

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0012b.1
(307 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
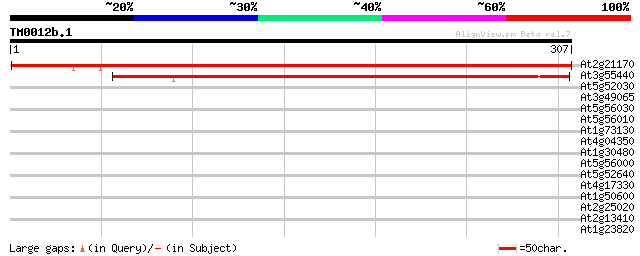
Score E
Sequences producing significant alignments: (bits) Value
At2g21170 putative triosephosphate isomerase 496 e-141
At3g55440 cytosolic triosephosphatisomerase 317 7e-87
At5g52030 unknown protein 40 0.002
At3g49065 unknown protein 33 0.25
At5g56030 HEAT SHOCK PROTEIN 81-2 (HSP81-2) (sp|P55737) 32 0.33
At5g56010 heat shock protein 90 32 0.33
At1g73130 hypothetical protein 31 0.74
At4g04350 putative leucyl tRNA synthetase 30 1.3
At1g30480 DNA damage repair protein, putative 30 1.6
At5g56000 heat shock protein (emb|CAA72514.1) 29 2.8
At5g52640 heat-shock protein 28 4.8
At4g17330 G2484-1 protein 28 6.2
At1g50600 scarecrow-like 5 (SCL5) 28 6.2
At2g25020 hypothetical protein 28 8.2
At2g13410 T26C18.1 28 8.2
At1g23820 spermidine synthase like protein 28 8.2
>At2g21170 putative triosephosphate isomerase
Length = 315
Score = 496 bits (1278), Expect = e-141
Identities = 255/313 (81%), Positives = 277/313 (88%), Gaps = 7/313 (2%)
Query: 2 ATSLASQLSV-GLRRPSPKLDSLNSQSTHSLFH---SNLRLPISSSKPSRS---VIAMAG 54
ATSL + S GLRR SPKLD+ S S FH S+ RL SSS RS V+AMAG
Sbjct: 3 ATSLTAPPSFSGLRRISPKLDAAAVSSHQSFFHRVNSSTRLVSSSSSSHRSPRGVVAMAG 62
Query: 55 SGKFFVGGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVS 114
SGKFFVGGNWKCNGTKDSI+KL++DLNSA LE DVDVVV+PPFVYIDQVK+S+TDRI++S
Sbjct: 63 SGKFFVGGNWKCNGTKDSIAKLISDLNSATLEADVDVVVSPPFVYIDQVKSSLTDRIDIS 122
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
QNSWV KGGAFTGEISVEQLKD G KWVILGHSERRH+IGEKDEFIGKKAAYAL+EGLG
Sbjct: 123 GQNSWVGKGGAFTGEISVEQLKDLGCKWVILGHSERRHVIGEKDEFIGKKAAYALSEGLG 182
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIACIGE LEEREAGKTFDVCF QLKA+ADAVPSWDNIV+AYEPVWAIGTGKVASP+QAQ
Sbjct: 183 VIACIGEKLEEREAGKTFDVCFAQLKAFADAVPSWDNIVVAYEPVWAIGTGKVASPQQAQ 242
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVH A+R WL+KNVS EVASKTRIIYGGSVNGGNS+ELAK+EDIDGFLVGGASLKGPEFA
Sbjct: 243 EVHVAVRGWLKKNVSEEVASKTRIIYGGSVNGGNSAELAKEEDIDGFLVGGASLKGPEFA 302
Query: 295 TIINSVTSKKVAA 307
TI+NSVTSKKVAA
Sbjct: 303 TIVNSVTSKKVAA 315
>At3g55440 cytosolic triosephosphatisomerase
Length = 254
Score = 317 bits (811), Expect = 7e-87
Identities = 159/252 (63%), Positives = 188/252 (74%), Gaps = 3/252 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKLEPD--VDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + K+V LN A++ V+VVV+PP+V++ VK+++ V+
Sbjct: 4 KFFVGGNWKCNGTAEEVKKIVNTLNEAQVPSQDVVEVVVSPPYVFLPLVKSTLRSDFFVA 63
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGAFTGE+S E L + + WVILGHSERR I+ E EF+G K AYAL +GL
Sbjct: 64 AQNCWVKKGGAFTGEVSAEMLVNLDIPWVILGHSERRAILNESSEFVGDKVAYALAQGLK 123
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIAC+GE LEEREAG T DV Q KA AD V +W N+VIAYEPVWAIGTGKVASP QAQ
Sbjct: 124 VIACVGETLEEREAGSTMDVVAAQTKAIADRVTNWSNVVIAYEPVWAIGTGKVASPAQAQ 183
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVH +R WL KNVSA+VA+ TRIIYGGSVNGGN EL + D+DGFLVGGASLK PEF
Sbjct: 184 EVHDELRKWLAKNVSADVAATTRIIYGGSVNGGNCKELGGQADVDGFLVGGASLK-PEFI 242
Query: 295 TIINSVTSKKVA 306
II + KK A
Sbjct: 243 DIIKAAEVKKSA 254
>At5g52030 unknown protein
Length = 391
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/72 (33%), Positives = 34/72 (46%), Gaps = 2/72 (2%)
Query: 206 VPSWDNIVIAYEPVWAIGTGKVASPEQAQEVHAAIRDWLQKNVSAEVA-SKTRIIYGGSV 264
VP+W + E +W +GT + SPE A V +R NV+ E+ S+ I+Y SV
Sbjct: 81 VPAWRKEFVEPEAIWLVGTSHI-SPESASIVERVVRTVKPDNVAVELCRSRAGIMYTSSV 139
Query: 265 NGGNSSELAKKE 276
G L E
Sbjct: 140 GGEVDQNLKSGE 151
>At3g49065 unknown protein
Length = 805
Score = 32.7 bits (73), Expect = 0.25
Identities = 20/81 (24%), Positives = 36/81 (43%), Gaps = 1/81 (1%)
Query: 88 DVDVVVAPPFVYIDQVKNSITDRI-EVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILG 146
DV V+ +D++ NS + E Q + G E VE + H +KW+++G
Sbjct: 72 DVKVIERVEKPKVDELMNSYLQLLSETEIQTDKLCIAGQNIEECIVELIARHKIKWLVMG 131
Query: 147 HSERRHIIGEKDEFIGKKAAY 167
+ +H + + KKA +
Sbjct: 132 AASDKHYSWKMTDLKSKKAIF 152
>At5g56030 HEAT SHOCK PROTEIN 81-2 (HSP81-2) (sp|P55737)
Length = 699
Score = 32.3 bits (72), Expect = 0.33
Identities = 22/95 (23%), Positives = 44/95 (46%), Gaps = 5/95 (5%)
Query: 99 YIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKD 158
Y+ ++K D ++ ++ + F +E+LK G++ + + + + IG+
Sbjct: 451 YVTRMKEGQNDIFYITGESKKAVENSPF-----LEKLKKKGIEVLYMVDAIDEYAIGQLK 505
Query: 159 EFIGKKAAYALTEGLGVIACIGELLEEREAGKTFD 193
EF GKK A EGL + E ++ E + F+
Sbjct: 506 EFEGKKLVSATKEGLKLDETEDEKKKKEELKEKFE 540
>At5g56010 heat shock protein 90
Length = 699
Score = 32.3 bits (72), Expect = 0.33
Identities = 22/95 (23%), Positives = 44/95 (46%), Gaps = 5/95 (5%)
Query: 99 YIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKD 158
Y+ ++K D ++ ++ + F +E+LK G++ + + + + IG+
Sbjct: 451 YVTRMKEGQNDIFYITGESKKAVENSPF-----LEKLKKKGIEVLYMVDAIDEYAIGQLK 505
Query: 159 EFIGKKAAYALTEGLGVIACIGELLEEREAGKTFD 193
EF GKK A EGL + E ++ E + F+
Sbjct: 506 EFEGKKLVSATKEGLKLDETEDEKKKKEELKEKFE 540
>At1g73130 hypothetical protein
Length = 646
Score = 31.2 bits (69), Expect = 0.74
Identities = 26/80 (32%), Positives = 41/80 (50%), Gaps = 8/80 (10%)
Query: 72 SISKLVADLNSAKLEPDVDVVVAPPFVYIDQ--VKNSITDRIEVSAQNSWVSKGGAFTGE 129
S S L+ + A L D + P + D+ + NS+T + + SA NSWV F+GE
Sbjct: 385 SKSPLLEEHCDANLTSDTTLGDEKPIITDDESWISNSLTSQ-KSSAGNSWV-----FSGE 438
Query: 130 ISVEQLKDHGVKWVILGHSE 149
SVE++K + V+ S+
Sbjct: 439 DSVEEVKVKSCRDVVSTESQ 458
>At4g04350 putative leucyl tRNA synthetase
Length = 973
Score = 30.4 bits (67), Expect = 1.3
Identities = 37/151 (24%), Positives = 66/151 (43%), Gaps = 18/151 (11%)
Query: 148 SERRHIIGEKDEFIGKKAAYALTEGLGVIACIGELLEEREAGKTFDVCFQQLKAYADAVP 207
+E++ + E +F +K+ TE L++ + G F C+ + A DA+P
Sbjct: 361 AEQKQQVEEYKDFASRKSDLERTE-----------LQKDKTG-VFTGCYAKNPANGDAIP 408
Query: 208 SW--DNIVIAY--EPVWAIGTGKVASPEQAQEVHAAIRDWLQKNVSAEVASKTRIIYGGS 263
W D ++ +Y + A+ E A + + I+ W+ +N A + + +Y G
Sbjct: 409 IWVADYVLASYGTGAIMAVPAHDTRDNEFALKYNIPIK-WVVRN-EANSSDDAKQVYPGL 466
Query: 264 VNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
NSS L DI+ A+LK E+A
Sbjct: 467 GIIENSSTLETGLDINQLSSKEAALKVIEWA 497
>At1g30480 DNA damage repair protein, putative
Length = 387
Score = 30.0 bits (66), Expect = 1.6
Identities = 14/44 (31%), Positives = 21/44 (46%)
Query: 247 NVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKG 290
N+S E A K R G +GG + ++DGF +G + G
Sbjct: 163 NISGEEAWKRRAAMSGGGSGGKGRSSSPPGNVDGFSIGKSETSG 206
>At5g56000 heat shock protein (emb|CAA72514.1)
Length = 699
Score = 29.3 bits (64), Expect = 2.8
Identities = 21/95 (22%), Positives = 43/95 (45%), Gaps = 5/95 (5%)
Query: 99 YIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKD 158
Y+ ++K + ++ ++ + F +E+LK G + + + + + IG+
Sbjct: 451 YVTRMKEGQNEIFYITGESKKAVENSPF-----LEKLKKKGYEVLYMVDAIDEYAIGQLK 505
Query: 159 EFIGKKAAYALTEGLGVIACIGELLEEREAGKTFD 193
EF GKK A EGL + E ++ E + F+
Sbjct: 506 EFEGKKLVSATKEGLKLEETDDEKKKKEELKEKFE 540
>At5g52640 heat-shock protein
Length = 705
Score = 28.5 bits (62), Expect = 4.8
Identities = 21/92 (22%), Positives = 42/92 (44%), Gaps = 5/92 (5%)
Query: 99 YIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKD 158
Y+ ++K D ++ ++ + F +E+LK G + + + + + +G+
Sbjct: 457 YVTRMKEGQKDIFYITGESKKAVENSPF-----LERLKKRGYEVLYMVDAIDEYAVGQLK 511
Query: 159 EFIGKKAAYALTEGLGVIACIGELLEEREAGK 190
E+ GKK A EGL + E ++RE K
Sbjct: 512 EYDGKKLVSATKEGLKLEDETEEEKKKREEKK 543
>At4g17330 G2484-1 protein
Length = 2037
Score = 28.1 bits (61), Expect = 6.2
Identities = 17/56 (30%), Positives = 35/56 (62%), Gaps = 3/56 (5%)
Query: 223 GTGKVASPEQAQEVHAAIRDWLQKNVSAEVASKTRIIYG--GSVNGGNSSELAKKE 276
G+ + ++ EQ+ E+ D Q ++++ S++ ++ G G V+GG++SELA+ E
Sbjct: 466 GSARTSNLEQSMELPVNANDRDQDVKNSQILSES-VVSGSVGYVSGGSTSELAESE 520
>At1g50600 scarecrow-like 5 (SCL5)
Length = 597
Score = 28.1 bits (61), Expect = 6.2
Identities = 21/77 (27%), Positives = 39/77 (50%), Gaps = 5/77 (6%)
Query: 18 PKLDSLNSQSTHSLFHSNLRLPISSSKPSR----SVIAMAGSGKFFVGGNWKCNGTKDSI 73
P L++ N+ S+ + F SN PIS + + + + + + G+ N + +
Sbjct: 120 PCLNNKNNSSSTTSFSSN-ESPISQANNNNLSRFNNHSPEENNNSPLSGSSATNTNETEL 178
Query: 74 SKLVADLNSAKLEPDVD 90
S ++ DL +A +EPDVD
Sbjct: 179 SLMLKDLETAMMEPDVD 195
>At2g25020 hypothetical protein
Length = 476
Score = 27.7 bits (60), Expect = 8.2
Identities = 17/46 (36%), Positives = 26/46 (55%), Gaps = 1/46 (2%)
Query: 236 VHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSE-LAKKEDIDG 280
V+ A+ + + KN +S +I+GGS G NS + LAK +DG
Sbjct: 58 VYLALEENIVKNDLTLRSSLKHVIFGGSSGGSNSGKGLAKDLYLDG 103
>At2g13410 T26C18.1
Length = 862
Score = 27.7 bits (60), Expect = 8.2
Identities = 15/62 (24%), Positives = 28/62 (44%)
Query: 209 WDNIVIAYEPVWAIGTGKVASPEQAQEVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGN 268
W++ + E AI + + E + +LQ V + S+T ++ GGS +G
Sbjct: 177 WNSFTLDRESARAIAARRKTLSKSIVEACEVNKSFLQSTVDKKEYSRTLLVDGGSADGTR 236
Query: 269 SS 270
S+
Sbjct: 237 SA 238
>At1g23820 spermidine synthase like protein
Length = 334
Score = 27.7 bits (60), Expect = 8.2
Identities = 15/50 (30%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Query: 243 WLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPE 292
WL ++ ++ S R I+ GSVN +S + GF++ S +GP+
Sbjct: 239 WLHMDIIEDIVSNCREIFKGSVNYAWTSVPTYPSGVIGFML--CSTEGPD 286
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,762,591
Number of Sequences: 26719
Number of extensions: 288981
Number of successful extensions: 590
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 578
Number of HSP's gapped (non-prelim): 18
length of query: 307
length of database: 11,318,596
effective HSP length: 99
effective length of query: 208
effective length of database: 8,673,415
effective search space: 1804070320
effective search space used: 1804070320
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0012b.1


