWKRGRVESTFVAKRLDRVLCRPQTRLKWQEASVTHLPFF 691
Query: 80 -ADHRPVSLALGSHDFGP---KPFCFFNYWLMEDGFKKMVEEWWSSAVVEGWSNFALMQK 135
+DH P+ + L +PF F WL GFK +++ W++ EG + A
Sbjct: 692 ASDHAPIYIQLEPEVRSNPLRRPFRFEAAWLTHSGFKDLLQASWNT---EGETPVA---- 744
Query: 136 LKGLKVKIREWKENRGIRG 154
L LK K+++W NR + G
Sbjct: 745 LAALKSKLKKW--NREVFG 761
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 44.3 bits (103), Expect = 5e-05
Identities = 44/175 (25%), Positives = 69/175 (39%), Gaps = 13/175 (7%)
Query: 3 GDFNAIL-NDGERKGLNSNTYGDVHFKHFVETYALIDLPLGNDEFTWGSTRGD-GLWSKI 60
GDFN IL NDG+ G + FK+ + + + + FTWG TR D + K+
Sbjct: 98 GDFNDILSNDGKLGGPSRLISSFQPFKNMLLNCDMHQMGSSGNSFTWGGTRNDQWIQCKL 157
Query: 61 DRWLVNDEAILQFDGATQ------SADHRPVSL-ALGSHDFGPKPFCFFNYWLME-DGFK 112
DR N E F + Q + HRPV + + + FC+ + +
Sbjct: 158 DRCFGNSEWFTMFSNSHQWFLEKLGSHHRPVLVNFVNDQEVFRGQFCYDKRFAEDPQCAA 217
Query: 113 KMVEEWWSSAVVEGWSNFALMQKLKGLKVKIREWKENRGIRGSEKIKGLEDNLHE 167
+ W + + + S+ M K + I WK+N +I L L E
Sbjct: 218 STLSSWIGNGISDVSSSMLRMVK---CRKAISGWKKNSDFNAQNRILRLRSELDE 269
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 43.5 bits (101), Expect = 9e-05
Identities = 39/176 (22%), Positives = 74/176 (41%), Gaps = 11/176 (6%)
Query: 3 GDFNAILNDGERKGLNSNTYGDV-HFKHFVETYALIDLPLGNDEFTW-GSTRGDGLWSKI 60
GDFN ILN E+KG + G + +F + + + DL + ++W G + + + S +
Sbjct: 497 GDFNEILNLNEKKGGRRRSIGSLQNFTNMINCCNMKDLKSKGNPYSWVGKRQNETIESCL 556
Query: 61 DRWLVNDEAILQFDG------ATQSADHRPVSLALGSHDFGPK-PFCFFNYWLMEDGFKK 113
DR +N + F +DH PV + + + F + + F
Sbjct: 557 DRVFINSDWQASFPAFETEFLPIAGSDHAPVIIDIAEEVCTKRGQFRYDRRHFQFEDFVD 616
Query: 114 MVEEWWSSAVVEGWSNFALMQKLKGLKVKIREWKENRGIRGSEKIKGLEDNLHEGE 169
V+ W+ + S+ +KL + ++ +WK +EKI+ L+ + E
Sbjct: 617 SVQRGWNRGRSD--SHGGYYEKLHCCRQELAKWKRRTKTNTAEKIETLKYRVDAAE 670
>At1g10160 putative reverse transcriptase
Length = 1118
Score = 42.4 bits (98), Expect = 2e-04
Identities = 50/192 (26%), Positives = 74/192 (38%), Gaps = 18/192 (9%)
Query: 3 GDFNAILNDGERKGLNS---NTYGDVHFKHFVETYALIDLPLGNDEFTWGSTRGDG-LWS 58
GDFN I E +N N G + + L DLP FTW + + D +
Sbjct: 141 GDFNQIAAASEHYSINQSLLNLRGMEDLQCCLRDSQLSDLPSRGVFFTWSNHQQDNPILR 200
Query: 59 KIDRWLVNDEAILQFDGATQ------SADHRPVSLALGSHDF-GPKPFCFFNYWLMEDGF 111
K+DR L N E F A +DH P + + + K F +F++ +
Sbjct: 201 KLDRALANGEWFAVFPSALAVFDPPGDSDHAPCIILIDNQPPPSKKSFKYFSFLSSHPSY 260
Query: 112 KKMVEEWWSSAVVEGWSNFALMQKLKGLKVKIREWK----ENRGIRGSEKIKGLEDNLHE 167
+ W + G F+L Q LK K+ R N R ++ + LED E
Sbjct: 261 LAALSTAWEENTLVGSHMFSLRQHLKVAKLCCRTLNRLRFSNIQQRTAQSLTRLEDIQVE 320
Query: 168 GECLVTKIKDSL 179
L+T D+L
Sbjct: 321 ---LLTSPSDTL 329
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.329 0.145 0.479
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,094,618
Number of Sequences: 26719
Number of extensions: 224971
Number of successful extensions: 646
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 611
Number of HSP's gapped (non-prelim): 43
length of query: 219
length of database: 11,318,596
effective HSP length: 95
effective length of query: 124
effective length of database: 8,780,291
effective search space: 1088756084
effective search space used: 1088756084
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0016.9


