

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
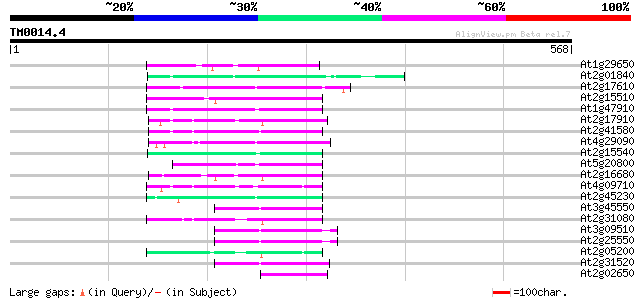
Query= TM0014.4
(568 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g29650 reverse transcriptase, putative 79 5e-15
At2g01840 putative non-LTR retroelement reverse transcriptase 77 2e-14
At2g17610 putative non-LTR retroelement reverse transcriptase 76 6e-14
At2g15510 putative non-LTR retroelement reverse transcriptase 74 2e-13
At1g47910 reverse transcriptase, putative 73 5e-13
At2g17910 putative non-LTR retroelement reverse transcriptase 72 8e-13
At2g41580 putative non-LTR retroelement reverse transcriptase 67 3e-11
At4g29090 putative protein 64 2e-10
At2g15540 putative non-LTR retroelement reverse transcriptase 62 7e-10
At5g20800 putative protein 60 3e-09
At2g16680 putative non-LTR retroelement reverse transcriptase 59 6e-09
At4g09710 RNA-directed DNA polymerase -like protein 58 1e-08
At2g45230 putative non-LTR retroelement reverse transcriptase 57 2e-08
At3g45550 putative protein 57 3e-08
At2g31080 putative non-LTR retroelement reverse transcriptase 57 4e-08
At3g09510 putative non-LTR reverse transcriptase 55 1e-07
At2g25550 putative non-LTR retroelement reverse transcriptase 55 1e-07
At2g05200 putative non-LTR retroelement reverse transcriptase 53 4e-07
At2g31520 putative non-LTR retroelement reverse transcriptase 53 5e-07
At2g02650 putative reverse transcriptase 52 9e-07
>At1g29650 reverse transcriptase, putative
Length = 1557
Score = 79.3 bits (194), Expect = 5e-15
Identities = 52/182 (28%), Positives = 91/182 (49%), Gaps = 15/182 (8%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+ +W DNW+ + P Q+ +RV DL + WN L++ FF ++ +
Sbjct: 1206 ISVWMDNWIYDNGPRIPLQKHFSVSLNLRVKDLINLDGRCWNRDLLQDLFF-----QVDI 1260
Query: 199 TLIRHQ----DVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQ---W 251
LI + D+ D W+ + Y+VK+GY W + N P + +P+
Sbjct: 1261 DLICKKKPVVDMDDFWVWLHSKSGEYSVKSGY-W--LVFQSNKPELLFKACRQPSTNGLK 1317
Query: 252 EFFWKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNV 311
E W +FP+++ WR+ + A+PV ++ RRG++VDP +CG E E + H+ +C+V
Sbjct: 1318 EKIWSTSTFPKIKLFLWRILSAALPVADQILRRGMNVDPCCQICGQEGESINHVLFTCSV 1377
Query: 312 SR 313
+R
Sbjct: 1378 AR 1379
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 77.4 bits (189), Expect = 2e-14
Identities = 62/262 (23%), Positives = 105/262 (39%), Gaps = 24/262 (9%)
Query: 140 RIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILVT 199
+IWED WLP P + DE ++VADL+ N W+ + E P
Sbjct: 1277 KIWEDPWLPTLPPRPARGPILDED--MKVADLWRENKREWDPVIFEGVLNPEDQQLAKSL 1334
Query: 200 LIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWKAPS 259
+ + D W T + YTV++GY W N N + + + W+
Sbjct: 1335 YLSNYAARDSYKWAYTRNTQYTVRSGY-WVATHVNLTEEEIINPLEGDVPLKQEIWRLKI 1393
Query: 260 FPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCWFAR 319
P+++ WR + A+ +L R + DP+ C DE + H+ +C+ ++ W +
Sbjct: 1394 TPKIKHFIWRCLSGALSTTTQLRNRNIPADPTCQRCCNADETINHIIFTCSYAQVVWRSA 1453
Query: 320 NLGLRLLSGSVLRLCSVIDSAEVLNQ*VNGRTNKSLGGSCLPCGRLRIN*LFRRKLGLWL 379
N SGS RLC + E + + G+ N++L + + W+
Sbjct: 1454 N-----FSGS-NRLCFTDNLEENIRLILQGKKNQNLP-------------ILNGLMPFWI 1494
Query: 380 --RFWHGQQRLFAPRLCRSPWR 399
R W + +L R PW+
Sbjct: 1495 MWRLWKSRNEYLFQQLDRFPWK 1516
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 75.9 bits (185), Expect = 6e-14
Identities = 60/212 (28%), Positives = 90/212 (42%), Gaps = 10/212 (4%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+++W+DNWLP P ++V+DL + G WN L+ + I
Sbjct: 387 IQLWKDNWLPLNPPRPPVGTCDSIYSQLKVSDLLI--EGRWNEDLLCKLIHQNDIPHIRA 444
Query: 199 TLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPS--SSNEVGLEPAQWEFFWK 256
D + W+ T D Y+VK+GY +L Q S S NEV + + WK
Sbjct: 445 IRPSITGANDAITWIYTHDGNYSVKSGYHLLRKLSQQQHASLPSPNEVSAQTV-FTNIWK 503
Query: 257 APSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
+ P+++ WR A+P L RR + D + CG E V HL C VS+ W
Sbjct: 504 QNAPPKIKHFWWRSAHNALPTAGNLKRRRLITDDTCQRCGEASEDVNHLLFQCRVSKEIW 563
Query: 317 FARNLGLRLLSGSVLRLCSV---IDSAEVLNQ 345
++L G L S ++S + LNQ
Sbjct: 564 --EQAHIKLCPGDSLMSNSFNQNLESIQKLNQ 593
>At2g15510 putative non-LTR retroelement reverse transcriptase
Length = 1138
Score = 73.9 bits (180), Expect = 2e-13
Identities = 49/181 (27%), Positives = 81/181 (44%), Gaps = 7/181 (3%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+ +W D W+ + +P Q ++V DL W L+E F+PT V I
Sbjct: 708 LNVWMDPWIFDIAPRLPLQRHFSVNLDLKVNDLINFEDRCWKRDLLEELFYPTDVELIT- 766
Query: 199 TLIRHQDVV---DCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFW 255
+ VV D W+ Y+VK+GY Q + +N + E W
Sbjct: 767 ---KRNPVVNMDDFWVWLHAKTGEYSVKSGYWLAFQSNKLDLIQQANLLPSTNGLKEQVW 823
Query: 256 KAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGC 315
S P+++ WR+ + A+PV ++ RRG+ +DP +CG E E H+ +C+++R
Sbjct: 824 STKSSPKIKMFLWRILSAALPVADQIIRRGMSIDPRCQICGDEGESTNHVLFTCSMARQV 883
Query: 316 W 316
W
Sbjct: 884 W 884
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 72.8 bits (177), Expect = 5e-13
Identities = 48/180 (26%), Positives = 76/180 (41%), Gaps = 7/180 (3%)
Query: 139 VRIWEDNWLPNG--SPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARI 196
+ +W D W+P P K+ + D ++V L WN L++ F P V I
Sbjct: 716 ISVWNDPWIPAQFPRPAKYGGSIVDPS--LKVKSLIDSRSNFWNIDLLKELFDPEDVPLI 773
Query: 197 LVTLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWK 256
I + ++ D L W T YTVK+GY N+ ++ A + WK
Sbjct: 774 SALPIGNPNMEDTLGWHFTKAGNYTVKSGYHTARLDLNEGTTLIGPDLTTLKA---YIWK 830
Query: 257 APSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
P++R W++ + VPV L +RG+ D CG +E + H C+ +R W
Sbjct: 831 VQCPPKLRHFLWQILSGCVPVSENLRKRGILCDKGCVSCGASEESINHTLFQCHPARQIW 890
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 72.0 bits (175), Expect = 8e-13
Identities = 48/187 (25%), Positives = 84/187 (44%), Gaps = 13/187 (6%)
Query: 141 IWEDNWLPNGS---PIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARIL 197
+W D WL +GS P+ + + + ++V+ L P WN ++ FP I+
Sbjct: 912 VWTDKWLHDGSNRRPLNRRRFINVD---LKVSQLIDPTSRNWNLNMLR-DLFPWKDVEII 967
Query: 198 VTLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFF--- 254
+ D W+ + + +Y+VKTGYE+ L Q E ++P+ F
Sbjct: 968 LKQRPLFFKEDSFCWLHSHNGLYSVKTGYEF---LSKQVHHRLYQEAKVKPSVNSLFDKI 1024
Query: 255 WKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRG 314
W + P++R W+ A+PV +L RG+ D +C E+E + H+ C ++R
Sbjct: 1025 WNLHTAPKIRIFLWKALHGAIPVEDRLRTRGIRSDDGCLMCDTENETINHILFECPLARQ 1084
Query: 315 CWFARNL 321
W +L
Sbjct: 1085 VWAITHL 1091
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 66.6 bits (161), Expect = 3e-11
Identities = 47/179 (26%), Positives = 80/179 (44%), Gaps = 7/179 (3%)
Query: 141 IWEDNWLPNGSPIKFP--QEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+W DNW+ + P + Q + D Q ++V+ L P WN ++ FP +I+
Sbjct: 663 VWMDNWIFDDKPRRPESLQIMVDIQ--LKVSQLIDPFSRNWNLNMLR-DLFPWKEIQIIC 719
Query: 199 TLIRHQDVVDCLFWMGTVDDVYTVKTGYEW-H*QLENQNCPSSSNEVGLEPAQWEFFWKA 257
D W GT +YTVK+ Y+ Q+ Q + + L P + W
Sbjct: 720 QQRPMASRQDSFCWFGTNHGLYTVKSEYDLCSRQVHKQMFKEAEEQPSLNPLFGKI-WNL 778
Query: 258 PSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
S P+++ W+V AV V +L RGV ++ +C ++E + H+ C ++R W
Sbjct: 779 NSAPKIKVFLWKVLKGAVAVEDRLRTRGVLIEDGCSMCPEKNETLNHILFQCPLARQVW 837
>At4g29090 putative protein
Length = 575
Score = 63.9 bits (154), Expect = 2e-10
Identities = 51/195 (26%), Positives = 89/195 (45%), Gaps = 13/195 (6%)
Query: 141 IWEDNWL---PNGSPIKF----PQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMV 193
IW WL P + ++ PQE +++V+DL + W ++E FP V
Sbjct: 137 IWRHKWLDSKPASAALRMQRVPPQEYASVSSILKVSDLIDESGREWRKDVIE-MLFPE-V 194
Query: 194 ARILVTLIRH--QDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQ-NCPSSSNEVGLEPAQ 250
R L+ +R + ++D W T YTVK+GY Q+ N+ + P +E L P
Sbjct: 195 ERKLIGELRPGGRRILDSYTWDYTSSGDYTVKSGYWVLTQIINKRSSPQEVSEPSLNPI- 253
Query: 251 WEFFWKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCN 310
++ WK+ + P+++ W+ + ++PV L R + + + C E V HL C
Sbjct: 254 YQKIWKSQTSPKIQHFLWKCLSNSLPVAGALAYRHLSKESACIRCPSCKETVNHLLFKCT 313
Query: 311 VSRGCWFARNLGLRL 325
+R W ++ + L
Sbjct: 314 FARLTWAISSIPIPL 328
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 62.4 bits (150), Expect = 7e-10
Identities = 46/179 (25%), Positives = 67/179 (36%), Gaps = 5/179 (2%)
Query: 140 RIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILVT 199
R+WED W+P+ P + + V DL P W ++ P+ + IL
Sbjct: 813 RVWEDPWIPSFPPRPAKSILNIRDTHLYVNDLIDPVTKQWKLGRLQELVDPSDIPLILGI 872
Query: 200 LIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNC--PSSSNEVGLEPAQWEFFWKA 257
D W T YTVK+GY L C P V AQ WK
Sbjct: 873 RPSRTYKSDDFSWSFTKSGNYTVKSGYWAARDLSRPTCDLPFQGPSVSALQAQ---VWKI 929
Query: 258 PSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
+ + + W+ + + +L R + + P CG E+E + HL C SR W
Sbjct: 930 KTTRKFKHFEWQCLSGCLATNQRLFSRHIGTEKVCPRCGAEEESINHLLFLCPPSRQIW 988
>At5g20800 putative protein
Length = 359
Score = 60.1 bits (144), Expect = 3e-09
Identities = 39/153 (25%), Positives = 66/153 (42%), Gaps = 6/153 (3%)
Query: 166 VRVADLFLPNHGGWNTPLVE*TFFPTMVARILVTLIRHQDVVDCLFWMGTVDDVYTVKTG 225
++V++L W ++ F P + I + + D L W T YT K+G
Sbjct: 41 LKVSNLIDRRSNFWKMDILHEIFDPNDIPLISALHVGAPNKEDTLGWHFTKSGKYTFKSG 100
Query: 226 YEWH*QLE--NQNCPSSSNEVGLEPAQWEFFWKAPSFPRVREVAWRVCTWAVPVRVKLHR 283
Y ++E + N E+ L A + WK P++R W++ VPVR L +
Sbjct: 101 YHTS-RIEYIDTNLSFIGPEITLLKA---YSWKVQCPPKLRHFLWQILASCVPVRENLRK 156
Query: 284 RGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
RG++ D CG ++ + H C+ +R W
Sbjct: 157 RGINCDIGCARCGATEKTINHTLFQCHPARQIW 189
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 59.3 bits (142), Expect = 6e-09
Identities = 47/184 (25%), Positives = 75/184 (40%), Gaps = 17/184 (9%)
Query: 141 IWEDNWLPNGSPIKFPQEVGDEQRL-VRVADLFLPNHGGWNTPLVE*TFFPTMVARILVT 199
+W D WL +G + P + + ++V+ L P WN + F V
Sbjct: 886 VWIDKWLFDGHSRR-PMNLHSLMNIHMKVSHLIDPLTRNWNLKKLTELFHEKDVQ----- 939
Query: 200 LIRHQDVV----DCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFF- 254
LI HQ + D W GT + +YTVK+GYE + + + E + P+ F
Sbjct: 940 LIMHQRPLISSEDSYCWAGTNNGLYTVKSGYE---RSSRETFKNLFKEADVYPSVNPLFD 996
Query: 255 --WKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVS 312
W + P+++ W+ A+ V +L RG+ C E E + HL C +
Sbjct: 997 KVWSLETVPKIKVFMWKALKGALAVEDRLRSRGIRTADGCLFCKEEIETINHLLFQCPFA 1056
Query: 313 RGCW 316
R W
Sbjct: 1057 RQVW 1060
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 58.2 bits (139), Expect = 1e-08
Identities = 45/181 (24%), Positives = 74/181 (40%), Gaps = 15/181 (8%)
Query: 139 VRIWEDNWLPNGSP---IKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVAR 195
+ +W + WL SP I P E + + V DL + WN + P +
Sbjct: 867 INVWTEAWLSPSSPQTPIGPPTETNKD---LSVHDLICHDVKSWNVEAIR-KHLPQYEDQ 922
Query: 196 ILVTLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFW 255
I I + D L W+ YT KTGY + + P+S + + + W
Sbjct: 923 IRKITINALPLQDSLVWLPVKSGEYTTKTGYAL---AKLNSFPASQLDFNWQ----KNIW 975
Query: 256 KAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGC 315
K + P+V+ W+ A+PV L RR ++ + + CG + E HL + C ++
Sbjct: 976 KIHTSPKVKHFLWKAMKGALPVGEALSRRNIEAEVTCKRCG-QTESSLHLMLLCPYAKKV 1034
Query: 316 W 316
W
Sbjct: 1035 W 1035
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 57.4 bits (137), Expect = 2e-08
Identities = 49/187 (26%), Positives = 72/187 (38%), Gaps = 11/187 (5%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVA--------DLFLPNHGGWNTPLVE*TFFP 190
+ +W D W+ P K Q V + + A DL LP+ WN LV F
Sbjct: 932 INVWTDPWI-GAKPAKAAQAVKRSHLVSQYAANSIHVVKDLLLPDGRDWNWNLVSLLFPD 990
Query: 191 TMVARILVTLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQ-NCPSSSNEVGLEPA 249
IL ++ D W + Y+VK+GY ++ NQ N P + L+P
Sbjct: 991 NTQENILALRPGGKETRDRFTWEYSRSGHYSVKSGYWVMTEIINQRNNPQEVLQPSLDPI 1050
Query: 250 QWEFFWKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSC 309
++ WK P++ WR + V L R + + S C E V HL C
Sbjct: 1051 -FQQIWKLDVPPKIHHFLWRCVNNCLSVASNLAYRHLAREKSCVRCPSHGETVNHLLFKC 1109
Query: 310 NVSRGCW 316
+R W
Sbjct: 1110 PFARLTW 1116
>At3g45550 putative protein
Length = 851
Score = 57.0 bits (136), Expect = 3e-08
Identities = 29/109 (26%), Positives = 49/109 (44%), Gaps = 1/109 (0%)
Query: 208 DCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWKAPSFPRVREVA 267
D L W YTV++GY + P+ + G + + W P P+++
Sbjct: 622 DRLIWSYNSTGDYTVRSGYWLSTHDPSNTIPTMAKPHGSVDLKTKI-WNLPIMPKLKHFL 680
Query: 268 WRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
WR+ + A+P +L RG+ +DP P C E+E + H +C + W
Sbjct: 681 WRILSKALPTTDRLTTRGMRIDPGCPRCRRENESINHALFTCPFATMAW 729
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 56.6 bits (135), Expect = 4e-08
Identities = 46/184 (25%), Positives = 76/184 (41%), Gaps = 19/184 (10%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVA-DLFLPNHGGWNTPLVE*TFFPTMVARIL 197
+R W D WL ++ ++ E ++VA D +LP GWN ++ + P V R L
Sbjct: 798 IRFWLDRWLLQEPLVELGTDMIPEGERIKVAADYWLPG-SGWNLEILG-LYLPETVKRRL 855
Query: 198 VTLIRHQDVV--DCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFF- 254
++++ + D + W GT D +TV++ Y +VG P FF
Sbjct: 856 LSVVVQVFLGNGDEISWKGTQDGAFTVRSAYSL-----------LQGDVGDRPNMGSFFN 904
Query: 255 --WKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVS 312
WK + RVR W V + V+ RR + + +C +E + H+ C
Sbjct: 905 RIWKLITPERVRVFIWLVSQNVIMTNVERVRRHLSENAICSVCNGAEETILHVLRDCPAM 964
Query: 313 RGCW 316
W
Sbjct: 965 EPIW 968
>At3g09510 putative non-LTR reverse transcriptase
Length = 484
Score = 54.7 bits (130), Expect = 1e-07
Identities = 33/125 (26%), Positives = 54/125 (42%), Gaps = 8/125 (6%)
Query: 208 DCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWKAPSFPRVREVA 267
D + W YTV++GY + N P+ + G + W P P+++
Sbjct: 117 DKIIWNYNTTGEYTVRSGYWLLTHDPSTNIPAINPPHGSIDLKTRI-WNLPIMPKLKHFL 175
Query: 268 WRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCWFARNLGLRLLS 327
WR + A+ +L RG+ +DPS P C E+E + H +C + W RL
Sbjct: 176 WRALSQALATTERLTTRGMRIDPSCPRCHRENESINHALFTCPFATMAW-------RLSD 228
Query: 328 GSVLR 332
S++R
Sbjct: 229 SSLIR 233
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 54.7 bits (130), Expect = 1e-07
Identities = 33/125 (26%), Positives = 54/125 (42%), Gaps = 8/125 (6%)
Query: 208 DCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWKAPSFPRVREVA 267
D + W YTV++GY + N P+ + G + W P P+++
Sbjct: 1383 DKIIWNYNTTGEYTVRSGYWLLTHDPSTNIPAINPPHGSIDLKTRI-WNLPIMPKLKHFL 1441
Query: 268 WRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCWFARNLGLRLLS 327
WR + A+ +L RG+ +DPS P C E+E + H +C + W RL
Sbjct: 1442 WRALSQALATTERLTTRGMRIDPSCPRCHRENESINHALFTCPFATMAW-------RLSD 1494
Query: 328 GSVLR 332
S++R
Sbjct: 1495 SSLIR 1499
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 53.1 bits (126), Expect = 4e-07
Identities = 44/183 (24%), Positives = 73/183 (39%), Gaps = 19/183 (10%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPN--HGGWNTPLVE*TFFPTMVARI 196
V IW D WL P+ E + +RV+ L N WN V + ++ ++
Sbjct: 827 VSIWNDPWLSISKPLVPIGPALREHQDLRVSALINQNTLQWDWNKIAVILPNYENLIKQL 886
Query: 197 LVTLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEF--- 253
R VD L W+ YT ++GY +S + + Q+ +
Sbjct: 887 PAPSSRG---VDKLAWLPVKSGQYTSRSGYGI----------ASVASIPIPQTQFNWQSN 933
Query: 254 FWKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSR 313
WK + P+++ + W+ A+PV ++L RR + + CG E HLF C +
Sbjct: 934 LWKLQTLPKIKHLMWKAAMEALPVGIQLVRRHISPSAACHRCG-APESTTHLFFHCEFAA 992
Query: 314 GCW 316
W
Sbjct: 993 QVW 995
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 52.8 bits (125), Expect = 5e-07
Identities = 29/116 (25%), Positives = 50/116 (43%), Gaps = 1/116 (0%)
Query: 208 DCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWKAPSFPRVREVA 267
D + W YTV++GY + N P+ + G + W P P+++
Sbjct: 1157 DKIIWNYNTTGEYTVRSGYWLLTHDPSTNIPAINPPHGSIDLKTRI-WNLPIMPKLKHFL 1215
Query: 268 WRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCWFARNLGL 323
WR + A+ +L RG+ +DP P C E+E + H +C + W+ + L
Sbjct: 1216 WRALSQALATTERLTTRGMRIDPICPRCHRENESINHALFTCPFATMAWWLSDSSL 1271
>At2g02650 putative reverse transcriptase
Length = 365
Score = 52.0 bits (123), Expect = 9e-07
Identities = 19/67 (28%), Positives = 34/67 (50%)
Query: 255 WKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRG 314
WK P+++ WR T A+ +L R +D DP C +E+E + H+ +C ++
Sbjct: 38 WKLHVAPKIKHFLWRCVTGALATNTRLRSRNIDADPICQRCCIEEETIHHIMFNCPYTQS 97
Query: 315 CWFARNL 321
W + N+
Sbjct: 98 VWRSANI 104
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.345 0.155 0.589
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,111,773
Number of Sequences: 26719
Number of extensions: 545456
Number of successful extensions: 1705
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 1612
Number of HSP's gapped (non-prelim): 69
length of query: 568
length of database: 11,318,596
effective HSP length: 105
effective length of query: 463
effective length of database: 8,513,101
effective search space: 3941565763
effective search space used: 3941565763
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0014.4


