

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0013.6
(937 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
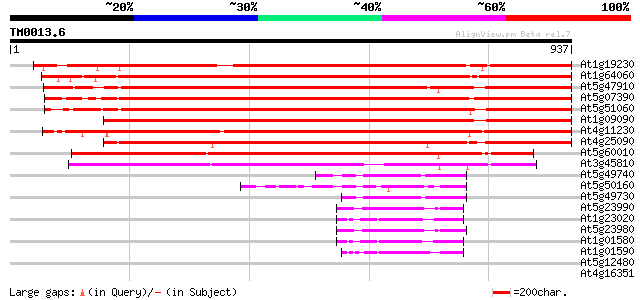
Score E
Sequences producing significant alignments: (bits) Value
At1g19230 hypothetical protein 1137 0.0
At1g64060 cytochrome b245 beta chain homolog RbohAp108 1011 0.0
At5g47910 respiratory burst oxidase protein 872 0.0
At5g07390 respiratory burst oxidase protein A 850 0.0
At5g51060 respiratory burst oxidase protein 843 0.0
At1g09090 respiratory burst oxidase protein B like 842 0.0
At4g11230 respiratory burst oxidase homolog F - like protein 831 0.0
At4g25090 respiratory burst oxidase - like protein 800 0.0
At5g60010 respiratory burst oxidase protein - like 691 0.0
At3g45810 respiratory burst oxidase - like protein 664 0.0
At5g49740 FRO1-like protein; NADPH oxidase-like 103 5e-22
At5g50160 FRO1 and FRO2-like protein 101 2e-21
At5g49730 FRO2-like protein; NADPH oxidase-like 99 9e-21
At5g23990 FRO2 homolog 99 1e-20
At1g23020 putative superoxide-generating NADPH oxidase flavocyto... 96 1e-19
At5g23980 FRO2 homolog 95 2e-19
At1g01580 hypothetical protein 92 1e-18
At1g01590 hypothetical protein 88 2e-17
At5g12480 calcium-dependent protein kinase - like protein 37 0.040
At4g16351 SOS3-like calcium binding protein 37 0.068
>At1g19230 hypothetical protein
Length = 926
Score = 1137 bits (2941), Expect = 0.0
Identities = 592/939 (63%), Positives = 706/939 (75%), Gaps = 89/939 (9%)
Query: 41 SMLPVFLNDLRTNH---------HKELVEITLELE--DDAVVLCNVAPASSGAGTSSSSS 89
+MLPVFLNDL N +ELVE+TLEL+ DD++++C ++ A+S
Sbjct: 35 AMLPVFLNDLSRNSGESGSGSSWERELVEVTLELDVGDDSILVCGMSEAAS--------- 85
Query: 90 HATGDEGTSASGAGAVARSLSKNLSITS-RIRRKF-PWLRSMSMRTSASSTESVEDP--- 144
+ A V LS+NLS S RIR+K LRS S +T+ SST D
Sbjct: 86 -----VDSRARSVDLVTARLSRNLSNASTRIRQKLGKLLRSESWKTTTSSTAGERDRDLE 140
Query: 145 --------VESARNARRMQAKLERTRSSAQRALKGLRFISKSGEAG----------EELW 186
+ +AR+ R+ AKL+R+ SSAQRALKGL+FI+K+ +++W
Sbjct: 141 RQTAVTLGILTARDKRKEDAKLQRSTSSAQRALKGLQFINKTTRGNSCVCDWDCDCDQMW 200
Query: 187 RKVEERFGSLAKDGLLAREDFGGCIGMDDSKEFAVGVFDALARRKGRKASKITKEELHHF 246
+KVE+RF SL+K+GLLAR+DFG C+GM DSK+FAV VFDALARR+ +K KITK+ELH F
Sbjct: 201 KKVEKRFESLSKNGLLARDDFGECVGMVDSKDFAVSVFDALARRRRQKLEKITKDELHDF 260
Query: 247 WQQISDQSFDARLQIFFDMADSNEDGRITREEVQELIMLSASANKLSKLKEQAEGYAALI 306
W QISDQSFDARLQIFFDMADSNEDG+ITREE++EL+MLSASANKL+KLKEQAE YA+LI
Sbjct: 261 WLQISDQSFDARLQIFFDMADSNEDGKITREEIKELLMLSASANKLAKLKEQAEEYASLI 320
Query: 307 MEELDPENLGYIELWQLEMLLLQKDRYMNYSRQLSTSSVSWSQNMTGLKHKNEIQRLCRT 366
MEELDPEN GYIELWQLE LLLQ+D YMNYSR LST+S
Sbjct: 321 MEELDPENFGYIELWQLETLLLQRDAYMNYSRPLSTTSGG-------------------- 360
Query: 367 LQCLALEYWRRGWVLFLWLVTTASLFAWKFYQYRNRTSFQVMGYCLPVAKGAAETLKLNM 426
+ W+R WVL +W++ A LF WKF +YR + +F+VMGYCL AKGAAETLKLNM
Sbjct: 361 -----VNNWQRSWVLLVWVMLMAILFVWKFLEYREKAAFKVMGYCLTTAKGAAETLKLNM 415
Query: 427 ALILLPVCRNTLTWLRSTRARKIVPFDDNINFHKIIACAIVVGVVVHAGNHLACDFPLLI 486
AL+LLPVCRNTLTWLRSTRAR VPFDDNINFHKIIACAI +G++VHAG HLACDFP +I
Sbjct: 416 ALVLLPVCRNTLTWLRSTRARACVPFDDNINFHKIIACAIAIGILVHAGTHLACDFPRII 475
Query: 487 NSSPEEFSLIASDFNNKKPTYKTLLTGVEGVTGISMVTLMAISFTLATHHFRRNAVRLPP 546
NSSPE+F LIAS FN KPT+K L+TG EG+TGISMV L I+FTLA+ HFRRN VRLP
Sbjct: 476 NSSPEQFVLIASAFNGTKPTFKDLMTGAEGITGISMVILTTIAFTLASTHFRRNRVRLPA 535
Query: 547 PLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERTL 606
PL+RLTGFNAFWY+HHLL VVYI+L +HG+FL +WY KTTWMYISVPL+LY+AER+L
Sbjct: 536 PLDRLTGFNAFWYTHHLLVVVYIMLIVHGTFLFFADKWYQKTTWMYISVPLVLYVAERSL 595
Query: 607 RTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITS 666
R RSKHY+VKI KVS+LPG V SLIMSKP GFKYKSGQYIFLQCP IS FEWHPFSITS
Sbjct: 596 RACRSKHYSVKILKVSMLPGEVLSLIMSKPPGFKYKSGQYIFLQCPTISRFEWHPFSITS 655
Query: 667 APGDEYLSVHIRTVGDWTQELKLLFT--EDDIPPVNCNATFGELVQMDQREHPRLFVDGP 724
APGD+ LSVHIRT+GDWT+EL+ + T +D V + F +D P+L VDGP
Sbjct: 656 APGDDQLSVHIRTLGDWTEELRRVLTVGKDLSTCVIGRSKFSAYCNIDMINRPKLLVDGP 715
Query: 725 YGAPAQDFKNFDVLLLIGLGIGATPFISILRDLLNNTRASDESMDQSNTETTRSDESWNS 784
YGAPAQD++++DVLLLIGLGIGATPFISIL+DLLNN+R +Q++ E +RSD SWNS
Sbjct: 716 YGAPAQDYRSYDVLLLIGLGIGATPFISILKDLLNNSRD-----EQTDNEFSRSDFSWNS 770
Query: 785 FTSS------NLTPGGNKRSQRTTNAYFYWVTREPGSFEWFKGVMDEVAEMDHRGQIELH 838
TSS T GG K++ A+FYWVTREPGS EWF+GVM+E+++MD RGQIELH
Sbjct: 771 CTSSYTTATPTSTHGGKKKA---VKAHFYWVTREPGSVEWFRGVMEEISDMDCRGQIELH 827
Query: 839 NYLTSVYEEGDARSTLITMIQALNHAKHGVDILSGTRVRTHFARPNWREVFIKIASKHPY 898
NYLTSVY+EGDARSTLI M+QALNHAKHGVDILSGTRVRTHFARPNW+EVF IA KHP
Sbjct: 828 NYLTSVYDEGDARSTLIKMVQALNHAKHGVDILSGTRVRTHFARPNWKEVFSSIARKHPN 887
Query: 899 STVGVFYCGMPVLAKELKKLSLELSHKTTTRFDFHKEYF 937
STVGVFYCG+ +AKELKK + ++S KTTTRF+FHKE+F
Sbjct: 888 STVGVFYCGIQTVAKELKKQAQDMSQKTTTRFEFHKEHF 926
>At1g64060 cytochrome b245 beta chain homolog RbohAp108
Length = 944
Score = 1011 bits (2613), Expect = 0.0
Identities = 519/913 (56%), Positives = 658/913 (71%), Gaps = 34/913 (3%)
Query: 53 NHHKELVEITLELEDD-AVVLCNVAPASS------------GAGTSSSSSHATGDEGTSA 99
N +E VE+T++L+DD +VL +V PA++ G T S S + + TS+
Sbjct: 38 NGDQEFVEVTIDLQDDDTIVLRSVEPATAINVIGDISDDNTGIMTPVSISRSPTMKRTSS 97
Query: 100 S-----GAGAVARSLSKNLSITSRIRRKFPWLRSMSMRTSASSTESVE---------DPV 145
+ A +++K ++ ++R F W RS S + +ST + + +
Sbjct: 98 NRFRQFSQELKAEAVAKAKQLSQELKR-FSWSRSFSGNLTTTSTAANQSGGAGGGLVNSA 156
Query: 146 ESARNARRMQAKLERTRSSAQRALKGLRFISKSGEAGEELWRKVEERFGSLAKDGLLARE 205
AR R+ +A+L+RTRSSAQRAL+GLRFIS + + W V+ F K+G + R
Sbjct: 157 LEARALRKQRAQLDRTRSSAQRALRGLRFISNK-QKNVDGWNDVQSNFEKFEKNGYIYRS 215
Query: 206 DFGGCIGMDDSKEFAVGVFDALARRKGRKASKITKEELHHFWQQISDQSFDARLQIFFDM 265
DF CIGM DSKEFA+ +FDAL+RR+ K KI +EL+ +W QI+D+SFD+RLQIFFD+
Sbjct: 216 DFAQCIGMKDSKEFALELFDALSRRRRLKVEKINHDELYEYWSQINDESFDSRLQIFFDI 275
Query: 266 ADSNEDGRITREEVQELIMLSASANKLSKLKEQAEGYAALIMEELDPENLGYIELWQLEM 325
D NEDGRIT EEV+E+IMLSASANKLS+LKEQAE YAALIMEELDPE LGYIELWQLE
Sbjct: 276 VDKNEDGRITEEEVKEIIMLSASANKLSRLKEQAEEYAALIMEELDPERLGYIELWQLET 335
Query: 326 LLLQKDRYMNYSRQLSTSSVSWSQNMTGLKHKNEIQRLCRTLQCLALEYWRRGWVLFLWL 385
LLLQKD Y+NYS+ LS +S + SQN+ GL+ K+ I R+ + E W+R WVL LW+
Sbjct: 336 LLLQKDTYLNYSQALSYTSQALSQNLQGLRGKSRIHRMSSDFVYIMQENWKRIWVLSLWI 395
Query: 386 VTTASLFAWKFYQYRNRTSFQVMGYCLPVAKGAAETLKLNMALILLPVCRNTLTWLRSTR 445
+ LF WKF+QY+ + +F VMGYCL AKGAAETLK NMALIL PVCRNT+TWLRSTR
Sbjct: 396 MIMIGLFLWKFFQYKQKDAFHVMGYCLLTAKGAAETLKFNMALILFPVCRNTITWLRSTR 455
Query: 446 ARKIVPFDDNINFHKIIACAIVVGVVVHAGNHLACDFPLLINSSPEEFS-LIASDFNNKK 504
VPFDDNINFHK IA AIVV V++H G+HLACDFP ++ ++ +++ + F K+
Sbjct: 456 LSYFVPFDDNINFHKTIAGAIVVAVILHIGDHLACDFPRIVRATEYDYNRYLFHYFQTKQ 515
Query: 505 PTYKTLLTGVEGVTGISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLL 564
PTY L+ G EG+TGI MV LM ISFTLAT FRRN V+LP P +RLTGFNAFWYSHHL
Sbjct: 516 PTYFDLVKGPEGITGILMVILMIISFTLATRWFRRNLVKLPKPFDRLTGFNAFWYSHHLF 575
Query: 565 GVVYILLFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVL 624
+VYILL +HG FL WY +TTWMY++VP+LLY ERTLR RS Y+V++ KV++
Sbjct: 576 VIVYILLILHGIFLYFAKPWYVRTTWMYLAVPVLLYGGERTLRYFRSGSYSVRLLKVAIY 635
Query: 625 PGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWT 684
PGNV +L MSKP F+YKSGQY+F+QCP +SPFEWHPFSITSAP D+Y+S+HIR +GDWT
Sbjct: 636 PGNVLTLQMSKPTQFRYKSGQYMFVQCPAVSPFEWHPFSITSAPEDDYISIHIRQLGDWT 695
Query: 685 QELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDVLLLIGLG 744
QELK +F+E PPV + + ++ P+L +DGPYGAPAQD++ +DVLLL+GLG
Sbjct: 696 QELKRVFSEVCEPPVGGKSGLLRADETTKKSLPKLLIDGPYGAPAQDYRKYDVLLLVGLG 755
Query: 745 IGATPFISILRDLLNNTRASDESMDQSNTETTRSDESWNSFTSSNLTPGGNKRSQRTTNA 804
IGATPFISIL+DLLNN +E D S ++ +RS E T SN KR +TTNA
Sbjct: 756 IGATPFISILKDLLNNIVKMEEHAD-SISDFSRSSE---YSTGSNGDTPRRKRILKTTNA 811
Query: 805 YFYWVTREPGSFEWFKGVMDEVAEMDHRGQIELHNYLTSVYEEGDARSTLITMIQALNHA 864
YFYWVTRE GSF+WFKGVM+EVAE+D RG IE+HNYLTSVYEEGDARS LITM+QALNHA
Sbjct: 812 YFYWVTREQGSFDWFKGVMNEVAELDQRGVIEMHNYLTSVYEEGDARSALITMVQALNHA 871
Query: 865 KHGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKELKKLSLELSH 924
K+GVDI+SGTRVRTHFARPNW++V K++SKH + +GVFYCG+PVL KEL KL +
Sbjct: 872 KNGVDIVSGTRVRTHFARPNWKKVLTKLSSKHCNARIGVFYCGVPVLGKELSKLCNTFNQ 931
Query: 925 KTTTRFDFHKEYF 937
K +T+F+FHKE+F
Sbjct: 932 KGSTKFEFHKEHF 944
>At5g47910 respiratory burst oxidase protein
Length = 921
Score = 872 bits (2254), Expect = 0.0
Identities = 460/889 (51%), Positives = 592/889 (65%), Gaps = 50/889 (5%)
Query: 57 ELVEITLELEDDAVVLCNVAPASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSIT 116
E VEITL++ DD+V + +V A+ G G A + T S SLS S +
Sbjct: 75 EYVEITLDIRDDSVAVHSVQQAAGGGGHLEDPELALLTKKTLESSLNNTT-SLSFFRSTS 133
Query: 117 SRIRRKFPWLRSMSMRTSASSTESVEDPVESARNARRMQAKLERTRSSAQRALKGLRFIS 176
SRI+ LR + R + + + +RT S+A ALKGL+FI+
Sbjct: 134 SRIKNASRELRRVFSRRPSPAVR-----------------RFDRTSSAAIHALKGLKFIA 176
Query: 177 KSGEAGEELWRKVEERFGSLAKD--GLLAREDFGGCIGMD-DSKEFAVGVFDALARRKGR 233
A W V++RF L+ D GLL F C+GM+ +SK+FA +F ALARR
Sbjct: 177 TKTAA----WPAVDQRFDKLSADSNGLLLSAKFWECLGMNKESKDFADQLFRALARRNNV 232
Query: 234 KASKITKEELHHFWQQISDQSFDARLQIFFDMADSNEDGRITREEVQELIMLSASANKLS 293
ITKE+L FW+QISD+SFDA+LQ+FFDM D +EDGR+T EEV E+I LSASANKLS
Sbjct: 233 SGDAITKEQLRIFWEQISDESFDAKLQVFFDMVDKDEDGRVTEEEVAEIISLSASANKLS 292
Query: 294 KLKEQAEGYAALIMEELDPENLGYIELWQLEMLLLQKDRYMNYSRQLSTSSVSWSQNMTG 353
+++QA+ YAALIMEELDP+N G+I + LEMLLLQ S SQ +
Sbjct: 293 NIQKQAKEYAALIMEELDPDNAGFIMIENLEMLLLQAPNQSVRMGDSRILSQMLSQKLRP 352
Query: 354 LKHKNEIQRLCRTLQCLALEYWRRGWVLFLWLVTTASLFAWKFYQYRNRTSFQVMGYCLP 413
K N + R ++ L+ W+R W++ LWL LF +KF QY+N+ ++ VMGYC+
Sbjct: 353 AKESNPLVRWSEKIKYFILDNWQRLWIMMLWLGICGGLFTYKFIQYKNKAAYGVMGYCVC 412
Query: 414 VAKGAAETLKLNMALILLPVCRNTLTWLRS-TRARKIVPFDDNINFHKIIACAIVVGVVV 472
VAKG AETLK NMALILLPVCRNT+TWLR+ T+ +VPFDD++NFHK+IA IVVGV++
Sbjct: 413 VAKGGAETLKFNMALILLPVCRNTITWLRNKTKLGTVVPFDDSLNFHKVIASGIVVGVLL 472
Query: 473 HAGNHLACDFPLLINSSPEEFSLIASDFNNKKPTYKTLLTGVEGVTGISMVTLMAISFTL 532
HAG HL CDFP LI + + + + F ++ +Y + GVEG TGI MV LMAI+FTL
Sbjct: 473 HAGAHLTCDFPRLIAADEDTYEPMEKYFGDQPTSYWWFVKGVEGWTGIVMVVLMAIAFTL 532
Query: 533 ATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTTWMY 592
AT FRRN + LP L +LTGFNAFWY+HHL +VY LL +HG L LT WY KTTWMY
Sbjct: 533 ATPWFRRNKLNLPNFLKKLTGFNAFWYTHHLFIIVYALLIVHGIKLYLTKIWYQKTTWMY 592
Query: 593 ISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCP 652
++VP+LLY +ER LR RS VK+ KV+V PGNV SL M+KP GFKYKSGQ++ + C
Sbjct: 593 LAVPILLYASERLLRAFRSSIKPVKMIKVAVYPGNVLSLHMTKPQGFKYKSGQFMLVNCR 652
Query: 653 KISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMD 712
+SPFEWHPFSITSAPGD+YLSVHIRT+GDWT++L+ +F+E PP A L++ D
Sbjct: 653 AVSPFEWHPFSITSAPGDDYLSVHIRTLGDWTRKLRTVFSEVCKPP---TAGKSGLLRAD 709
Query: 713 QRE----HPRLFVDGPYGAPAQDFKNFDVLLLIGLGIGATPFISILRDLLNNTRASDESM 768
+ P++ +DGPYGAPAQD+K +DV+LL+GLGIGATP ISIL+D++NN + D
Sbjct: 710 GGDGNLPFPKVLIDGPYGAPAQDYKKYDVVLLVGLGIGATPMISILKDIINNMKGPDRDS 769
Query: 769 DQSNTETTRSDESWNSFTSSNLTPGGNKRSQRTTNAYFYWVTREPGSFEWFKGVMDEVAE 828
D N + N + +T AYFYWVTRE GSFEWFKG+MDE++E
Sbjct: 770 DIENNNS-----------------NNNSKGFKTRKAYFYWVTREQGSFEWFKGIMDEISE 812
Query: 829 MDHRGQIELHNYLTSVYEEGDARSTLITMIQALNHAKHGVDILSGTRVRTHFARPNWREV 888
+D G IELHNY TSVYEEGDAR LI M+Q+L HAK+GVD++SGTRV++HFA+PNWR+V
Sbjct: 813 LDEEGIIELHNYCTSVYEEGDARVALIAMLQSLQHAKNGVDVVSGTRVKSHFAKPNWRQV 872
Query: 889 FIKIASKHPYSTVGVFYCGMPVLAKELKKLSLELSHKTTTRFDFHKEYF 937
+ KIA +HP +GVFYCGMP + KELK L+L+ S KTTT+FDFHKE F
Sbjct: 873 YKKIAVQHPGKRIGVFYCGMPGMIKELKNLALDFSRKTTTKFDFHKENF 921
>At5g07390 respiratory burst oxidase protein A
Length = 902
Score = 850 bits (2197), Expect = 0.0
Identities = 451/889 (50%), Positives = 593/889 (65%), Gaps = 42/889 (4%)
Query: 59 VEITLELEDDAVVLCNV-APASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNL--SI 115
VEITL++ DD+V + + +P GAG++ D+ G + S+ K L S+
Sbjct: 46 VEITLDIHDDSVSVYGLKSPNHRGAGSNYE------DQSLLRQGRSGRSNSVLKRLASSV 99
Query: 116 TSRIRRKFPWLRSMSMRTSASSTESVEDPVESARNARRMQAKLERTRSSAQRALKGLRFI 175
++ I R + +S SS+ + + P R AKL R++S A+ ALKGL+FI
Sbjct: 100 STGITR---------VASSVSSSSARKPP-------RPQLAKLRRSKSRAELALKGLKFI 143
Query: 176 SKSGEAGEELWRKVEERFG--SLAKDGLLAREDFGGCIGMDDSKEFAVGVFDALARRKGR 233
+K+ G W +VE+RF ++ +GLL R FG CIGM S EFA+ +FDALARR+
Sbjct: 144 TKTD--GVTGWPEVEKRFYVMTMTTNGLLHRSRFGECIGMK-STEFALALFDALARRENV 200
Query: 234 KASKITKEELHHFWQQISDQSFDARLQIFFDMADSNEDGRITREEVQELIMLSASANKLS 293
I EL FW+QI+DQ FD+RL+ FF M D + DGR+ EV+E+I LSASAN+L
Sbjct: 201 SGDSININELKEFWKQITDQDFDSRLRTFFAMVDKDSDGRLNEAEVREIITLSASANELD 260
Query: 294 KLKEQAEGYAALIMEELDPENLGYIELWQLEMLLLQKDRYMNYSRQLSTSSVSWSQNMTG 353
++ QA+ YAALIMEELDP + GYI + LE+LLLQ + S SQN+
Sbjct: 261 NIRRQADEYAALIMEELDPYHYGYIMIENLEILLLQAPMQDVRDGEGKKLSKMLSQNLMV 320
Query: 354 LKHKNEIQRLCRTLQCLALEYWRRGWVLFLWLVTTASLFAWKFYQYRNRTSFQVMGYCLP 413
+ +N R CR ++ + W+R WV+ LW+ A LF WKF +YR R++++VMG C+
Sbjct: 321 PQSRNLGARFCRGMKYFLFDNWKRVWVMALWIGAMAGLFTWKFMEYRKRSAYEVMGVCVC 380
Query: 414 VAKGAAETLKLNMALILLPVCRNTLTWLRS-TRARKIVPFDDNINFHKIIACAIVVGVVV 472
+AKGAAETLKLNMA+ILLPVCRNT+TWLR+ T+ IVPFDD++NFHK+IA I VGV +
Sbjct: 381 IAKGAAETLKLNMAMILLPVCRNTITWLRTKTKLSAIVPFDDSLNFHKVIAIGISVGVGI 440
Query: 473 HAGNHLACDFPLLINSSPEEFSLIASDFNNKKPTYKTLLTGVEGVTGISMVTLMAISFTL 532
HA +HLACDFP LI + +++ + F + Y + VEGVTGI MV LM I+FTL
Sbjct: 441 HATSHLACDFPRLIAADEDQYEPMEKYFGPQTKRYLDFVQSVEGVTGIGMVVLMTIAFTL 500
Query: 533 ATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLT-HRWYNKTTWM 591
AT FRRN + LP PL ++TGFNAFWYSHHL +VY LL +HG ++ L WY KTTWM
Sbjct: 501 ATTWFRRNKLNLPGPLKKITGFNAFWYSHHLFVIVYSLLVVHGFYVYLIIEPWYKKTTWM 560
Query: 592 YISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQC 651
Y+ VP++LY+ ER +R RS AV + KV+VLPGNV SL +S+P+ F+YKSGQY++L C
Sbjct: 561 YLMVPVVLYLCERLIRAFRSSVEAVSVLKVAVLPGNVLSLHLSRPSNFRYKSGQYMYLNC 620
Query: 652 PKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFTE--DDIPPVNCNATFGELV 709
+S EWHPFSITSAPGD+YLSVHIR +GDWT++L+ LF+E PP +
Sbjct: 621 SAVSTLEWHPFSITSAPGDDYLSVHIRVLGDWTKQLRSLFSEVCKPRPPDEHRLNRADSK 680
Query: 710 QMDQ-REHPRLFVDGPYGAPAQDFKNFDVLLLIGLGIGATPFISILRDLLNNTRASDESM 768
D + PR+ +DGPYGAPAQD+K F+V+LL+GLGIGATP ISI+ D++NN + +E
Sbjct: 681 HWDYIPDFPRILIDGPYGAPAQDYKKFEVVLLVGLGIGATPMISIVSDIINNLKGVEEG- 739
Query: 769 DQSNTETTRSDESWNSFTSSNLTPGGNKRSQRTTNAYFYWVTREPGSFEWFKGVMDEVAE 828
+ R ++ + ++P + RT AYFYWVTRE GSF+WFK VMDEV E
Sbjct: 740 ------SNRRQSPIHNMVTPPVSPSRKSETFRTKRAYFYWVTREQGSFDWFKNVMDEVTE 793
Query: 829 MDHRGQIELHNYLTSVYEEGDARSTLITMIQALNHAKHGVDILSGTRVRTHFARPNWREV 888
D + IELHNY TSVYEEGDARS LITM+Q+LNHAKHGVD++SGTRV +HFARPNWR V
Sbjct: 794 TDRKNVIELHNYCTSVYEEGDARSALITMLQSLNHAKHGVDVVSGTRVMSHFARPNWRSV 853
Query: 889 FIKIASKHPYSTVGVFYCGMPVLAKELKKLSLELSHKTTTRFDFHKEYF 937
F +IA HP + VGVFYCG L KEL+ LSL+ SHKT+T+F FHKE F
Sbjct: 854 FKRIAVNHPKTRVGVFYCGAAGLVKELRHLSLDFSHKTSTKFIFHKENF 902
>At5g51060 respiratory burst oxidase protein
Length = 905
Score = 843 bits (2179), Expect = 0.0
Identities = 452/895 (50%), Positives = 603/895 (66%), Gaps = 60/895 (6%)
Query: 59 VEITLELEDDAVVLCNVAPASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSITSR 118
V++T++++DD V S H+ EG S+ V S L +R
Sbjct: 55 VDLTVDVQDDTV-----------------SVHSLKMEGGSS-----VEESPELTLLKRNR 92
Query: 119 IRRKFPWLRSM-SMRTSASSTESVEDPVESARNARRMQAKLERTRSSAQRALKGLRFISK 177
+ +K ++ + S+ SV + + R AKL+RT+S+A +ALKGL+FISK
Sbjct: 93 LEKKTTVVKRLASVSHELKRLTSVSGGIGGRKPPR--PAKLDRTKSAASQALKGLKFISK 150
Query: 178 S-GEAGEELWRKVEERFGSLAKD--GLLAREDFGGCIGMDDSKEFAVGVFDALARRKGRK 234
+ G AG W VE+RF + GLL R FG CIGM SK+FA+ +FDALARR+
Sbjct: 151 TDGGAG---WSAVEKRFNQITATTGGLLLRTKFGECIGMT-SKDFALELFDALARRRNIT 206
Query: 235 ASKITKEELHHFWQQISDQSFDARLQIFFDMADSNEDGRITREEVQELIMLSASANKLSK 294
I ++L FW+QI+DQSFD+RL+ FFDM D + DGR+T +EV+E+I LSASAN LS
Sbjct: 207 GEVIDGDQLKEFWEQINDQSFDSRLKTFFDMVDKDADGRLTEDEVREIISLSASANNLST 266
Query: 295 LKEQAEGYAALIMEELDPENLGYIELWQLEMLLLQK--DRYMNYSRQLSTSSVSWSQNMT 352
++++A+ YAALIMEELDP+N+GYI L LE LLLQ + + + S SQ +
Sbjct: 267 IQKRADEYAALIMEELDPDNIGYIMLESLETLLLQAATQSVITSTGERKNLSHMMSQRLK 326
Query: 353 GLKHKNEIQRLCRTLQCLALEYWRRGWVLFLWLVTTASLFAWKFYQYRNRTSFQVMGYCL 412
++N ++R R L+ L+ W+R WV+ LW + A LF +K+ QYR + VMG C+
Sbjct: 327 PTFNRNPLKRWYRGLRFFLLDNWQRCWVIVLWFIVMAILFTYKYIQYRRSPVYPVMGDCV 386
Query: 413 PVAKGAAETLKLNMALILLPVCRNTLTWLRS-TRARKIVPFDDNINFHKIIACAIVVGVV 471
+AKGAAET+KLNMALILLPVCRNT+TWLR+ TR ++VPFDDN+NFHK+IA I+VGV
Sbjct: 387 CMAKGAAETVKLNMALILLPVCRNTITWLRNKTRLGRVVPFDDNLNFHKVIAVGIIVGVT 446
Query: 472 VHAGNHLACDFPLLINSSPEEFSLIASDFNNKKP-TYKTLLTGVEGVTGISMVTLMAISF 530
+HAG HLACDFP L++++PE + + F +++P +Y + VEG+TG+ MV LMAI+F
Sbjct: 447 MHAGAHLACDFPRLLHATPEAYRPLRQFFGDEQPKSYWHFVNSVEGITGLVMVLLMAIAF 506
Query: 531 TLATHHFRRNAVR-LPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTT 589
TLAT FRR + LP PL +L FNAFWY+HHL +VYILL HG +L LT W+NKTT
Sbjct: 507 TLATPWFRRGKLNYLPGPLKKLASFNAFWYTHHLFVIVYILLVAHGYYLYLTRDWHNKTT 566
Query: 590 WMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFL 649
WMY+ VP++LY ER +R RS AV I+KV+V PGNV ++ +S+P FKYKSGQY+F+
Sbjct: 567 WMYLVVPVVLYACERLIRAFRSSIKAVTIRKVAVYPGNVLAIHLSRPQNFKYKSGQYMFV 626
Query: 650 QCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFTEDDIPP---VNCNATFG 706
C +SPFEWHPFSITSAP D+YLSVHIR +GDWT+ LK +F+E PP V+
Sbjct: 627 NCAAVSPFEWHPFSITSAPQDDYLSVHIRVLGDWTRALKGVFSEVCKPPPAGVSGLLRAD 686
Query: 707 ELVQMDQREHPRLFVDGPYGAPAQDFKNFDVLLLIGLGIGATPFISILRDLLNNTRASDE 766
L + + P++ +DGPYGAPAQD+K ++V+LL+GLGIGATP ISI++D++NN +A ++
Sbjct: 687 MLHGANNPDFPKVLIDGPYGAPAQDYKKYEVVLLVGLGIGATPMISIVKDIVNNIKAKEQ 746
Query: 767 S----MDQSNTETTRSDESWNSFTSSNLTPGGNKRSQRTTNAYFYWVTREPGSFEWFKGV 822
+ M+ +E RS K S RT AYFYWVTRE GSF+WFK +
Sbjct: 747 AQLNRMENGTSEPQRS----------------KKESFRTRRAYFYWVTREQGSFDWFKNI 790
Query: 823 MDEVAEMDHRGQIELHNYLTSVYEEGDARSTLITMIQALNHAKHGVDILSGTRVRTHFAR 882
M+EVAE D IE+HNY TSVYEEGDARS LI M+Q+LNHAK+GVDI+SGTRV +HFA+
Sbjct: 791 MNEVAERDANRVIEMHNYCTSVYEEGDARSALIHMLQSLNHAKNGVDIVSGTRVMSHFAK 850
Query: 883 PNWREVFIKIASKHPYSTVGVFYCGMPVLAKELKKLSLELSHKTTTRFDFHKEYF 937
PNWR V+ +IA HP + VGVFYCG P L KEL+ L+L+ +HKT+TRF FHKE F
Sbjct: 851 PNWRNVYKRIAMDHPNTKVGVFYCGAPALTKELRHLALDFTHKTSTRFSFHKENF 905
>At1g09090 respiratory burst oxidase protein B like
Length = 843
Score = 842 bits (2174), Expect = 0.0
Identities = 433/791 (54%), Positives = 545/791 (68%), Gaps = 31/791 (3%)
Query: 157 KLERTRS-SAQRALKGLRFISKSGEAGEELWRKVEERFGSLAKDGLLAREDFGGCIGMDD 215
+L+R++S A AL+GLRFI+K+ G W +V RF LA +G L + FG CIGM +
Sbjct: 74 RLDRSKSFGAMFALRGLRFIAKNDAVGRG-WDEVAMRFDKLAVEGKLPKSKFGHCIGMVE 132
Query: 216 SKEFAVGVFDALARRKGRKASKITKEELHHFWQQISDQSFDARLQIFFDMADSNEDGRIT 275
S EF +F+AL RR+G +S ITK EL FW+QI+ SFD RLQIFFDM D N DGRIT
Sbjct: 133 SSEFVNELFEALVRRRGTTSSSITKTELFEFWEQITGNSFDDRLQIFFDMVDKNLDGRIT 192
Query: 276 REEVQELIMLSASANKLSKLKEQAEGYAALIMEELDPENLGYIELWQLEMLLLQKDRYMN 335
+EV+E+I LSASANKLSK+KE + YAALIMEELD +NLGYIEL LE LLLQ N
Sbjct: 193 GDEVKEIIALSASANKLSKIKENVDEYAALIMEELDRDNLGYIELHNLETLLLQVPSQSN 252
Query: 336 YSRQLSTSSV---SWSQNMTGLKHKNEIQRLCRTLQCLALEYWRRGWVLFLWLVTTASLF 392
S + SQ + K +N ++R + LE W+R WVL LW+ +LF
Sbjct: 253 NSPSSANKRALNKMLSQKLIPTKDRNPVKRFAMNISYFFLENWKRIWVLTLWISICITLF 312
Query: 393 AWKFYQYRNRTSFQVMGYCLPVAKGAAETLKLNMALILLPVCRNTLTWLR--STRARKIV 450
WKF QY+ +T F+VMGYC+ VAKG+AETLK NMALILLPVCRNT+TWLR S +V
Sbjct: 313 TWKFLQYKRKTVFEVMGYCVTVAKGSAETLKFNMALILLPVCRNTITWLRTKSKLIGSVV 372
Query: 451 PFDDNINFHKIIACAIVVGVVVHAGNHLACDFPLLINSSPEEFSLIASDFNNKKP-TYKT 509
PFDDNINFHK++A I VG+ +HA +HLACDFP L+++ EF + F +++P Y
Sbjct: 373 PFDDNINFHKVVAFGIAVGIGLHAISHLACDFPRLLHAKNVEFEPMKKFFGDERPENYGW 432
Query: 510 LLTGVEGVTGISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYI 569
+ G +G TG++MV LM +++ LA FRRN LP L RLTGFNAFWYSHHL +VY+
Sbjct: 433 FMKGTDGWTGVTMVVLMLVAYVLAQSWFRRNRANLPKSLKRLTGFNAFWYSHHLFVIVYV 492
Query: 570 LLFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVF 629
LL +HG F+ L+ WY+KTTWMY++VP+LLY ER +R R AVK+ KV+V PGNV
Sbjct: 493 LLIVHGYFVYLSKEWYHKTTWMYLAVPVLLYAFERLIRAFRPGAKAVKVLKVAVYPGNVL 552
Query: 630 SLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKL 689
SL MSKP GFKY SGQYI++ C +SP +WHPFSITSA GD+YLSVHIRT+GDWT +LK
Sbjct: 553 SLYMSKPKGFKYTSGQYIYINCSDVSPLQWHPFSITSASGDDYLSVHIRTLGDWTSQLKS 612
Query: 690 LFTE-DDIPPVNCNATF-GELVQMDQ-REHPRLFVDGPYGAPAQDFKNFDVLLLIGLGIG 746
L+++ +P + + F ++ Q + PRL +DGPYGAPAQD++N+DVLLL+GLGIG
Sbjct: 613 LYSKVCQLPSTSQSGLFIADIGQANNITRFPRLLIDGPYGAPAQDYRNYDVLLLVGLGIG 672
Query: 747 ATPFISILRDLLNNTRASDESMDQSNTETTRSDESWNSFTSSNLTPGGNKRSQRTTNAYF 806
ATP ISI+RD+LNN + + S+++ + K T AYF
Sbjct: 673 ATPLISIIRDVLNNIK-NQNSIERGTNQHI-------------------KNYVATKRAYF 712
Query: 807 YWVTREPGSFEWFKGVMDEVAEMDHRGQIELHNYLTSVYEEGDARSTLITMIQALNHAKH 866
YWVTRE GS EWF VM+EVAE D G IELHNY TSVYEEGDARS LITM+Q+L+HAK
Sbjct: 713 YWVTREQGSLEWFSEVMNEVAEYDSEGMIELHNYCTSVYEEGDARSALITMLQSLHHAKS 772
Query: 867 GVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKELKKLSLELSHKT 926
G+DI+SGTRVRTHFARPNWR VF +A H VGVFYCG + ELK+L+ + S KT
Sbjct: 773 GIDIVSGTRVRTHFARPNWRSVFKHVAVNHVNQRVGVFYCGNTCIIGELKRLAQDFSRKT 832
Query: 927 TTRFDFHKEYF 937
TT+F+FHKE F
Sbjct: 833 TTKFEFHKENF 843
>At4g11230 respiratory burst oxidase homolog F - like protein
Length = 942
Score = 831 bits (2147), Expect = 0.0
Identities = 456/909 (50%), Positives = 586/909 (64%), Gaps = 40/909 (4%)
Query: 56 KELVEITLELEDDAVVLCNVAPASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSI 115
+EL+E+T+E ++ + + +G GT S T E TS S +G+ +RSLS S
Sbjct: 47 EELLEVTIEFPSGVIINID---SVTGTGTDISG---TDLEITSCSDSGSGSRSLSLGWSA 100
Query: 116 TSRI----RRKFPWLRSMSMRTSASSTESVEDPVESARNARRMQAKLER------TR--- 162
+S ++ +S R S+ S +PV R L R TR
Sbjct: 101 SSERLTAGTNSKQQIQKISRRGYGYSSRSAPEPVVPHRGEITDSVNLPRALSQRPTRPNR 160
Query: 163 --SSAQRALKGLRFISKSGEAGEELWRKVEERFGSLAKDGLLAREDFGGCIGMDD--SKE 218
S +RA+ GL+FIS S E G W V+ F L+KDG L + DF CIG+++ SKE
Sbjct: 161 DGSGTERAIHGLKFIS-SKENGIVDWNDVQNNFAHLSKDGYLFKSDFAHCIGLENENSKE 219
Query: 219 FAVGVFDALARRKGRKASKITKEELHHFWQQISDQSFDARLQIFFDMADSNEDGRITREE 278
FA +FDAL RR+ KI +EL+ FW QI+D+SFD+RLQIFF+M + +
Sbjct: 220 FADELFDALCRRRRIMVDKINLQELYEFWYQITDESFDSRLQIFFNMYCYQLSSNLVKHI 279
Query: 279 VQELIMLSASANKLSKLKEQAEGYAALIMEELDPENL--GYIELWQLEMLLLQKDRYMNY 336
Q +I+LSASAN LS+L+E+AE YAALIMEEL P+ L YIEL LE+LLL+KD +Y
Sbjct: 280 DQHIIILSASANNLSRLRERAEEYAALIMEELAPDGLYSQYIELKDLEILLLEKDISHSY 339
Query: 337 SRQLSTSSVSWSQNMTGLKHKNEIQRLCRTLQCLALEYWRRGWVLFLWLVTTASLFAWKF 396
S S +S + SQN+ K+ R+ R L + W+R WVL LW V A LF WK
Sbjct: 340 SLPFSQTSRALSQNL-----KDRRWRMSRNLLYSLQDNWKRIWVLTLWFVIMAWLFMWKC 394
Query: 397 YQYRNRTSFQVMGYCLPVAKGAAETLKLNMALILLPVCRNTLTWLRSTRARKIVPFDDNI 456
YQY+++ +F VMGYCL +AKGAAETLK NMALILLPVCRNT+T+LRST VPFDD I
Sbjct: 395 YQYKHKDAFHVMGYCLVMAKGAAETLKFNMALILLPVCRNTITYLRSTALSHSVPFDDCI 454
Query: 457 NFHKIIACAIVVGVVVHAGNHLACDFPLLINSSPEEFS-LIASDFNNKKPTYKTLLTGVE 515
NFHK I+ AI+ +++HA +HLACDFP ++ S+ ++ + F +PTY L+
Sbjct: 455 NFHKTISVAIISAMLLHATSHLACDFPRILASTDTDYKRYLVKYFGVTRPTYFGLVNTPV 514
Query: 516 GVTGISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHG 575
G+TGI MV M I+FTLA+ RRN +LP P ++LTG+NAFWYSHHLL VY+LL IHG
Sbjct: 515 GITGIIMVAFMLIAFTLASRRCRRNLTKLPKPFDKLTGYNAFWYSHHLLLTVYVLLVIHG 574
Query: 576 SFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFSLIMSK 635
L L H+WY KT WMY++VP+LLY+ ER R RS+ Y V+I KV + PGNV L MSK
Sbjct: 575 VSLYLEHKWYRKTVWMYLAVPVLLYVGERIFRFFRSRLYTVEICKVVIYPGNVVVLRMSK 634
Query: 636 PNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFTEDD 695
P F YKSGQY+F+QCP +S FEWHPFSITS+PGD+YLS+HIR GDWT+ +K F+
Sbjct: 635 PTSFDYKSGQYVFVQCPSVSKFEWHPFSITSSPGDDYLSIHIRQRGDWTEGIKKAFSVVC 694
Query: 696 IPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDVLLLIGLGIGATPFISILR 755
P + +QR P L +DGPYGAPAQD +DV+LL+GLGIGATPF+SILR
Sbjct: 695 HAPEAGKSGLLRADVPNQRSFPELLIDGPYGAPAQDHWKYDVVLLVGLGIGATPFVSILR 754
Query: 756 DLLNNTRASDE-------SMDQSNTETTRSDESWNSFTSSNLTPGGNKRSQRTTNAYFYW 808
DLLNN E S SN + S NS +S + P +++ T NAYFYW
Sbjct: 755 DLLNNIIKQQEQAECISGSCSNSNISSDHSFSCLNSEAASRI-PQTQRKTLNTKNAYFYW 813
Query: 809 VTREPGSFEWFKGVMDEVAEMDHRGQIELHNYLTSVYEEGDARSTLITMIQALNHAKHGV 868
VTRE GSF+WFK +M+E+A+ D +G IE+HNYLTSVYEEGD RS L+TMIQ LNHAK+GV
Sbjct: 814 VTREQGSFDWFKEIMNEIADSDRKGVIEMHNYLTSVYEEGDTRSNLLTMIQTLNHAKNGV 873
Query: 869 DILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKELKKLSLELSHKTTT 928
DI SGT+VRTHF RP W++V KI++KH + +GVFYCG+P L KEL L E + T
Sbjct: 874 DIFSGTKVRTHFGRPKWKKVLSKISTKHRNARIGVFYCGVPSLGKELSTLCHEFNQTGIT 933
Query: 929 RFDFHKEYF 937
RFDFHKE F
Sbjct: 934 RFDFHKEQF 942
>At4g25090 respiratory burst oxidase - like protein
Length = 863
Score = 800 bits (2066), Expect = 0.0
Identities = 406/801 (50%), Positives = 555/801 (68%), Gaps = 37/801 (4%)
Query: 157 KLERTRSSAQRALKGLRFISKSGEAGEELWRKVEERFGSLAK--DGLLAREDFGGCIGMD 214
+L+R++S+A +ALKGL+ ISK+ G W VE+R+ + DGLL R FG CIGM+
Sbjct: 80 RLDRSKSTAGQALKGLKIISKTD--GNAAWTVVEKRYLKITANTDGLLLRSKFGECIGMN 137
Query: 215 DSKEFAVGVFDALARRKGRKASKITKEELHHFWQQISDQSFDARLQIFFDMADSNEDGRI 274
SKEFA+ +FDALAR+ K IT+ EL FW+QI+D+SFD+RL FFD+ D + DGR+
Sbjct: 138 -SKEFALELFDALARKSHLKGDVITETELKKFWEQINDKSFDSRLITFFDLMDKDSDGRL 196
Query: 275 TREEVQELIMLSASANKLSKLKEQAEGYAALIMEELDPENLGYIELWQLEMLLLQKDRYM 334
T +EV+E+I LS+SAN LS ++ +A+ YAA+IMEELDP+++GYI + L+ LLLQ +
Sbjct: 197 TEDEVREIIKLSSSANHLSCIQNKADEYAAMIMEELDPDHMGYIMMESLKKLLLQAETKS 256
Query: 335 NY----SRQLSTSSVSWSQNMTGLKHKNEIQRLCRTLQCLALEYWRRGWVLFLWLVTTAS 390
S + S ++++ + N ++R L+ L+ W+R WV+ LWL A
Sbjct: 257 VSTDINSEERKELSDMLTESLKPTRDPNHLRRWYCQLRFFVLDSWQRVWVIALWLTIMAI 316
Query: 391 LFAWKFYQYRNRTSFQVMGYCLPVAKGAAETLKLNMALILLPVCRNTLTWLRS-TRARKI 449
LFA+K+ QY+NR ++V+G C+ +AKGAAETLKLNMALILLPVCRNT+TWLR+ TR
Sbjct: 317 LFAYKYIQYKNRAVYEVLGPCVCLAKGAAETLKLNMALILLPVCRNTITWLRNKTRLGVF 376
Query: 450 VPFDDNINFHKIIACAIVVGVVVHAGNHLACDFPLLINSSPEEFSLIASDFNNKKPT-YK 508
VPFDDN+NFHK+IA I +GV +H+ +HLACDFPLLI ++P E+ + F ++P Y
Sbjct: 377 VPFDDNLNFHKVIAVGIAIGVAIHSVSHLACDFPLLIAATPAEYMPLGKFFGEEQPKRYL 436
Query: 509 TLLTGVEGVTGISMVTLMAISFTLATHHFRRNAV--RLPPPLNRLTGFNAFWYSHHLLGV 566
+ EG+TG+ MV LM I+FTLA FRR + +LP PL +L FNAFWY+HHL +
Sbjct: 437 HFVKSTEGITGLVMVFLMVIAFTLAMPWFRRGKLEKKLPGPLKKLASFNAFWYTHHLFVI 496
Query: 567 VYILLFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPG 626
VYILL +HG ++ L WY KTTWMY++VP+ LY ER +R RS VK+ K++ PG
Sbjct: 497 VYILLVLHGYYIYLNKEWYKKTTWMYLAVPVALYAYERLIRAFRSSIRTVKVLKMAAYPG 556
Query: 627 NVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQE 686
V +L MSKP FKY SGQY+F+ CP +SPFEWHPFSITS P D+YLSVHI+ +GDWT+
Sbjct: 557 KVLTLQMSKPTNFKYMSGQYMFVNCPAVSPFEWHPFSITSTPQDDYLSVHIKALGDWTEA 616
Query: 687 LKLLFTEDD---------IPPVNCNATFGELVQ-MDQREHPRLFVDGPYGAPAQDFKNFD 736
++ +F+E I V+ G+++ + P++ +DGPYGAPAQD+K ++
Sbjct: 617 IQGVFSESREKIVEKHHVIFQVSKPPPVGDMLNGANSPRFPKIMIDGPYGAPAQDYKKYE 676
Query: 737 VLLLIGLGIGATPFISILRDLLNNTRASDESMDQSNTETTRSDESWNSFTSSNLTPGGNK 796
V+LLIGLGIGATP ISI++D++NNT E + Q + + + GNK
Sbjct: 677 VVLLIGLGIGATPMISIIKDIINNTETK-EQLSQMEKGSPQEQQ-------------GNK 722
Query: 797 RSQRTTNAYFYWVTREPGSFEWFKGVMDEVAEMDHRGQIELHNYLTSVYEEGDARSTLIT 856
+ +T AYFYWVT+E G+F+WFK +M+E+AE D IELHN+ TSVYEEGD RS LI
Sbjct: 723 ETFKTRRAYFYWVTKEQGTFDWFKNIMNEIAERDKSKVIELHNHCTSVYEEGDVRSALIR 782
Query: 857 MIQALNHAKHGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKELK 916
M+Q+LN+AK+G+DI++GTRV +HFARPNW+ V+ +IA HP + VGVFYCG PVL KEL+
Sbjct: 783 MLQSLNYAKNGLDIVAGTRVMSHFARPNWKNVYKQIAMDHPGANVGVFYCGAPVLTKELR 842
Query: 917 KLSLELSHKTTTRFDFHKEYF 937
+L+LE +HKT+TRF FHKE F
Sbjct: 843 QLALEFTHKTSTRFSFHKENF 863
>At5g60010 respiratory burst oxidase protein - like
Length = 839
Score = 691 bits (1783), Expect = 0.0
Identities = 366/793 (46%), Positives = 497/793 (62%), Gaps = 28/793 (3%)
Query: 103 GAVARSLSKNLSITSRIRRKFP--WLRSMSMRTSASSTESVEDPVESARNARRMQAKLER 160
G + ++ S+NL + S IR W +S ++ + ++ P +A + ++ER
Sbjct: 40 GGLKKNASRNLGVGSIIRTLSVSNWRKSGNLGSPSTRKSGNLGPPTNAVPKKTGPQRVER 99
Query: 161 TRSSAQRALKGLRFISKSGEAGE-ELWRKVEERFGSLAKDGLLAREDFGGCIGMDDSKEF 219
T SSA R L+ LRF+ ++ E + WR +E RF + DG L +E FG CIGM D+ EF
Sbjct: 100 TTSSAARGLQSLRFLDRTVTGRERDAWRSIENRFNQFSVDGKLPKEKFGVCIGMGDTMEF 159
Query: 220 AVGVFDALARRKGRKASK-ITKEELHHFWQQISDQSFDARLQIFFDMADSNEDGRITREE 278
A V++AL RR+ + I KE+L FW+ + + D RLQIFFDM D N DG++T EE
Sbjct: 160 AAEVYEALGRRRQIETENGIDKEQLKLFWEDMIKKDLDCRLQIFFDMCDKNGDGKLTEEE 219
Query: 279 VQELIMLSASANKLSKLKEQAEGYAALIMEELDPENLGYIELWQLEMLLLQKDRYMNYSR 338
V+E+I+LSASAN+L LK+ A YA+LIMEELDP++ GYIE+WQLE+LL N
Sbjct: 220 VKEVIVLSASANRLGNLKKNAAAYASLIMEELDPDHKGYIEMWQLEILLT--GMVTNADT 277
Query: 339 QLSTSSVSWSQNMTGLKHKNEIQRLCRTLQCLALEYWRRGWVLFLWLVTTASLFAWKFYQ 398
+ S + ++ M +++ + + L E W++ WVL LW + LF WK+ +
Sbjct: 278 EKMKKSQTLTRAMIPERYRTPMSKYVSVTAELMHENWKKLWVLALWAIINVYLFMWKYEE 337
Query: 399 YRNRTSFQVMGYCLPVAKGAAETLKLNMALILLPVCRNTLTWLRSTRARKIVPFDDNINF 458
+ + + G C+ AKGAAETLKLNMALIL+PVCR TLT LRST ++VPFDDNINF
Sbjct: 338 FMRNPLYNITGRCVCAAKGAAETLKLNMALILVPVCRKTLTILRSTFLNRVVPFDDNINF 397
Query: 459 HKIIACAIVVGVVVHAGNHLACDFPLLINSSPEEFSLIA-SDFNNKKPTYKTLLTGVEGV 517
HK+IA I ++H H+ C++P L + S + F A + N +P+Y L+ +
Sbjct: 398 HKVIAYMIAFQALLHTALHIFCNYPRLSSCSYDVFLTYAGAALGNTQPSYLGLMLTSVSI 457
Query: 518 TGISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGSF 577
TG+ M+ M SFTLA H+FRRN V+LP P N L GFNAFWY+HHLL + YILL IHG +
Sbjct: 458 TGVLMIFFMGFSFTLAMHYFRRNIVKLPKPFNVLAGFNAFWYAHHLLVLAYILLIIHGYY 517
Query: 578 LNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKH-YAVKIQKVSVLPGNVFSLIMSKP 636
L + WY KTTWMY++VP+L Y +ER +H + V + K V GNV +L ++KP
Sbjct: 518 LIIEKPWYQKTTWMYLAVPMLFYASERLFSRLLQEHSHRVNVIKAIVYSGNVLALYVTKP 577
Query: 637 NGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFTEDDI 696
GFKYKSG Y+F++CP +S FEWHPFSITSAPGD+YLSVHIR +GDWT EL+ F +
Sbjct: 578 PGFKYKSGMYMFVKCPDLSKFEWHPFSITSAPGDDYLSVHIRALGDWTTELRSRFAKTCE 637
Query: 697 P-PVNCNATFGELVQMDQRE-------------HPRLFVDGPYGAPAQDFKNFDVLLLIG 742
P L++M+ R P++F+ GPYGAPAQ+++ FD+LLL+G
Sbjct: 638 PTQAAAKPKPNSLMRMETRAAGVNPHIEESQVLFPKIFIKGPYGAPAQNYQKFDILLLVG 697
Query: 743 LGIGATPFISILRDLLNNTRASDESMDQSNTETTRSDESWNSFTSSNLTPGGNKRSQRTT 802
LGIGATPFISIL+D+LN+ + Q + + S GG K QR
Sbjct: 698 LGIGATPFISILKDMLNHLKPGIPRSGQKYEGSVGGESIGGDSVSGG---GGKKFPQR-- 752
Query: 803 NAYFYWVTREPGSFEWFKGVMDEVAEMDHRGQIELHNYLTSVYEEGDARSTLITMIQALN 862
AYF+WVTRE SF+WFKGVMD++AE D IE+HNYLTS+YE GDARS LI M+Q L
Sbjct: 753 -AYFFWVTREQASFDWFKGVMDDIAEYDKTHVIEMHNYLTSMYEAGDARSALIAMVQKLQ 811
Query: 863 HAKHGVDILSGTR 875
HAK+GVDI+S +R
Sbjct: 812 HAKNGVDIVSESR 824
>At3g45810 respiratory burst oxidase - like protein
Length = 835
Score = 664 bits (1714), Expect = 0.0
Identities = 366/815 (44%), Positives = 488/815 (58%), Gaps = 67/815 (8%)
Query: 98 SASGAGAVARSLSKNLSITSRIRRKF--PWLRSMSMRT-SASSTESVEDPVESARNARRM 154
S+ G + +++SKNL++ S IR W +S ++ + S + ++ P+ ++ R
Sbjct: 44 SSGAGGGILKNVSKNLAVGSIIRSMSVNKWRKSGNLGSPSTRKSGNLGPPLPVSQVKRPG 103
Query: 155 QAKLERTRSSAQRALKGLRFISKSGEAGE-ELWRKVEERFGSLAKDGLLAREDFGGCIGM 213
++ERT SSA R L+ LRF+ ++ E + WR +E RF A DG L ++ FG CIGM
Sbjct: 104 PQRVERTTSSAARGLQSLRFLDRTVTGRERDSWRSIENRFNQFAVDGRLPKDKFGVCIGM 163
Query: 214 DDSKEFAVGVFDALARRKGRKASK-ITKEELHHFWQQISDQSFDARLQIFFDMADSNEDG 272
D+ EFA V++AL RR+ K I KE+L FW+ + + D RLQIFFDM D + DG
Sbjct: 164 GDTLEFAAKVYEALGRRRQIKTENGIDKEQLKLFWEDMIKKDLDCRLQIFFDMCDKDGDG 223
Query: 273 RITREEVQELIMLSASANKLSKLKEQAEGYAALIMEELDPENLGYIELWQLEMLLLQKDR 332
++T EEV+E+I+LSASAN+L LK+ A YA+LIMEELDP GYIE+WQLE+LL
Sbjct: 224 KLTEEEVKEVIVLSASANRLVNLKKNAASYASLIMEELDPNEQGYIEMWQLEVLLTGIVS 283
Query: 333 YMNYSRQLSTSSVSWSQNMTGLKHKNEIQRLCRTLQCLALEYWRRGWVLFLWLVTTASLF 392
+ S ++ S ++ M +++ + L E+W++ WV+ LWL LF
Sbjct: 284 NAD-SHKVVRKSQQLTRAMIPKRYRTPTSKYVVVTAELMYEHWKKIWVVTLWLAVNVVLF 342
Query: 393 AWKFYQYRNRTSFQVMGYCLPVAKGAAETLKLNMALILLPVCRNTLTWLRSTRARKIVPF 452
WK+ ++ + + G CL AKG AE LKLNMALIL+PV R TLT+LRST ++PF
Sbjct: 343 MWKYEEFTTSPLYNITGRCLCAAKGTAEILKLNMALILVPVLRRTLTFLRSTFLNHLIPF 402
Query: 453 DDNINFHKIIACAIVVGVVVHAGNHLACDFPLLINSSPEEFSLIASD-FNNKKPTYKTLL 511
DDNINFHK+IA AI V ++H H+ C++P L + +S A + K+PTY L+
Sbjct: 403 DDNINFHKLIAVAIAVISLLHTALHMLCNYPRLSSCPYNFYSDYAGNLLGAKQPTYLGLM 462
Query: 512 TGVEGVTGISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILL 571
VTG+ M+ M ISFTLA H+FRRN V+LP P NRL GFN+FWY+HHLL + Y LL
Sbjct: 463 LTPVSVTGVLMIIFMGISFTLAMHYFRRNIVKLPIPFNRLAGFNSFWYAHHLLVIAYALL 522
Query: 572 FIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFSL 631
IHG L + WY KT +Y GNV +L
Sbjct: 523 IIHGYILIIEKPWYQKTAIVY--------------------------------SGNVLAL 550
Query: 632 IMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLF 691
M+KP GFKYKSG Y+F++CP IS FEWHPFSITSAPGDEYLSVHIR +GDWT EL+ F
Sbjct: 551 YMTKPQGFKYKSGMYMFVKCPDISKFEWHPFSITSAPGDEYLSVHIRALGDWTSELRNRF 610
Query: 692 TEDDIPPVNCNATFGELVQMDQREH-------------PRLFVDGPYGAPAQDFKNFDVL 738
E P + +L++M+ R PR+F+ GPYGAPAQ ++ FD+L
Sbjct: 611 AETCEPHQKSKPSPNDLIRMETRARGANPHVEESQALFPRIFIKGPYGAPAQSYQKFDIL 670
Query: 739 LLIGLGIGATPFISILRDLLNNTR-------------ASDESMDQSNTETTRSDESWNSF 785
LLIGLGIGATPFISIL+D+LNN + ES+ S+ S S
Sbjct: 671 LLIGLGIGATPFISILKDMLNNLKPGIPKTGQKYEGSVGGESLGGSSVYGGSSVNGGGSV 730
Query: 786 TSSNLTPGGNKRSQRTTNAYFYWVTREPGSFEWFKGVMDEVAEMDHRGQIELHNYLTSVY 845
GG ++ + AYFYWVTRE SFEWFKGVMD++A D IE+HNYLTS+Y
Sbjct: 731 NGGGSVSGGGRKFPQ--RAYFYWVTREQASFEWFKGVMDDIAVYDKTNVIEMHNYLTSMY 788
Query: 846 EEGDARSTLITMIQALNHAKHGVDILSGTRVRTHF 880
E GDARS LI M+Q L HAK+GVDI+S +RV + F
Sbjct: 789 EAGDARSALIAMVQKLQHAKNGVDIVSESRVCSFF 823
>At5g49740 FRO1-like protein; NADPH oxidase-like
Length = 739
Score = 103 bits (257), Expect = 5e-22
Identities = 69/254 (27%), Positives = 124/254 (48%), Gaps = 32/254 (12%)
Query: 512 TGVEGVTGISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILL 571
TG+ + G+ + + + + H R+N F F+Y+H L V + L
Sbjct: 253 TGIAVLPGVISLVAGLLMWVTSLHTVRKNY------------FELFFYTHQLYIVFVVFL 300
Query: 572 FIH-GSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFS 630
+H G +L + ++ + L+I +R LR +S+ V + LP
Sbjct: 301 ALHVGDYL-----------FSIVAGGIFLFILDRFLRFYQSRR-TVDVISAKSLPCGTLE 348
Query: 631 LIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAP--GDEYLSVHIRTVGDWTQELK 688
L++SKP +Y + +IFLQ ++S +WHPFS++S+P G+ +++V I+ +G WT +L+
Sbjct: 349 LVLSKPPNMRYNALSFIFLQVKELSWLQWHPFSVSSSPLDGNHHVAVLIKVLGGWTAKLR 408
Query: 689 LLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDVLLLIGLGIGAT 748
D + + +L+ + V+GPYG + ++ L+L+ GIG T
Sbjct: 409 -----DQLSTLYEAENQDQLISPESYPKITTCVEGPYGHESPYHLAYENLVLVAGGIGIT 463
Query: 749 PFISILRDLLNNTR 762
PF +IL D+L+ R
Sbjct: 464 PFFAILSDILHRKR 477
>At5g50160 FRO1 and FRO2-like protein
Length = 728
Score = 101 bits (251), Expect = 2e-21
Identities = 100/385 (25%), Positives = 160/385 (40%), Gaps = 83/385 (21%)
Query: 386 VTTASLFAWKFYQYRNRTSFQVMGYCLPVAKGAAETLKLNMALILLPVCRNTLTWLRSTR 445
V T +L W+ YR T F ++ ++L+L PV R L+ R
Sbjct: 140 VKTMNLNLWQLKYYRVATRFGLLAEAC-------------LSLLLFPVLRG-LSMFRLLN 185
Query: 446 ARKIVPFDDNINFHKIIACAIVVGVVVHAGNHLACDFPLLINSSPEEFSLIASDFNNKKP 505
+ F ++ +H ++ +VH G+ L F I EE
Sbjct: 186 ----IEFAASVKYHVWFGTGLIFFSLVHGGSTL---FIWTITHHIEE------------E 226
Query: 506 TYKTLLTGVEGVTG-ISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLL 564
+K TG V G IS+VT + + T R+N F F+Y+HHL
Sbjct: 227 IWKWQRTGRVYVAGLISLVTGLLMWITSLPQIRRKN-------------FEVFYYTHHLY 273
Query: 565 GVVYILLFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVL 624
V + H + + ++ + L+ ++ LR +S+ + +L
Sbjct: 274 IVFLVAFLFHAGDRH----------FYWVLPGMFLFGLDKILRIVQSR------SESCIL 317
Query: 625 PGNVFS-----LIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPG-DEY-LSVHI 677
N+FS L++ K Y +IFL P +S F+WHPFSI S+ D++ LS+ +
Sbjct: 318 SANLFSCKAIELVLPKDPMLNYAPSSFIFLNIPLVSRFQWHPFSIISSSSVDKHSLSIMM 377
Query: 678 RTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDV 737
+ GDWT + E NC ++ + V+GPYG + DF +D
Sbjct: 378 KCEGDWTNSVYNKIEE----AANCENKINNII---------VRVEGPYGPASVDFLRYDN 424
Query: 738 LLLIGLGIGATPFISILRDLLNNTR 762
L L+ GIG TPF+SIL++L + R
Sbjct: 425 LFLVAGGIGITPFLSILKELASKNR 449
>At5g49730 FRO2-like protein; NADPH oxidase-like
Length = 738
Score = 99.4 bits (246), Expect = 9e-21
Identities = 62/212 (29%), Positives = 109/212 (51%), Gaps = 20/212 (9%)
Query: 554 FNAFWYSHHLLGVVYILLFIH-GSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSK 612
F F+Y+H L V + L +H G ++ + ++ + L+I +R LR +S+
Sbjct: 282 FELFFYTHQLYIVFIVFLALHVGDYM-----------FSIVAGGIFLFILDRFLRFCQSR 330
Query: 613 HYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAP--GD 670
V + LP L++SKP +Y + +IFLQ ++S +WHPFS++S+P G+
Sbjct: 331 R-TVDVISAKSLPCGTLELVLSKPPNMRYNALSFIFLQVRELSWLQWHPFSVSSSPLDGN 389
Query: 671 EYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQ 730
+++V I+ +G WT +L+ D + + +L+ V+GPYG +
Sbjct: 390 HHVAVLIKVLGGWTAKLR-----DQLSNLYEAENQDQLISPQSYPKITTCVEGPYGHESP 444
Query: 731 DFKNFDVLLLIGLGIGATPFISILRDLLNNTR 762
++ L+L+ GIG TPF +IL D+L+ R
Sbjct: 445 YHLAYENLVLVAGGIGITPFFAILSDILHRKR 476
>At5g23990 FRO2 homolog
Length = 707
Score = 98.6 bits (244), Expect = 1e-20
Identities = 66/215 (30%), Positives = 101/215 (46%), Gaps = 34/215 (15%)
Query: 547 PLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTTWMYISVP-LLLYIAERT 605
P R F F+Y+HHL G+ + IH +W + +P + L+ +R
Sbjct: 260 PSFRRKKFEIFFYTHHLYGLYIVFYAIHVG-----------DSWFCMILPNIFLFFIDRY 308
Query: 606 LRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSIT 665
LR +S + ++ +LP + L +K +G Y +FL P IS +WHPF+IT
Sbjct: 309 LRFLQSTKRS-RLVSAKILPSDNLELTFAKTSGLHYTPTSILFLHVPSISKLQWHPFTIT 367
Query: 666 SAPG--DEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDG 723
S+ + LSV IR G WTQ+L + +D E + +G
Sbjct: 368 SSSNLEKDTLSVVIRKQGSWTQKLYTHLSS----------------SIDSLE---VSTEG 408
Query: 724 PYGAPAQDFKNFDVLLLIGLGIGATPFISILRDLL 758
PYG + D D L+L+G G G TPFIS++R+L+
Sbjct: 409 PYGPNSFDVSRHDSLILVGGGSGVTPFISVIRELI 443
>At1g23020 putative superoxide-generating NADPH oxidase
flavocytochrome
Length = 693
Score = 95.5 bits (236), Expect = 1e-19
Identities = 68/215 (31%), Positives = 107/215 (49%), Gaps = 32/215 (14%)
Query: 547 PLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTTWMYISVP-LLLYIAERT 605
P R F F+Y+H+L +V++L F+ ++ ++ IS P +++ +R
Sbjct: 269 PAIRRRFFEVFFYTHYLY-MVFMLFFV----------FHVGISYALISFPGFYIFMVDRF 317
Query: 606 LRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSIT 665
LR +S++ VK+ VLP L SK Y +F+ P IS +WHPF+IT
Sbjct: 318 LRFLQSRNN-VKLVSARVLPCETVELNFSKNPMLMYSPTSILFVNIPSISKLQWHPFTIT 376
Query: 666 SAPGDE--YLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDG 723
S+ E LSV I++ G W+ +L + L +Q +H + V+G
Sbjct: 377 SSSKLEPKKLSVMIKSQGKWSSKLHHM-----------------LASSNQIDHLAVSVEG 419
Query: 724 PYGAPAQDFKNFDVLLLIGLGIGATPFISILRDLL 758
PYG + D+ D L+++ G G TPFISI+RDLL
Sbjct: 420 PYGPASTDYLRHDSLVMVSGGSGITPFISIIRDLL 454
>At5g23980 FRO2 homolog
Length = 699
Score = 95.1 bits (235), Expect = 2e-19
Identities = 65/219 (29%), Positives = 102/219 (45%), Gaps = 34/219 (15%)
Query: 547 PLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTTWMYISVP-LLLYIAERT 605
P R F F+Y+HHL G+ + IH +W + +P + L+ +R
Sbjct: 252 PSFRRKKFEIFFYTHHLYGLYIVFYVIHVG-----------DSWFCMILPNIFLFFIDRY 300
Query: 606 LRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSIT 665
LR +S + ++ +LP + L SK G Y +FL P IS +WHPF+IT
Sbjct: 301 LRFLQSTKRS-RLVSARILPSDNLELTFSKTPGLHYTPTSILFLHVPSISKIQWHPFTIT 359
Query: 666 SAPG--DEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDG 723
S+ + LSV IR G WTQ+L + +D E + +G
Sbjct: 360 SSSNLEKDTLSVVIRRQGSWTQKLYTHLSS----------------SIDSLE---VSTEG 400
Query: 724 PYGAPAQDFKNFDVLLLIGLGIGATPFISILRDLLNNTR 762
PYG + D + L+L+ G G TPFIS++R+L++ ++
Sbjct: 401 PYGPNSFDVSRHNSLILVSGGSGITPFISVIRELISQSQ 439
>At1g01580 hypothetical protein
Length = 725
Score = 92.0 bits (227), Expect = 1e-18
Identities = 62/215 (28%), Positives = 106/215 (48%), Gaps = 32/215 (14%)
Query: 547 PLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTTWMYISVP-LLLYIAERT 605
P R F F+Y+H+L +V++L F+ + ++ +I++P +++ +R
Sbjct: 279 PKIRRRFFEVFFYTHYLY-IVFMLFFV----------LHVGISFSFIALPGFYIFLVDRF 327
Query: 606 LRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSIT 665
LR +S+ V++ +LP + L SK + Y +F+ P IS +WHPF+IT
Sbjct: 328 LRFLQSREN-VRLLAARILPSDTMELTFSKNSKLVYSPTSIMFVNIPSISKLQWHPFTIT 386
Query: 666 SAPG--DEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDG 723
S+ E LS+ I+ G W+ +L L DQ + + V+G
Sbjct: 387 SSSKLEPEKLSIVIKKEGKWSTKLHQ-----------------RLSSSDQIDRLAVSVEG 429
Query: 724 PYGAPAQDFKNFDVLLLIGLGIGATPFISILRDLL 758
PYG + DF + L+++ G G TPFIS++RDL+
Sbjct: 430 PYGPASADFLRHEALVMVCGGSGITPFISVIRDLI 464
>At1g01590 hypothetical protein
Length = 704
Score = 88.2 bits (217), Expect = 2e-17
Identities = 64/207 (30%), Positives = 103/207 (48%), Gaps = 30/207 (14%)
Query: 554 FNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTTWMYISVP-LLLYIAERTLRTSRSK 612
F F+YSH+L +V++L F+ + ++H I +P +++ +R LR +S+
Sbjct: 277 FEVFFYSHYLY-IVFMLFFVF--HVGISHA--------LIPLPGFYIFLVDRFLRFLQSR 325
Query: 613 HYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPG--D 670
+ VK+ VLP + L SK Y +F+ P IS +WHPF+I S+
Sbjct: 326 NN-VKLVSARVLPCDTVELNFSKNPMLMYSPTSTMFVNIPSISKLQWHPFTIISSSKLEP 384
Query: 671 EYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQ 730
E LSV I++ G W+ +L + + +N A + V+GPYG +
Sbjct: 385 ETLSVMIKSQGKWSTKLYDMLSSSSSDQINRLA---------------VSVEGPYGPSST 429
Query: 731 DFKNFDVLLLIGLGIGATPFISILRDL 757
DF + L+++ G G TPFISI+RDL
Sbjct: 430 DFLRHESLVMVSGGSGITPFISIVRDL 456
>At5g12480 calcium-dependent protein kinase - like protein
Length = 535
Score = 37.4 bits (85), Expect = 0.040
Identities = 30/112 (26%), Positives = 46/112 (40%), Gaps = 8/112 (7%)
Query: 216 SKEFAVGVFDALARRKGRKASKITKEELHHFWQQISDQSFDARLQIFFDMADSNEDGRIT 275
S E A G+ +A K KI EEL + Q+ Q D LQI + D + DG +
Sbjct: 358 SVEEAAGIKEAFEMMDVNKRGKINLEELKYGLQKAGQQIADTDLQILMEATDVDGDGTL- 416
Query: 276 REEVQELIMLSASANKLSKLKEQAEGYAALIMEELDPENLGYIELWQLEMLL 327
E + +S K++ + + + D GYIE+ +L L
Sbjct: 417 --NYSEFVAVSVHLKKMANDEHLHKAF-----NFFDQNQSGYIEIDELREAL 461
>At4g16351 SOS3-like calcium binding protein
Length = 226
Score = 36.6 bits (83), Expect = 0.068
Identities = 34/142 (23%), Positives = 68/142 (46%), Gaps = 6/142 (4%)
Query: 191 ERFGSLAKDGLLAREDFGGCI-GMDDSKE-FAVGVFDAL-ARRKGRKASKITKEELHHFW 247
E F S++K+GL+ +E F + M+ ++ FA VFD + G + L F
Sbjct: 50 ELFKSISKNGLIDKEQFQLVLFKMNTTRSLFADRVFDLFDTKNTGILDFEAFARSLSVFH 109
Query: 248 QQISDQSFDARLQIFFDMADSNEDGRITREEVQELIMLSASANKLSKLKEQAEGYAALIM 307
+ F+ +++ F + D N+ G I R+EV+++++ + + + ++ E
Sbjct: 110 ---PNAKFEDKIEFSFKLYDLNQQGYIKRQEVKQMVVRTLAESGMNLSDHVIESIIDKTF 166
Query: 308 EELDPENLGYIELWQLEMLLLQ 329
EE D + G I+ + L+L+
Sbjct: 167 EEADTKLDGKIDKEEWRSLVLR 188
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,770,757
Number of Sequences: 26719
Number of extensions: 879908
Number of successful extensions: 3033
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 2884
Number of HSP's gapped (non-prelim): 84
length of query: 937
length of database: 11,318,596
effective HSP length: 109
effective length of query: 828
effective length of database: 8,406,225
effective search space: 6960354300
effective search space used: 6960354300
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0013.6


