

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0016.18
(931 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
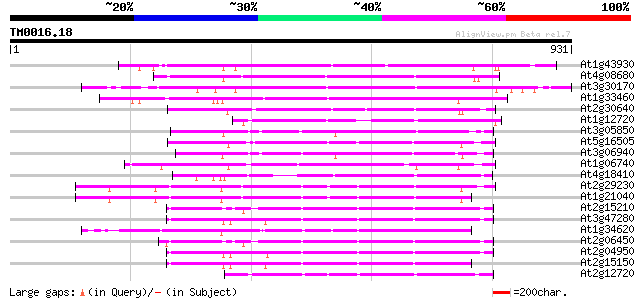
Score E
Sequences producing significant alignments: (bits) Value
At1g43930 hypothetical protein 270 2e-72
At4g08680 putative MuDR-A-like transposon protein 251 1e-66
At3g30170 unknown protein 249 4e-66
At1g33460 mutator transposase MUDRA, putative 241 1e-63
At2g30640 Mutator-like transposase 219 4e-57
At1g12720 hypothetical protein 207 2e-53
At3g05850 unknown protein 207 3e-53
At5g16505 mutator-like transposase-like protein (MQK4.25) 206 6e-53
At3g06940 putative mudrA protein 202 5e-52
At1g06740 mudrA-like protein 202 5e-52
At4g18410 MuDR transposable element - like protein 192 6e-49
At2g29230 Mutator-like transposase 182 6e-46
At1g21040 hypothetical protein 174 3e-43
At2g15210 putaive transposase of transposon FARE2.7 (cds1) 171 2e-42
At3g47280 putative protein 167 3e-41
At1g34620 Mutator-like protein 165 1e-40
At2g06450 putative transposase of FARE2.11 164 3e-40
At2g04950 putative transposase of transposon FARE2.9 (cds1) 163 5e-40
At2g15150 putative transposase of FARE2.2 (CDS1) 156 4e-38
At2g12720 putative protein 156 4e-38
>At1g43930 hypothetical protein
Length = 946
Score = 270 bits (691), Expect = 2e-72
Identities = 232/814 (28%), Positives = 347/814 (42%), Gaps = 105/814 (12%)
Query: 181 DVQQDGDGRDDVNRNSGPNNGVQDGD--ESDNNVGF-------------IPSEVAAQHNI 225
+V G+ +VN G V +G+ E + N G +E + N
Sbjct: 134 EVNNGGNAEAEVNDGGGVEAEVNNGESVEEEGNGGLDMEEEDCEDFEAEFGAEAREEENE 193
Query: 226 DMEYESDDLLSD--------YSCDEEDNGSRVRYQRFRQEDFDKDYKFRLGMEFASTEEF 277
DD+ D +S DE DN Q D D + RL F + E+F
Sbjct: 194 SDGDSGDDIWDDERIPDPLVHSDDEGDNDDE---DGAPQTD-DPEESLRLAKTFNNPEDF 249
Query: 278 KKAILAHSVMNGKAVIYKKNDKNRVRFACKQEE--CRFIAYCANIKGTSTYRIKTLNPDH 335
K A+L +S+ + ++ RV C + C++ YC+ K ++K H
Sbjct: 250 KIAVLRYSLKTRYDIKLYRSQSLRVGAKCTDTDVNCQWRCYCSYDKKKQKMQVKVYKDGH 309
Query: 336 TCGRTFKNRAATAKWIA-------KNARKIAKEAVEGDALKQFTL--------------- 373
+C R+ ++ + IA + KI K+ + + +++ L
Sbjct: 310 SCVRSGYSKMLKQRTIAWLFRERLRKNPKITKQEMVDEIKREYKLTVTEDQCSKAKTMVM 369
Query: 374 -------------ITSYAAELMRVNHENTAAIILRYIPT--TLQPRFGCFYMCLEGCKSA 418
I Y A++ R N T I PT +LQ RF ++C + K +
Sbjct: 370 RERQATHQEHFARIWDYQAKIFRSNPGTTFEIETILGPTIGSLQ-RFLRLFICFKSQKES 428
Query: 419 FKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLM 478
+K CRP IGIDG LK G LL A GRD +++ P+A+AVVE E ++W WF+ L
Sbjct: 429 WKQTCRPIIGIDGAFLKWDIKGHLLAATGRDGDNRIVPIAWAVVEIENDDNWDWFVRMLS 488
Query: 479 RDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLM 538
R + + ISDKQ GL++ P EHR C RH+ N+K+ R L
Sbjct: 489 RTLDLQDGRNVAIISDKQSGLVKAIHSVIPQAEHRQCARHIMENWKRNSHDMELQR--LF 546
Query: 539 WASKATYKE-LWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAF 597
W +Y E + M+ +K N +A+ L+ + W R F C+ +NNL+E+F
Sbjct: 547 WKIVRSYTEGEFGAHMRALKSYNASAFELLLKTLPVTWSRAFFKIGSCCNDNLNNLSESF 606
Query: 598 NCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIK--YSGN 655
N TI AR KPL+ M E IR+ M R EK + + KR EI+ +G+
Sbjct: 607 NRTIREARRKPLLDMLEDIRRQCMVR----NEKRYVIADRLRTRFTKRAHAEIEKMIAGS 662
Query: 656 WVA-TWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPEN 714
V W+A+ G D F VD++ +C C W++ GIPC HA + K K E+
Sbjct: 663 QVCQRWMARHNKHEIKVGPVDSFTVDMNNNTCGCMKWQMTGIPCIHAASVIIGKRQKVED 722
Query: 715 YVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRG-PGRPKKLRRR----E 769
YV +Y + ++ Y I P+ G+ WP +++ +LPP +RG PGRPK RR E
Sbjct: 723 YVSDWYTTSMWKQTYNDGIGPVQGKLLWPTVNKVGVLPPPWRRGNPGRPKNHARRKGVFE 782
Query: 770 PDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCP----LPPAK-------KQPTPSQPA 818
+ T+L R TC C +GHN++ C PPAK K P
Sbjct: 783 SSTASSSSTTELSRLHRVMTCSNCQGEGHNKQGCKNETVAPPAKRPRGRPRKDKDPRMVF 842
Query: 819 AAAIATNQAPQ---PPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAAR 875
T Q PQ P S A ++ H AP P+ A + A Q Q + ++
Sbjct: 843 FGQGQTWQEPQAQGAPSSHGAPSQVFHGAPLSQPSQEAPSQGAPSQGAQ-------SLSQ 895
Query: 876 ARHQAPQPPPPSPAAAARA--RHEGEPAARSASQ 907
+AP P+ R+ R E + R SQ
Sbjct: 896 PSQEAPSQAAPTETRGTRSVRRGESQRGGRGGSQ 929
>At4g08680 putative MuDR-A-like transposon protein
Length = 761
Score = 251 bits (642), Expect = 1e-66
Identities = 177/624 (28%), Positives = 282/624 (44%), Gaps = 57/624 (9%)
Query: 239 SCDEEDNGSRVRYQRFRQEDFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKND 298
S DE+++ VR +R R+ DK F +G F + EFK+AIL ++ +G V+ + +
Sbjct: 86 SSDEDEDPILVRDRRIRRAMGDK---FVIGSTFFTGIEFKEAILEVTLKHGHNVVQDRWE 142
Query: 299 KNRVRFACKQE-ECRFIAYCANIKGTSTYRIKTLNPDHTCGRTFKNRAATAKWIAK---- 353
K ++ F C +C++ YC+ + +KT H+C K + + IA+
Sbjct: 143 KEKISFKCGMGGKCKWRVYCSYDPDRQLFVVKTSCTWHSCSPNGKCQILKSPVIARLFLD 202
Query: 354 -------------------------------NARKIAKEAVEGDALKQFTLITSYAAELM 382
R +A + ++ + +QF I Y E+
Sbjct: 203 KLRLNEKFMPMDIQEYIKERWKMVSTIPQCQRGRLLALKMLKKEYEEQFAHIRGYVEEIH 262
Query: 383 RVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGIL 442
N + A I Y + F FY+C ++ + +CRP IG+DG LK G+L
Sbjct: 263 SQN-PGSVAFIDTYRNEKGEDVFNRFYVCFNILRTQWAGSCRPIIGLDGTFLKVVVKGVL 321
Query: 443 LIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQV 502
L AVG DPN+Q +P+A+AVV++E E+W WF+ + +D+ ++V +SD+ KGLL
Sbjct: 322 LTAVGHDPNNQIYPIAWAVVQSENAENWLWFVQQIKKDLNLEDGSRFVILSDRSKGLLSA 381
Query: 503 FAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKATYKEL-WEEKMQEIKKINE 561
++ P EHR CV+H+ N K K ++ L+W +Y E + + + ++ +E
Sbjct: 382 VKQELPNAEHRMCVKHIVENLK-KNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRCYDE 440
Query: 562 AAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLM 621
A Y ++ W R + C+ NN E+FN TI AR K LI M E IR+ M
Sbjct: 441 ALYNDVLNEEPHTWSRCFYKLGSCCEDVDNNATESFNSTITKARAKSLIPMLETIRRQGM 500
Query: 622 TRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDI 681
TR +KS + +G K L E + G+F+V E+ VDI
Sbjct: 501 TRIVKRNKKSLRHEGRFTKYALKMLALEKTDADRSKVYRCTHGVFEVYID--ENGHRVDI 558
Query: 682 DKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENK 741
K C+C W+++GIPC H+ + GL ENY+ +++ + Y P+ G
Sbjct: 559 PKTQCSCGKWQISGIPCEHSYGAMIEAGLDAENYISEFFSTDLWRDSYETATMPLRGPKY 618
Query: 742 WPKMSQDDILPPEIKRGPGRPKKLRRREP--------DEPDKK-----NPTKLKRGGSSY 788
W S + P PGR K+ ++E + P KK KL + G
Sbjct: 619 WLNSSYGLVTAPPEPILPGRKKEKSKKEKFARIKGKNESPKKKKRKKNEVEKLGKKGKII 678
Query: 789 TCGRCHQKGHNQRKCPLPPAKKQP 812
C C + GHN +C P +K P
Sbjct: 679 HCKSCGEAGHNALRCKKFPKEKVP 702
>At3g30170 unknown protein
Length = 995
Score = 249 bits (637), Expect = 4e-66
Identities = 224/866 (25%), Positives = 360/866 (40%), Gaps = 88/866 (10%)
Query: 120 DAVLDDEDDVFVEVTEVEVIPMTQEAPTVAVPEVEASNIDSEDEICKNLTNDGFDDVDEP 179
DAV D+D E + E + +A + EA D + T+DG +D D
Sbjct: 104 DAVGGDDDATGGEGNDDEAVGGGDDATGCGGNDDEALGYDDD------ATSDGGNDDD-- 155
Query: 180 FDVQQDGDGRDDVNRNSGPNNGVQDGDESDNNVGFIPSEVAAQHNIDMEYESDDLLSDYS 239
DG +D N N G E D + P +V +E D + +S
Sbjct: 156 -----DGGVEEDANLNLAEEFPEFAGVEEDCSDDDPPDDV---------WEEDKIPDPFS 201
Query: 240 CDEEDNGSRVRYQRFRQEDFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDK 299
D+ED SR R ++ +++ L + + ++FK A+L +S+ + +++
Sbjct: 202 SDDEDE-SRTREVHGHRDAANEEVLLELKKTYNTPDDFKLAVLRYSLKTRYDIKLFRSEA 260
Query: 300 NRVRFACKQEE-----CRFIAYCANIKGTSTYRIKTLNPDHTCGRT----FKNRAATAKW 350
V C + C + YC+ K +I+T +H C R+ R++ A
Sbjct: 261 RIVAAKCSYVDKDGVQCPWKVYCSYEKTMHKMQIRTYVNNHICVRSGHSKMLKRSSIAYL 320
Query: 351 IAKNAR---KIAKEAVEGDALKQFTL---------ITSYAAELMRVNHENTAAIILRYIP 398
+ R K+ K + + L+++ L + + +HE A + Y
Sbjct: 321 FEERLRVNPKLTKYEMAAEILREYNLEVTPDQCAKAKTKVVKARNASHEAHFARVWDYQA 380
Query: 399 TTLQPRFGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLA 458
++ + + RPFIG+DG LK G LL AVGRD +++ P+A
Sbjct: 381 EVIKQNPWTEFEIETTAGAVIGAKQRPFIGLDGAFLKWDIKGHLLAAVGRDGDNKIVPIA 440
Query: 459 FAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRH 518
+AVVE E ++W WFL HL +G +SDKQ GL++ P EHR C +H
Sbjct: 441 WAVVEIENDDNWDWFLRHLSASLGLCQMANIAILSDKQSGLVKAIHTILPHAEHRQCSKH 500
Query: 519 LYANFKQKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRH 578
+ N+K+ E++ L +++ E + M E+++ N AY L + W R
Sbjct: 501 IMDNWKRD-SHDLELQRLFWKIARSYTVEEFNNHMMELQQYNRQAYESLQLTSPVTWSRA 559
Query: 579 AFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNV 638
F C+ +NNL+E+FN TI AR KP++ M E IR+ M R A ++K +
Sbjct: 560 FFRIGTCCNDNLNNLSESFNRTIRQARRKPILDMLEDIRRQCMVRSAKRFLIAEKLQTRF 619
Query: 639 MPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPC 698
+ + +++ I + L ++ + VD+++++C C ++ GIPC
Sbjct: 620 TKRAHEEIEKMIVGLRQCERYMARENLHEIYVNNV--SYFVDMEQKTCDCRKCQMVGIPC 677
Query: 699 RHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRG 758
HA K K E+YV YY + + Y I P+ G W + ++ +LPP +RG
Sbjct: 678 IHATCVIIGKKEKVEDYVSDYYTKVRWRETYLKGIRPVQGMPLWCRTNRLPVLPPPWRRG 737
Query: 759 -PGRPKKLRRRE--PDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLP--------- 806
GRP RR+ + NP K+ R TC CHQ+GHN+++C +P
Sbjct: 738 NAGRPSNYARRKGRNEAAAPSNPNKMSREKRIMTCSNCHQEGHNKQRCNIPTVLNPTKRP 797
Query: 807 ---PAKKQPTPSQPAAAAIATNQAPQP------PPSPAAAARARHQAPPPPP----ASPA 853
P K Q S + A +Q P P+AA R P S
Sbjct: 798 RGRPRKNQQPQSHQGSQADLESQGPSAAGRGLGSQGPSAAGRGLGSQGPSAAGRGLGSQG 857
Query: 854 AAARARHQAPQPPPAS--------PAAAARARHQAPQPPPPSPAAAARARHEGEPAARSA 905
+A R Q P A+ P+AA R R P AA R R G R+A
Sbjct: 858 PSAAGRGLGSQEPSAAGRGRGAQGPSAAGRGR------VAQGPRAAGRGR--GAQGQRAA 909
Query: 906 SQPKKPAVQPSKPAAQPTKKPATKPS 931
+ Q + + ++ A PS
Sbjct: 910 GRGCGAQGQTASGGGRGAQRQAGGPS 935
>At1g33460 mutator transposase MUDRA, putative
Length = 826
Score = 241 bits (616), Expect = 1e-63
Identities = 192/748 (25%), Positives = 320/748 (42%), Gaps = 79/748 (10%)
Query: 150 VPEVEASNIDSEDE-ICKNLTNDGFDDVDEPFDVQQDGDGR--DDVNRNSGPNNGV---- 202
V E EA+ ++ +D I + +DG ++ ++ D+ DGD DD +R+
Sbjct: 40 VEEDEATGVEGDDVGIDGDRESDGEENREDDGDIASDGDINLEDDGDRDESQKKRTREEK 99
Query: 203 ---------------QDGDESDNNVG----FIPSEVAAQHNIDMEYESDDLLSDYSCDEE 243
Q DE + ++ F E A +++ +++ + +E+
Sbjct: 100 RKEEEAEEGQKKRRKQKKDEPEEDLAERFEFEIEEAVAMWYDELKIRRNEIPESDNEEED 159
Query: 244 DNGSRVRYQRFRQEDFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVR 303
+R R R +D + +G F + EFK+ +L +++ + + +K+++
Sbjct: 160 HVITRDRKIRLASDD-----RLAIGRTFFTGFEFKEVVLHYAMKHRINAKQNRWEKDKIS 214
Query: 304 FACKQ-EECRFIAYCANIKGTSTYRIKTLNPDHTC--------------GRTFKN----- 343
F C Q +EC + Y + + +KT DH+C GR F +
Sbjct: 215 FRCAQRKECEWYVYASYSHERQLWVLKTKCLDHSCTSNGKCKLLKRRVIGRLFMDKLRLQ 274
Query: 344 ---------RAATAKWI-------AKNARKIAKEAVEGDALKQFTLITSYAAELMRVNHE 387
R +W ++ R +A + ++ + +QF + Y AE++ N
Sbjct: 275 PNFMPLDIQRHIKEQWKLVSTIGQVQDGRLLALKWLKEEYAQQFAHLRGYVAEILSTNKG 334
Query: 388 NTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVG 447
+TA I+ F Y+C K+AF CRP IGIDG LK G LL A+
Sbjct: 335 STA-IVDTIRDANENDVFNRIYVCFGAMKNAFYF-CRPLIGIDGTFLKHAVKGCLLTAIA 392
Query: 448 RDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDF 507
D N+Q +P+A+A V+ E E+W WFLN L D+ +V ISD+ KG++
Sbjct: 393 HDANNQIYPVAWATVQFENAENWLWFLNQLKHDLELKDGSGYVVISDRCKGIISAVKNAL 452
Query: 508 PGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKATYKEL-WEEKMQEIKKINEAAYWW 566
P EHR CV+H+ N K++ G ++ +W +Y + ++ + E++ + Y
Sbjct: 453 PNAEHRPCVKHIVENLKKRH-GSLDLLKKFVWNLAWSYSDTQYKANLNEMRAYIMSLYED 511
Query: 567 LMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFAT 626
+M WCR F C+ NN E+FN TI+ AR K L+ M E IR+ M R +
Sbjct: 512 VMKEEPNTWCRSWFRIGSYCEDVDNNATESFNATIVKARAKALVPMMETIRRQAMARISK 571
Query: 627 LREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSC 686
+ K KWK + + L E + + T F+V G + + ++ C
Sbjct: 572 RKAKIGKWKKKISEYVSEILKEEWELAVKCEVTKGTHEKFEVWFDGNSNAVSLKTEEWDC 631
Query: 687 TCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENKWP--- 743
+C W++ GIPC HA A ND G E++V + + Y P+ G+ WP
Sbjct: 632 SCCKWQVTGIPCEHAYAAINDVGKDVEDFVIPMFYTIAWREQYDTGPDPVRGQMYWPTGL 691
Query: 744 ---KMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRG--GSSYTCGRCHQKGH 798
D +PP K+G + + + P KK P +LK G G C C + GH
Sbjct: 692 GLITAPLQDPVPPGRKKGEKKNYHRIKGPNESPKKKGPQRLKVGKKGVVMHCKSCGEAGH 751
Query: 799 NQRKCPLPPAKKQPTPSQPAAAAIATNQ 826
N C P +K+ + T+Q
Sbjct: 752 NAAGCKKFPKEKKGRKKKNQEIKAGTSQ 779
>At2g30640 Mutator-like transposase
Length = 754
Score = 219 bits (559), Expect = 4e-57
Identities = 164/595 (27%), Positives = 257/595 (42%), Gaps = 68/595 (11%)
Query: 262 DYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIK 321
++ +GMEF+ ++A+ ++ + K+DK R C E C + +CA +
Sbjct: 177 EHNLIVGMEFSDVLACRRALRDAAIALRFEMQTVKSDKTRFTAKCNSEGCPWRIHCAKLP 236
Query: 322 GTSTYRIKTLNPDHTCGRT--FKNRAATAKWIAKNARKIAK------------------- 360
G T+ I+T++ HTCG + A+ +W+A + K
Sbjct: 237 GLPTFTIRTIHGSHTCGGISHLGHHQASVQWVADAVTERLKVNPHCKPKEILEQIHQVHG 296
Query: 361 ----------------EAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPR 404
AV G + + L+ Y ++ R N + A + + + Q
Sbjct: 297 ITLTYKQAWRGKERIMAAVRGSYEEDYRLLPRYCDQIRRTNPGSVAVVHGSPVDGSFQQ- 355
Query: 405 FGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEA 464
F++ + F ACRP IG+D LK+KY G LL+A G D FPLAFA++
Sbjct: 356 ---FFISFQASICGFLNACRPLIGLDRTVLKSKYVGTLLLATGFDGEGAVFPLAFAIISE 412
Query: 465 ECKESWRWFLNHLMRDIGDNGEK--KWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYAN 522
E SW+WFL+ L + + N E K +S + + ++ +FP H CV L +
Sbjct: 413 ENDSSWQWFLSELRQLLEVNSENMPKLTILSSRDQSIVDGVDTNFPTAFHGLCVHCLTES 472
Query: 523 FKQKFGGGTEVRDLLMW-ASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFS 581
+ +F V L+W A+K +E KM EI +I+ A W+ I W + F
Sbjct: 473 VRTQFNNSILVN--LVWEAAKCLTDFEFEGKMGEIAQISPEAASWIRNIQHSQWATYCFE 530
Query: 582 YFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPK 641
+T N++E+ N + A P+I M E IR+ LMT F RE S +W G ++P
Sbjct: 531 GTRFGHLTA-NVSESLNSWVQDASGLPIIQMLESIRRQLMTLFNERRETSMQWSGMLVPS 589
Query: 642 PRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHA 701
+ + I+ + + F+V + E ++IVDI ++C C WEL G+PC HA
Sbjct: 590 AERHVLEAIEECRLYPVHKANEAQFEVMTS--EGKWIVDIRCRTCYCRGWELYGLPCSHA 647
Query: 702 VACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKM-----SQDD-----IL 751
VA + Y+ A Y Y I P+ + +W S +D I
Sbjct: 648 VAALLACRQNVYRFTESYFTVANYRRTYAETIHPVPDKTEWKTTEPAGESGEDGEEIVIR 707
Query: 752 PPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLP 806
PP R P RPKK R + D +K + C RC+Q GH + C P
Sbjct: 708 PPRDLRQPPRPKKRRSQGEDRGRQKRVVR---------CSRCNQAGHFRTTCTAP 753
>At1g12720 hypothetical protein
Length = 585
Score = 207 bits (527), Expect = 2e-53
Identities = 146/461 (31%), Positives = 206/461 (44%), Gaps = 50/461 (10%)
Query: 371 FTLITSYAAELMRVNH------ENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACR 424
F+ I Y AE++ N E TA + + RF Y+C K ++K CR
Sbjct: 62 FSRIWDYQAEVLNRNPNSDFDIETTARTFIGS-----KQRFFRLYICFNSQKVSWKQHCR 116
Query: 425 PFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDN 484
P IGIDG LK G LL AVGRD +++ PLA+AVVE E ++W WFL L +G
Sbjct: 117 PVIGIDGAFLKWDIKGHLLAAVGRDGDNRIVPLAWAVVEIENDDNWDWFLKKLSESLGLC 176
Query: 485 GEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKAT 544
ISDKQ GL++ P EHR C +H+ N+K+ E++ L S++
Sbjct: 177 EMVNLALISDKQSGLVKAIHNVLPQAEHRQCSKHIMDNWKRD-SHDMELQRLFWKISRSY 235
Query: 545 YKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVA 604
E + M +K N AY L + W TI A
Sbjct: 236 TIEEFNTHMANLKSYNPQAYASLQLTSPMTW------------------------TIRQA 271
Query: 605 RDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQG 664
R KPL+ M E IR+ M R A +++ K P+ +++ I S
Sbjct: 272 RRKPLLDMLEDIRRQCMVRTAKRFIIAERLKSRFTPRAHAEIEKMIAGSAGCERHLARNN 331
Query: 665 LFQ--VEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLR 722
L + V G + VD+DK++C C WE+ GIPC H + K E+YV YY +
Sbjct: 332 LHEIYVNDVG----YFVDMDKKTCGCRKWEMVGIPCVHTPCVIIGRKEKVEDYVSDYYTK 387
Query: 723 ATYEICYGHMISPINGENKWPKMSQDDILPPEIKRG-PGRPKKLRRREP--DEPDKKNPT 779
+ Y I P+ G WP+MS+ +LPP +RG PGR R++ + N
Sbjct: 388 VRWRETYRDGIRPVQGMPLWPRMSRLPVLPPPWRRGNPGRQSNYARKKGRYETASSSNKN 447
Query: 780 KLKRGGSSYTCGRCHQKGHNQRKCP-----LPPAKKQPTPS 815
K+ R TC C Q+GHN+ C LPP + + PS
Sbjct: 448 KMSRANRIMTCSNCKQEGHNKSSCKNATVLLPPPRPRGRPS 488
>At3g05850 unknown protein
Length = 777
Score = 207 bits (526), Expect = 3e-53
Identities = 163/589 (27%), Positives = 263/589 (43%), Gaps = 76/589 (12%)
Query: 267 LGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTY 326
+G F + EF++A+ +++ N YKKND +RV CK E C + + + + T
Sbjct: 206 VGQRFKNVGEFREALRKYAIANQFGFRYKKNDSHRVTVKCKAEGCPWRIHASRLSTTQLI 265
Query: 327 RIKTLNPDHTC-GRTFKNRAATAK-WIAK------------------------------- 353
IK +NP HTC G N T++ W+A
Sbjct: 266 CIKKMNPTHTCEGAGGINGLQTSRSWVASIIKEKLKVFPNYKPKDIVSDIKEEYGIQLNY 325
Query: 354 ----NARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFY 409
++IA+E ++G + + + ++M N + A + ++ F F+
Sbjct: 326 FQAWRGKEIAREQLQGSYKDGYKQLPLFCEKIMETNPGSLATFTTKE-DSSFHRVFVSFH 384
Query: 410 MCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKES 469
+ G F ACRP + +D LK+KY G LL A D +D+ FPLAFAVV+AE ++
Sbjct: 385 ASVHG----FLEACRPLVFLDSMQLKSKYQGTLLAATSVDGDDEVFPLAFAVVDAETDDN 440
Query: 470 WRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGG 529
W WFL L + + F++D+QK L + + F H YC+R+L + G
Sbjct: 441 WEWFLLQLRSLL--STPCYITFVADRQKNLQESIPKVFEKSFHAYCLRYLTDELIKDLKG 498
Query: 530 --GTEVRDLLM----WASKATYKELWEEKMQEIKKINEAAYWWLM-AIPTIHWCRHAFSY 582
E++ L++ A+ A + +E ++ IK ++ AY W++ HW +A+
Sbjct: 499 PFSHEIKRLIVDDFYSAAYAPRADSFERHVENIKGLSPEAYDWIVQKSQPDHWA-NAYFR 557
Query: 583 FPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKP 642
+ + ++ E F A D P+ M + IR +M R + + GN+ P
Sbjct: 558 GARYNHMTSHSGEPFFSWASDANDLPITQMVDVIRGKIMGLIHVRRISANEANGNLTPSM 617
Query: 643 RKRLDRE-IKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHA 701
+L++E ++ VA LFQV +E +V++ + C+C W+L G+PC HA
Sbjct: 618 EVKLEKESLRAQTVHVAPSADNNLFQVRGETYE---LVNMAECDCSCKGWQLTGLPCHHA 674
Query: 702 VACFNDKGLKPENYVHQYYLRATYEICYGHMISPI---NGE--NKWPKMSQDDILPPEIK 756
VA N G P +Y +Y+ A Y Y I+P+ GE + S + PP +
Sbjct: 675 VAVINYYGRNPYDYCSKYFTVAYYRSTYAQSINPVPLLEGEMCRESSGGSAVTVTPPPTR 734
Query: 757 RGPGRPKKLRRREPDEPDKKNPTK--LKRGGSSYTCGRCHQKGHNQRKC 803
R PGRP P KK P + +KR C RC GHN+ C
Sbjct: 735 RPPGRP----------PKKKTPAEEVMKR---QLQCSRCKGLGHNKSTC 770
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 206 bits (523), Expect = 6e-53
Identities = 152/598 (25%), Positives = 261/598 (43%), Gaps = 76/598 (12%)
Query: 262 DYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIK 321
++ +G EF E ++ + ++ + K+D++R C +E C + + A
Sbjct: 22 EHVLAVGKEFPDVETCRRTLKDLAIALHFDLRIVKSDRSRFIAKCSKEGCPWRIHAAKCP 81
Query: 322 GTSTYRIKTLNPDHTCG--RTFKNRAATAKWIAKNARKIAKE------------------ 361
G T+ ++TLN +HTC R ++ A+ W+A++ ++
Sbjct: 82 GVQTFTVRTLNSEHTCEGVRDLHHQQASVGWVARSVEARIRDNPQYKPKEILQDIRDEHG 141
Query: 362 -----------------AVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPR 404
A+ G + + + +Y ++ VN + A++ + L P
Sbjct: 142 VAVSYMQAWRGKERSMAALHGTYEEGYRFLPAYCEQIKLVNPGSFASV------SALGPE 195
Query: 405 FGCF---YMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAV 461
CF ++ C S F +CRP + +D HLK KY G +L A D +D FPLA A+
Sbjct: 196 -NCFQRLFIAYRACISGFFSSCRPLLELDRAHLKGKYLGAILCAAAVDADDGLFPLAIAI 254
Query: 462 VEAECKESWRWFLNHLMRDIGDNGEK--KWVFISDKQKGLLQVFAEDFPGIEHRYCVRHL 519
V+ E E+W WFL+ L + +G N + K +S++Q +++ FP H +C+R++
Sbjct: 255 VDNESDENWSWFLSELRKLLGMNTDSMPKLTILSERQSAVVEAVETHFPTAFHGFCLRYV 314
Query: 520 YANFKQKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKIN-EAAYWWLMAIPTIHWCRH 578
NF+ F T++ ++ A A +E K E+ +I+ + W+ + +P + W
Sbjct: 315 SENFRDTF-KNTKLVNIFWSAVYALTPAEFETKSNEMIEISQDVVQWFELYLPHL-W--- 369
Query: 579 AFSYFPKCDVTMNNL--AEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKG 636
A +YF L E L + P+I M E IR + + F RE S W
Sbjct: 370 AVAYFQGVRYGHFGLGITEVLYNWALECHELPIIQMMEHIRHQISSWFDNRRELSMGWNS 429
Query: 637 NVMPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGI 696
++P +R+ + + + + F++ E IVDI + C+C W++ G+
Sbjct: 430 ILVPSAERRITEAVADARCYQVLRANEVEFEI--VSTERTNIVDIRTRDCSCRRWQIYGL 487
Query: 697 PCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQD-------- 748
PC HA A G + + ++Y+ Y MI I + W ++
Sbjct: 488 PCAHAAAALISCGRNVHLFAEPCFTVSSYQQTYSQMIGEIPDRSLWKDEGEEAGGGVESL 547
Query: 749 DILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLP 806
I PP+ +R PGRPKK R + LKR CGRCH GH+Q+KC P
Sbjct: 548 RIRPPKTRRPPGRPKKKVVRVEN---------LKRPKRIVQCGRCHLLGHSQKKCTQP 596
>At3g06940 putative mudrA protein
Length = 749
Score = 202 bits (515), Expect = 5e-52
Identities = 152/579 (26%), Positives = 249/579 (42%), Gaps = 74/579 (12%)
Query: 276 EFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRIKTLNPDH 335
EF+ A+ +SV +G A YKKND +RV CK + C + + + T IK +NP H
Sbjct: 191 EFRDALHKYSVAHGFAYKYKKNDSHRVSVKCKAQGCPWRITASRLSTTQLICIKKMNPRH 250
Query: 336 TCGRTF--KNRAATAKWI-----------------------------------AKNARKI 358
TC R AT W+ A A++I
Sbjct: 251 TCERAVIKPGYRATRGWVRTILKEKLKAFPDYKPKDIAEDIKKEYGIQLNYSQAWRAKEI 310
Query: 359 AKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSA 418
A+E ++G + ++ + ++ N + A + + ++ F FY + G
Sbjct: 311 AREQLQGSYKEAYSQLPLICEKIKETNPGSIATFMTKE-DSSFHRLFISFYASISG---- 365
Query: 419 FKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLM 478
FK RP + +D L +KY G++L+A D D FP+AFA+V++E +E+W WFL L
Sbjct: 366 FKQGSRPLLFLDNAILNSKYQGVMLVATASDAEDGIFPVAFAIVDSETEENWLWFLEQLK 425
Query: 479 RDIGDNGEKKWVFISDKQKGLLQVFAEDF-PGIEHRYCVRHLYANFKQKFGG--GTEVRD 535
+ ++ + F++D Q GL A+ F H YC+ L G E R
Sbjct: 426 TALSES--RIITFVADFQNGLKNAIAQVFEKDAHHAYCLGQLAEKLNVDLKGQFSHEARR 483
Query: 536 LLM----WASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMN 591
++ + AT + ++ IK I+ AY W++ HW F + + +
Sbjct: 484 YMLNDFYSVAYATTPVGYYLALENIKSISPDAYNWVIESEPHHWANALFQ-GERYNKMNS 542
Query: 592 NLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIK 651
N + F + A + P+ M + R +M T + KS++W + P ++L +EI+
Sbjct: 543 NFGKDFYSWVSEAHEFPITQMIDEFRAKMMQSIYTRQVKSREWVTTLTPSNEEKLQKEIE 602
Query: 652 YSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLK 711
+ + LFQV + G IVDI++ C C W L G+PC HA+A
Sbjct: 603 LAQLLQVSSPEGSLFQV-NGGESSVSIVDINQCDCDCKTWRLTGLPCSHAIAVIGCIEKS 661
Query: 712 PENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDD-------ILPPEIKRGPGRPKK 764
P Y Y ++ + Y I P+ + +M DD + PP +R PGRP K
Sbjct: 662 PYEYCSTYLTVESHRLMYAESIQPVPNMD---RMMLDDPPEGLVCVTPPPTRRTPGRP-K 717
Query: 765 LRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKC 803
+++ EP + K+ C +C GHN++ C
Sbjct: 718 IKKVEPLDMMKR----------QLQCSKCKGLGHNKKTC 746
>At1g06740 mudrA-like protein
Length = 726
Score = 202 bits (515), Expect = 5e-52
Identities = 162/675 (24%), Positives = 278/675 (41%), Gaps = 87/675 (12%)
Query: 191 DVNRNSGPNNGVQDGDESDNNVGFIPSEVAAQHNIDMEYESDDLLSDYSCDEEDNGSRVR 250
DVN+N G + D D+S+ + + A H ++ + D L + S E + V
Sbjct: 79 DVNQNDGYRS-FPDNDDSEIDKSTVIEATGAIHKVESQDTDDKLELEMSQSTEFSQKSVM 137
Query: 251 ----YQRFRQEDFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFAC 306
Q + D++ +GMEF+ ++AI ++ + K+DK R C
Sbjct: 138 PSPSSQCWSMSGAGTDHEMVVGMEFSDAYACRRAIKNAAISLRFEMRTIKSDKTRFTAKC 197
Query: 307 KQEECRFIAYCANIKGTSTYRIKTLNPDHTCGRT--FKNRAATAKWIAKNARKIAKE--- 361
+ C + +CA + T+ I+T++ HTCG ++ A+ +W+A + KE
Sbjct: 198 NSKGCPWRIHCAKVSNAPTFTIRTIHGSHTCGGISHLGHQQASVQWVADVVAEKLKENPH 257
Query: 362 -------------------------------------AVEGDALKQFTLITSYAAELMRV 384
+ G +++ L+ Y E+ R
Sbjct: 258 FKPKEILEEIYRVHGISLSYKQAWRGKERIMATLRGSTLRGSFEEEYRLLPQYCDEIRRS 317
Query: 385 NHENTAAIILRYIPTTLQPRFGCF---YMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGI 441
N + A + + P GCF ++ + S F ACRP I +D LK+KY G
Sbjct: 318 NPGSVAVV-------HVNPIDGCFQHLFISFQASISGFLNACRPLIALDSTVLKSKYPGT 370
Query: 442 LLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQ 501
LL+A G D + FPLAFA+V E ++W FL+ L R I D K +S ++ ++
Sbjct: 371 LLLATGFDGDGAVFPLAFAIVNEENDDNWHRFLSEL-RKILDENMPKLTILSSGERPVVD 429
Query: 502 VFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKINE 561
+FP H +C+ +L F+++F V DL A+ ++ K+ +I++I+
Sbjct: 430 GVEANFPAAFHGFCLHYLTERFQREFQSSVLV-DLFWEAAHCLTVLEFKSKINKIEQISP 488
Query: 562 AAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLM 621
A W+ W F +T N + E+ + + P+I E I ++L+
Sbjct: 489 EASLWIQNKSPARWASSYFEGTRFGQLTANVITESLSNWVEDTSGLPIIQTMECIHRHLI 548
Query: 622 TRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGF-----EDQ 676
RE S W ++P K++ I+ S A +++ A F E
Sbjct: 549 NMLKERRETSLHWSNVLVPSAEKQMLAAIEQSR-------AHRVYRANEAEFEVMTCEGN 601
Query: 677 FIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPI 736
+V+I+ SC C W++ G+PC HAV Y + Y Y + PI
Sbjct: 602 VVVNIENCSCLCGRWQVYGLPCSHAVGALLSCEEDVYRYTESCFTVENYRRAYAETLEPI 661
Query: 737 NGENKW-----PKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCG 791
+ + +W + S++ I P+ +G R +++R + D R CG
Sbjct: 662 SDKVQWKENDSERDSENVIKTPKAMKGAPRKRRVRAEDRD-----------RVRRVVHCG 710
Query: 792 RCHQKGHNQRKCPLP 806
RC+Q GH + C P
Sbjct: 711 RCNQTGHFRTTCTAP 725
>At4g18410 MuDR transposable element - like protein
Length = 633
Score = 192 bits (489), Expect = 6e-49
Identities = 151/581 (25%), Positives = 248/581 (41%), Gaps = 95/581 (16%)
Query: 271 FASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQ-------EECRFIAYCANIKGT 323
F S FK+A++ +++ G + + DK+++ F C +C + Y A +
Sbjct: 73 FFSGVAFKEAVIDYALRTGHNLKQYRYDKDKIGFICVGCDGKEGGAKCEWKVYAAILPSD 132
Query: 324 STYRIKTLNPDHTC--------------GRTFKNRAATA---------------KW---I 351
+ ++I+ H+C R F ++ + +W +
Sbjct: 133 NMWKIRKFIDTHSCIPNGECEMFKVPHIARLFVDKIRDSPEFYLPAKIEEIILEQWKISV 192
Query: 352 AKN----ARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGC 407
+N ARK A + +E + QF+ + YAAE++ N ++ + + + F
Sbjct: 193 TRNQCQSARKKALQWIEKEYDDQFSRLRDYAAEILESNKDSHVEVEC-LVSDEGKDMFNR 251
Query: 408 FYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECK 467
FY+C + + +K +CRP IGIDGC LK K
Sbjct: 252 FYVCFDSLRRTWKESCRPLIGIDGCFLKNKL----------------------------- 282
Query: 468 ESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKF 527
L D + + ++ ISD+QKGL++ + P IEHR CV+H+Y N K+ +
Sbjct: 283 ---------LKEDFDLSDGEGFIMISDRQKGLIKAVQLELPKIEHRMCVQHIYGNLKKTY 333
Query: 528 GGGTEVRDLLMWASKATYKEL-WEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKC 586
G T ++ LL W +Y E +++ +++I+ + Y +M W R C
Sbjct: 334 GSKTMIKPLL-WNLAWSYNETEYKQHLEKIRCYDTKVYESVMKTNPRSWVRAFQKIGSFC 392
Query: 587 DVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRL 646
+ NN E+FN ++ AR+K + M E IR+ M R A +S G P K L
Sbjct: 393 EDVDNNSVESFNGSLNKAREKQFVAMLETIRRMAMVRIAKRSVESHTHTGVCTPYVMKFL 452
Query: 647 DREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFN 706
E K + G+++V H G D VD+ +CTC W++ GIPC HA
Sbjct: 453 AGEHKVASTAKVAPGTNGMYEVRHGG--DTHRVDLAAYTCTCIKWQICGIPCEHAYGVIL 510
Query: 707 DKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDDIL---PP---EIKRGPG 760
K L+PE++V Q++ A ++ Y + P G WP+ + + PP E K+
Sbjct: 511 HKKLQPEDFVCQWFRTAMWKKNYTDGLFPQRGPKFWPESNGPRVFAAEPPEGEEDKKMTK 570
Query: 761 RPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQR 801
KK ++ + P KK P KR CG C HN R
Sbjct: 571 AEKKRKKGVNESPTKKQPKAKKR---IMHCGVCGAADHNSR 608
>At2g29230 Mutator-like transposase
Length = 915
Score = 182 bits (463), Expect = 6e-46
Identities = 173/767 (22%), Positives = 313/767 (40%), Gaps = 91/767 (11%)
Query: 110 EVVRNEMQKEDAVLDDEDDVFVEVTEVEVIPMTQEA-PTVAVPEVEASNIDSEDE----- 163
E V M+ + + D V + V P + A P + ++ + D + E
Sbjct: 169 EAVLRWMRSTETEGQNVGDSSTNVQQHPVRPAVEAASPMLTTESMDTATEDGDQEEDSTV 228
Query: 164 ----ICKNLTNDGFDDVDEPFDVQQDGDGRDDVNRNSGPNNGVQDGDESDNNVGFI-PSE 218
+ KNL + E +D +G+ +D N G + + + P
Sbjct: 229 VEADLLKNLGLPSINLATESYDSLSEGEAYNDENDYPGGETIKECNFFGPSATNLVEPLG 288
Query: 219 VAAQHNIDMEYESDDLLSDYSC---------DEEDNGSRVRYQ-RFRQEDFDKDYKFRLG 268
V + + +D+ +++ DE+ RV + R ++ D F +G
Sbjct: 289 VHLDLAVPLHKALEDMCLNFASFNRDAAPTLDEQGEEGRVGMELAIRDVVYEGDELF-VG 347
Query: 269 MEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRI 328
F + ++ + H++ ++ K + C E C + Y ++ + Y+I
Sbjct: 348 RVFKNKQDCNVKLAVHALNRRFHFRRDRSCKKLMTLTCISETCPWRVYIVKLEDSDNYQI 407
Query: 329 KTLNPDHTCG---RTFKNRAATAK------------------------------------ 349
++ N +HTC R+ +RAAT +
Sbjct: 408 RSANLEHTCTVEERSNYHRAATTRVIGSIIQSKYAGNSRGPRAIDLQRILLTDYSVRISY 467
Query: 350 WIAKNARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFY 409
W A +R+IA ++ +G A FTL+ +Y L N + + + RF +
Sbjct: 468 WKAWKSREIAMDSAQGSAANSFTLLPAYLHVLREAN-PGSIVDLKTEVDGKGNHRFKYMF 526
Query: 410 MCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKES 469
+ F R + IDG HLK KYGG LL A G+D N Q FP+AF VV++E ++
Sbjct: 527 LAFAASIQGFSCMKRVIV-IDGAHLKGKYGGCLLTASGQDANFQVFPIAFGVVDSENDDA 585
Query: 470 WRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGG 529
W WF L I D F+SD+ + +P +H C+ HL N +
Sbjct: 586 WEWFFRVLSTAIPDG--DNLTFVSDRHSSIYTGLRRVYPKAKHGACIVHLQRNIATSY-- 641
Query: 530 GTEVRDLLMWASKATYKEL---WEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKC 586
+ + LL S+A + E+ +++ A +L ++ HW R A+ +
Sbjct: 642 --KKKHLLFHVSRAARAFRICEFHTYFNEVIRLDPACARYLESVGFCHWTR-AYFLGKRY 698
Query: 587 DVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRL 646
+V +N+AE+ N + AR+ P+I++ E+IR L++ FA RE ++ + PK R+ +
Sbjct: 699 NVMTSNVAESLNAVLKEARELPIISLLEFIRTTLISWFAMRREAARTEASPLPPKMREVV 758
Query: 647 DREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFN 706
R + S + + + +++ G + V + +++C+C ++L +PC HA+A
Sbjct: 759 HRNFEKSVRFAVHRLDRYDYEIREEG-ASVYHVKLMERTCSCRAFDLLHLPCPHAIAAAV 817
Query: 707 DKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQD----DILPPEIKRGPGRP 762
+G+ + + Y ++ + Y I P+ + + + PP +R GRP
Sbjct: 818 AEGVPIQGLMAPEYSVESWRMSYLGTIKPVPEVGDVFALPEPIASLRLFPPATRRPSGRP 877
Query: 763 KKLR---RREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLP 806
KK R R E P ++ C RC GHN+ C +P
Sbjct: 878 KKKRITSRGEFTCPQRQ----------VTRCSRCTGAGHNRATCKMP 914
>At1g21040 hypothetical protein
Length = 904
Score = 174 bits (440), Expect = 3e-43
Identities = 162/724 (22%), Positives = 298/724 (40%), Gaps = 78/724 (10%)
Query: 110 EVVRNEMQKEDAVLDDEDDVFVEVTEVEVIPMTQEA-PTVAVPEVEASNIDSEDE----- 163
E V M+ + + D V + V P + A P + ++ + D + E
Sbjct: 169 EAVLRWMRSTETEGQNVGDSSTNVQQHPVRPAVEAASPMLTTESMDTATEDGDQEEDSTV 228
Query: 164 ----ICKNLTNDGFDDVDEPFDVQQDGDGRDDVNRNSGPNNGVQDGDESDNNVGFI-PSE 218
+ KNL + E +D +G+ +D N G + + + P
Sbjct: 229 VEADLLKNLGLPSINLATESYDSLSEGEAYNDENDYPGGETIKECNFFGPSATNLVEPLG 288
Query: 219 VAAQHNIDMEYESDDLLSDYSC---------DEEDNGSRVRYQ-RFRQEDFDKDYKFRLG 268
V + + +D+ +++ DE+ RV + R ++ D F +G
Sbjct: 289 VHLDLAVPLHEALEDMCLNFASFNRDAAPTLDEQGEEGRVGMELAIRDVVYEGDELF-VG 347
Query: 269 MEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRI 328
F + ++ + H++ ++ K + C E C + Y ++ + Y+I
Sbjct: 348 RVFKNKQDCNVKLAVHALNRRFHFRRDRSCKKLMTLTCISETCPWRVYIVKLEDSDNYQI 407
Query: 329 KTLNPDHTCG---RTFKNRAATAK------------------------------------ 349
++ N +HTC R+ +RAAT +
Sbjct: 408 RSANLEHTCTVEERSNYHRAATTRVIGSIIQSKYAGNSRGPRAIDLQRILLTDYSVRISY 467
Query: 350 WIAKNARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFY 409
W A +R+IA ++ +G A FTL+ +Y L N + + + RF +
Sbjct: 468 WKAWKSREIAMDSAQGSAANSFTLLPAYLHVLREAN-PGSIVDLKTEVDGKGNHRFKYMF 526
Query: 410 MCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKES 469
+ F R + IDG HLK KYGG LL A G+D N Q FP+AF VV++E ++
Sbjct: 527 LAFAASIQGFSCMKRVIV-IDGAHLKGKYGGCLLTASGQDANFQVFPIAFGVVDSENDDA 585
Query: 470 WRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGG 529
W WF L I D F+SD+ + +P +H C+ HL N +
Sbjct: 586 WEWFFRVLSTAIPDG--DNLTFVSDRHSSIYTGLRRVYPKAKHGACIVHLQRNIATSY-- 641
Query: 530 GTEVRDLLMWASKATYKEL---WEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKC 586
+ + LL S+A + E+ +++ A +L ++ HW R A+ +
Sbjct: 642 --KKKHLLFHVSRAARAFRICEFHTYFNEVIRLDPACARYLESVGFCHWTR-AYFLGERY 698
Query: 587 DVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRL 646
+V +N+AE+ N + AR+ P+I++ E+IR L++ FA RE ++ + PK R+ +
Sbjct: 699 NVMTSNVAESLNAVLKEARELPIISLLEFIRTTLISWFAMRREAARTEASPLPPKMREVV 758
Query: 647 DREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFN 706
R + S + + + +++ G + V + +++C+C ++L +PC HA+A
Sbjct: 759 HRNFEKSVRFAVHRLDRYDYEIREEG-ASVYHVKLMERTCSCRAFDLLHLPCPHAIAAAV 817
Query: 707 DKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQD----DILPPEIKRGPGRP 762
+G+ + + Y ++ + Y I P+ + + + PP +R GRP
Sbjct: 818 AEGVPIQGLMAPEYSVESWRMSYLGTIKPVPEVGDVFALPEPIASLRLFPPATRRPSGRP 877
Query: 763 KKLR 766
KK R
Sbjct: 878 KKKR 881
>At2g15210 putaive transposase of transposon FARE2.7 (cds1)
Length = 580
Score = 171 bits (432), Expect = 2e-42
Identities = 146/563 (25%), Positives = 237/563 (41%), Gaps = 49/563 (8%)
Query: 260 DKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCAN 319
D D F +G F +E + +V + K+D R C E C +
Sbjct: 44 DADSLF-IGKNFKDKDEMVFTLRMFAVKHNFEFHTVKSDLTRYVLHCIDENCSWRLRATR 102
Query: 320 IKGTSTYRIKTLNPDHTCGRTFKN---RAATAKWIAKNARKIAKEAVEGDALKQFTLITS 376
G+ +Y I+ H+C + +N R A+A+ + + + EG L L
Sbjct: 103 AGGSESYVIRKYVSHHSCDSSLRNVSHRQASARTLGR----LISNHFEGGKLP---LRPK 155
Query: 377 YAAELMRVNH------------ENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACR 424
E+ R +H + AA + R +P E +KL R
Sbjct: 156 QLIEIFRKDHGVGINYSKAWRVQEHAAELARGLPDD----------SFEVLPRGYKLM-R 204
Query: 425 PFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDN 484
I IDG HL +K+ G LL A +D N +P+AFA+V++E SW WFL L+ I D
Sbjct: 205 KVISIDGAHLTSKFKGTLLGASAQDRNFNLYPIAFAIVDSENDASWDWFLKCLLNIIPD- 263
Query: 485 GEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKAT 544
E VF+S++ + + ++P H C HL N + F G + + + AS+
Sbjct: 264 -ENDLVFVSERAASIASGLSGNYPLAHHGLCTFHLQKNLETHFRGSSLI-PVYYAASRVY 321
Query: 545 YKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVA 604
K ++ EI ++ +L + W R A+S + ++ +NLAE+ N +
Sbjct: 322 TKTEFDSLFWEITNSDKKLAQYLWEVHVRKWSR-AYSPSNRYNIMTSNLAESVNALLKQN 380
Query: 605 RDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWV-ATWVAQ 663
R+ P++ +FE IR + F RE+S + V K++ S W+ V Q
Sbjct: 381 REYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKMKASYDTSTRWLEVCQVNQ 440
Query: 664 GLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRA 723
F+V+ G +V++DK++CTC ++++ PC H +A N L +V +++
Sbjct: 441 EEFEVK--GDTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASANHINLNENMFVDEFHSTY 498
Query: 724 TYEICYGHMISPINGENKW---PKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTK 780
+ Y I P W +S+ LPP + GR KK R E + P
Sbjct: 499 RWRQAYSESIHPNGDMEYWEIPETISEVICLPPSTRVPSGRRKKKRIPSVWEHGRSQPKP 558
Query: 781 LKRGGSSYTCGRCHQKGHNQRKC 803
+ C RC Q GHN+ C
Sbjct: 559 -----KLHKCSRCGQSGHNKSTC 576
>At3g47280 putative protein
Length = 739
Score = 167 bits (422), Expect = 3e-41
Identities = 145/581 (24%), Positives = 243/581 (40%), Gaps = 49/581 (8%)
Query: 260 DKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCAN 319
D D F +G F +E + +V + K+D R C E C +
Sbjct: 167 DADSLF-IGKIFKDKDEMVFTLRKFAVKHSFKFHTVKSDLTRYVLHCIDENCSWRLRATR 225
Query: 320 IKGTSTYRIKTLNPDHTCGRTFKN---RAATAKWIAK------NARKIA---KEAVE--- 364
G+ +Y I+ H+C + +N R A+A+ + + K+ K+ +E
Sbjct: 226 AGGSESYVIRKYVSHHSCDSSLRNVSHRQASARTLGRLISNHFEGGKLPLRPKQLIEIFR 285
Query: 365 -----GDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAF 419
G + + +AAEL R +++ ++ R+ G + + F
Sbjct: 286 KDHGVGINYSKAWRVQKHAAELARGLPDDSFEVLPRWFHRVQVTNPGSITFFKKDSANKF 345
Query: 420 KLA-------------CRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAEC 466
K A R I IDG HL +K+ G LL A +D N +P+AFA+V++E
Sbjct: 346 KYAFLAFGASIRGYKLMRKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSEN 405
Query: 467 KESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQK 526
SW WFL L+ I D E VF+SD+ + + ++P H C HL N +
Sbjct: 406 DASWDWFLKCLLNIIPD--ENDLVFVSDRAASIASRLSGNYPLAHHGLCTFHLQKNLETH 463
Query: 527 FGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKC 586
F G + + + AS+ K ++ EI ++ +L + W R A+S +
Sbjct: 464 FRGSSLI-PVYYAASRIYTKTEFDSLFWEITNSDKKLAQYLCEVDVRKWSR-AYSPSNRY 521
Query: 587 DVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRL 646
++ +NLAE+ N + R+ P++ +FE IR + F RE+S + V K++
Sbjct: 522 NIMTSNLAESVNALLKQNREYPVVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKM 581
Query: 647 DREIKYSGNWV-ATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACF 705
S W+ V Q F+V+ G +V++DK++CTC ++++ PC H +A
Sbjct: 582 KASYDTSTRWLEVCQVNQEEFEVK--GDTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 639
Query: 706 NDKGLKPENYVHQYYLRATYEICYGHMISPINGENKW---PKMSQDDILPPEIKRGPGRP 762
L +V +++ + Y I P W +S+ LPP + G+
Sbjct: 640 KHINLNENMFVDEFHSTYRWRQAYSESIHPNGDMEYWEIPETISEVICLPPSTRVPTGKR 699
Query: 763 KKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKC 803
KK R E + P + C RC Q GHN+ C
Sbjct: 700 KKKRIPSVWEHGRSQPKP-----KLHKCSRCGQSGHNKSTC 735
>At1g34620 Mutator-like protein
Length = 1034
Score = 165 bits (418), Expect = 1e-40
Identities = 167/699 (23%), Positives = 288/699 (40%), Gaps = 92/699 (13%)
Query: 120 DAVLDDEDDVFVEVTEVEVIPMT--QEAPTVAVPEVEASNIDSEDEICKNLTNDGFDDVD 177
D D +DD+ + +P T P + +PE N D E
Sbjct: 245 DTSTDSDDDLLI-------VPFTLPPNPPAITIPE----NADIEGA-------------- 279
Query: 178 EPFDVQQDGDGRDDVNRNSGPNNGVQDGDESDNNVGFIPSEVAAQHNIDMEYESDDLLSD 237
D + G N S N V + D N+ F A + I ++ D +L
Sbjct: 280 ---DSSRAAVGTPHGNHVSPIGNAVAETDLQRQNILFWGRAQEALNTILSDFSDDPILFG 336
Query: 238 YSCDEEDNGSRVRYQRFRQEDFDKDY---KFRLGMEFASTEEFKKAILAHSVMNGKAVIY 294
N + + FD Y K +G F S + K I H++
Sbjct: 337 RDAPPVFNDGKG--EGVDSAFFDVKYEGDKLFVGRVFKSKSDCKIKIAIHAINRKFHFRT 394
Query: 295 KKNDKNRVRFACKQEECRFIAYCANIKGTSTYRIKTLNPDHTCG---RTFKNRAATAK-- 349
++ + C + C + Y + + + +++++ N HTC R +R AT +
Sbjct: 395 ARSTPKFMVLKCISKTCPWRVYASKVDTSDSFQVRQANQRHTCTIDQRRRYHRLATTQVI 454
Query: 350 ----------------------------------WIAKNARKIAKEAVEGDALKQFTLIT 375
W A AR++A E G + LI
Sbjct: 455 GELMQSRFLGIKRGPNAAVIRKFLLDDYHVSISYWKAWRAREVAMEKSLGSMAGSYALIP 514
Query: 376 SYAAELMRVNHENTAAIILRYIPT---TLQPRFGCFYMCLEGCKSAFKLACRPFIGIDGC 432
+YA L + N + PT + +F F ++G AF R I +DG
Sbjct: 515 AYAGLLQQANPGSLCFTEYDDDPTGPRRFKYQFIAFAASIKGY--AFM---RKVIVVDGT 569
Query: 433 HLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFI 492
+K +YGG L+ A +D N Q FPLAF +V +E ++ WF L DN + +FI
Sbjct: 570 SMKGRYGGCLISACCQDGNFQIFPLAFGIVNSENDSAYEWFFQRLSIIAPDNPD--LMFI 627
Query: 493 SDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKATYKELWEEK 552
SD+ + + ++ + H C HL+ N + + + R L+ A++A + +
Sbjct: 628 SDRHESIYTGLSKVYTQANHAACTVHLWRNIRHLYKPKSLCR-LMSEAAQAFHVTDFNRI 686
Query: 553 MQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVARDKPLITM 612
+I+K+N +L+ + W R S + ++ +N+ E++N I AR+ PLI M
Sbjct: 687 FLKIQKLNPGCAAYLVDLGFSEWTR-VHSKGRRFNIMDSNICESWNNVIREAREYPLICM 745
Query: 613 FEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAG 672
E+IR LM FAT R +++ + P+ ++R++ + + + + FQV+
Sbjct: 746 LEYIRTTLMDWFATRRAQAEDCPTTLAPRVQERVEENYQSAMSMSVKPICNFEFQVQERT 805
Query: 673 FEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHM 732
E FIV +D+ +C+C ++ GIPC HA+A G+ ++ Y ++ Y
Sbjct: 806 GE-CFIVKLDESTCSCLEFQGLGIPCAHAIAAAARIGVPTDSLAANGYFNELVKLSYEEK 864
Query: 733 ISPIN--GENKWPKM---SQDDILPPEIKRGPGRPKKLR 766
I PI+ G P++ + ++ PP ++R PGRP+K+R
Sbjct: 865 IYPIHSVGGEVAPEIAAGTTGELQPPFVRRPPGRPRKIR 903
>At2g06450 putative transposase of FARE2.11
Length = 580
Score = 164 bits (414), Expect = 3e-40
Identities = 147/581 (25%), Positives = 241/581 (41%), Gaps = 54/581 (9%)
Query: 247 SRVRYQRFRQEDF-----DKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNR 301
S + QR+ F D D F +G F +E + +V + K+D R
Sbjct: 26 SSILDQRYSSSAFSTGLSDADSLF-IGKIFKDKDEMVFTLRMFAVKHNFKFHTVKSDLTR 84
Query: 302 VRFACKQEECRFIAYCANIKGTSTYRIKTLNPDHTCGRTFKN---RAATAKWIAKNARKI 358
C E C + G+ +Y I+ H+C + +N R A+A+ + + +
Sbjct: 85 YVLHCIDENCSWRLRATRAGGSESYVIRKYVSHHSCDSSLRNVSHRQASARTLGR----L 140
Query: 359 AKEAVEGDALKQFTLITSYAAELMRVNH------------ENTAAIILRYIPTTLQPRFG 406
EG L L E+ R +H + AA + R +P
Sbjct: 141 ISNHFEGGKLP---LRPKQLIEIFRKDHGVGINYSKAWRVQEHAAELARGLPDD------ 191
Query: 407 CFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAEC 466
E +KL R I IDG HL +K+ G LL A +D N +P+AFA+V++E
Sbjct: 192 ----SFEVLPRGYKLM-RKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSEN 246
Query: 467 KESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQK 526
SW WFL L+ I D E VF+SD+ + + ++P H C HL N +
Sbjct: 247 DASWDWFLKCLLNIIPD--ENDLVFVSDRAAFIASGLSGNYPLAHHGLCTFHLQKNLETH 304
Query: 527 FGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKC 586
F G + + + AS+ K ++ EI ++ +L + W R +S +
Sbjct: 305 FRGSSLI-PVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLWEVDVRKWSR-TYSPSNRY 362
Query: 587 DVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRL 646
++ +NLAE+ N + R+ P++ +FE IR + F RE+S + V K++
Sbjct: 363 NIMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKM 422
Query: 647 DREIKYSGNWV-ATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACF 705
S W+ V Q F+V+ G +V++DK++CTC ++++ PC H +A
Sbjct: 423 KASYDTSTRWLEVCQVNQEEFEVK--GDTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 480
Query: 706 NDKGLKPENYVHQYYLRATYEICYGHMISPINGENKW---PKMSQDDILPPEIKRGPGRP 762
L +V +++ + Y I P W +S+ LPP + GR
Sbjct: 481 KHINLNENMFVDEFHSTYRWRQAYSESIHPNGDMEYWEILETISEVICLPPSTRVPSGRR 540
Query: 763 KKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKC 803
+K R E + P + C RC + GHN+ C
Sbjct: 541 EKKRIPSVWEHGRSQPKP-----KLHKCSRCGRSGHNKSTC 576
>At2g04950 putative transposase of transposon FARE2.9 (cds1)
Length = 616
Score = 163 bits (412), Expect = 5e-40
Identities = 144/581 (24%), Positives = 242/581 (40%), Gaps = 49/581 (8%)
Query: 260 DKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCAN 319
D D F +G F +E + +V + K+D + C E C +
Sbjct: 44 DADSLF-IGKFFKDKDEMVFTLRMSAVKHSFEFHTVKSDLTKYVLHCIDENCSWRLRATR 102
Query: 320 IKGTSTYRIKTLNPDHTCGRTFKN---RAATAKWIAK------NARKIA---KEAVE--- 364
G+ +Y I+ H+C + +N R A+A+ + + K+ K+ +E
Sbjct: 103 AGGSESYVIRKYVSHHSCDSSLRNVSHRQASARTLGRLISNHFEGGKLPLRPKQLIEIFR 162
Query: 365 -----GDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAF 419
G + + +AAEL R +++ ++ R+ G + + F
Sbjct: 163 KDHGVGINYSKAWRVQEHAAELARGLPDDSFEVLPRWFHRVQVTNPGSITFFKKDSANKF 222
Query: 420 KLACRPF-------------IGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAEC 466
K A F I IDG HL +K+ G LL A +D N +P+AFA+V++E
Sbjct: 223 KYAFLAFGASIRGYKLMKKVISIDGAHLISKFKGTLLGASAQDGNFNLYPIAFAIVDSEN 282
Query: 467 KESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQK 526
SW WFL L+ I D E VF+SD+ + + ++P + C HL N +
Sbjct: 283 DASWDWFLKCLLNIIPD--ENDLVFVSDRAASIASGLSGNYPLAHNGLCTFHLQKNLETH 340
Query: 527 FGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKC 586
F G + + + AS+ K ++ EI ++ +L + W R A+S +
Sbjct: 341 FRGSSLI-PVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLWEVDVRKWSR-AYSPSNRY 398
Query: 587 DVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRL 646
++ +NLAE+ N + R+ P++ +FE IR + F RE+S + V K++
Sbjct: 399 NIMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKM 458
Query: 647 DREIKYSGNWV-ATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACF 705
S W+ V Q F+V+ G +V++DK++CTC ++++ PC H +A
Sbjct: 459 KASYDTSTRWLEVCQVNQEEFEVK--GDTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 516
Query: 706 NDKGLKPENYVHQYYLRATYEICYGHMISPINGENKW---PKMSQDDILPPEIKRGPGRP 762
L +V +++ + Y I P W +S+ LPP + GR
Sbjct: 517 KHINLNENMFVDEFHSTYKWRQAYSKSIHPNGDMEYWEIPETISEVICLPPSTRVPSGRR 576
Query: 763 KKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKC 803
KK R E + P + C RC Q GHN C
Sbjct: 577 KKKRIPSVWEHGRSQPKP-----KLHKCSRCGQSGHNNSTC 612
>At2g15150 putative transposase of FARE2.2 (CDS1)
Length = 607
Score = 156 bits (395), Expect = 4e-38
Identities = 136/544 (25%), Positives = 231/544 (42%), Gaps = 44/544 (8%)
Query: 260 DKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCAN 319
D D F +G F +E + +V + K+D R C E C +
Sbjct: 44 DADSLF-IGKIFKDKDEMVFTLRMFAVKHNFEFHTVKSDLTRYVLHCIDENCSWRLRATR 102
Query: 320 IKGTSTYRIKTLNPDHTCGRTFKN---RAATAKWIAK------NARKIA---KEAVE--- 364
G+ +Y I+ H+C + +N R A+A+ + + K+ K+ +E
Sbjct: 103 AGGSESYAIRKYVSHHSCDSSLRNVSHRQASARTLGRLISNHFEGGKLPLRPKQLIEIFR 162
Query: 365 -----GDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAF 419
G + + +AAEL R +++ ++ R+ G + + F
Sbjct: 163 KDHGVGINYSKAWRVQEHAAELARGLPDDSFEVLPRWFHRVQVTNPGSITFFKKDSANKF 222
Query: 420 KLA-------------CRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAEC 466
K A R I IDG HL +K+ G LL A +D N +P+AFA+V++E
Sbjct: 223 KYAFLAFGASIRGYKLMRKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSEN 282
Query: 467 KESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQK 526
SW WFL L+ I D E VF+S++ + + ++P H C HL N +
Sbjct: 283 DASWDWFLKCLLNIIPD--ENDLVFVSERAASIASGLSGNYPLAHHGLCTFHLQKNLETH 340
Query: 527 FGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKC 586
F G + + + AS+ K ++ EI ++ +L + W R A+S +
Sbjct: 341 FRGSSLI-PVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLWEVHVRKWSR-AYSPSNRY 398
Query: 587 DVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRL 646
++ +NLAE+ N + R+ P++ +FE IR + F RE+S + V K++
Sbjct: 399 NIMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKM 458
Query: 647 DREIKYSGNWV-ATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACF 705
S W+ V Q F+V+ G +V++DK++CTC ++++ PC H +A
Sbjct: 459 KASYDTSTRWLEVCQVNQEEFEVK--GDTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 516
Query: 706 NDKGLKPENYVHQYYLRATYEICYGHMISPINGENKW---PKMSQDDILPPEIKRGPGRP 762
N L +V +++ + Y I P W +S+ LPP + GR
Sbjct: 517 NHINLNENMFVDEFHSTYRWRQAYSESIHPNGDMEYWEIPETISEVICLPPSTRVPSGRR 576
Query: 763 KKLR 766
KK R
Sbjct: 577 KKKR 580
>At2g12720 putative protein
Length = 819
Score = 156 bits (395), Expect = 4e-38
Identities = 121/450 (26%), Positives = 193/450 (42%), Gaps = 21/450 (4%)
Query: 357 KIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCK 416
+ A+E G F + SY + R N A + + RF ++
Sbjct: 386 RFARELTLGTPDSSFEELPSYLYMIRRANPGTVARLQIDE-----SGRFNYMFIVFGASI 440
Query: 417 SAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNH 476
+ F R + +DG L Y G LL A+ +D N Q FPLAF VV+ E +SWRW
Sbjct: 441 AGFHYMRRVVV-VDGTFLHGSYKGTLLTALAQDGNFQIFPLAFGVVDTENDDSWRWLFTQ 499
Query: 477 LMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDL 536
L I D + ISD+ K + + E +P C HLY N KF ++ L
Sbjct: 500 LKVVIPDATD--LAIISDRHKSIGKAIGEVYPLAARGICTYHLYKNILVKF-KRKDLFPL 556
Query: 537 LMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFP--KCDVTMNNLA 594
+ A++ + EI++++ + +L W R ++FP + ++ N+A
Sbjct: 557 VKKAARCYRLNDFTNAFNEIEELDPLLHAYLQRAGVEMWAR---AHFPGDRYNLMTTNIA 613
Query: 595 EAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIKYSG 654
E+ N + A++ P++ M E IR+ + FA R+ + K + P K L + S
Sbjct: 614 ESMNRALSQAKNLPIVRMLEAIRQMMTRWFAERRDDASKQHTQLTPGVEKLLQTRVTSSR 673
Query: 655 NWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPEN 714
+ QV + +V++D++ CTC + +PC HA+A G+ +
Sbjct: 674 LLDVQTIDASRVQVAYEA--SLHVVNVDEKQCTCRLFNKEKLPCIHAIAAAEHMGVSRIS 731
Query: 715 YVHQYYLRATYEICYGHMISPINGENKWPKMSQDD-ILPPEIKRGPGRPKKLRRREPDEP 773
YY + Y I P + E P++ D LPP + PGRPKKLR + E
Sbjct: 732 LCSPYYKSSHLVNVYACAIMPSDTEVPVPQIVIDQPCLPPIVANQPGRPKKLRMKSALEV 791
Query: 774 DKKNPTKLKRGGSSYTCGRCHQKGHNQRKC 803
+ KR + C RC + GHN + C
Sbjct: 792 ----AVETKRPRKEHACSRCKETGHNVKTC 817
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,439,197
Number of Sequences: 26719
Number of extensions: 1172521
Number of successful extensions: 16888
Number of sequences better than 10.0: 553
Number of HSP's better than 10.0 without gapping: 278
Number of HSP's successfully gapped in prelim test: 292
Number of HSP's that attempted gapping in prelim test: 5930
Number of HSP's gapped (non-prelim): 3004
length of query: 931
length of database: 11,318,596
effective HSP length: 109
effective length of query: 822
effective length of database: 8,406,225
effective search space: 6909916950
effective search space used: 6909916950
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0016.18


