

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0012a.8
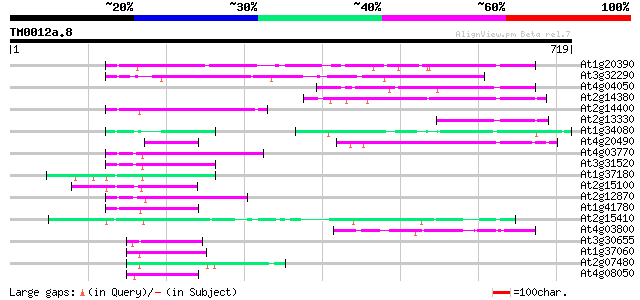
(719 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 148 1e-35
At3g32290 hypothetical protein 102 6e-22
At4g04050 putative transposon protein 99 1e-20
At2g14380 putative retroelement pol polyprotein 93 5e-19
At2g14400 putative retroelement pol polyprotein 79 9e-15
At2g13330 F14O4.9 79 1e-14
At1g34080 putative protein 79 1e-14
At4g20490 putative protein 76 8e-14
At4g03770 hypothetical protein 74 2e-13
At3g31520 hypothetical protein 70 4e-12
At1g37180 hypothetical protein 69 7e-12
At2g15100 putative retroelement pol polyprotein 66 6e-11
At2g12870 putative retroelement pol polyprotein 65 1e-10
At1g41780 hypothetical protein 65 1e-10
At2g15410 putative retroelement pol polyprotein 64 2e-10
At4g03800 64 3e-10
At3g30655 hypothetical protein, 3' partial 51 3e-06
At1g37060 Athila retroelment ORF 1, putative 50 3e-06
At2g07480 F9A16.15 50 4e-06
At4g08050 50 6e-06
>At1g20390 hypothetical protein
Length = 1791
Score = 148 bits (373), Expect = 1e-35
Identities = 152/585 (25%), Positives = 250/585 (41%), Gaps = 87/585 (14%)
Query: 123 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKM----VIIAASDAVKCRMFPS 178
PF+ + +V+I K + L+SY+G +DPK+ L FN + + I DA +C++F
Sbjct: 156 PFTRRITNVSIRGAQK-IKLESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIE 214
Query: 179 TFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYM 238
A WF+ L SI +F +S FL ++ + DL++I Q ESL+ ++
Sbjct: 215 HLTGPAHNWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFV 274
Query: 239 ARYS--AASVKVEDEEPRACALAFKNGL-LPGGLNSKLTRKPARSMGEIRARASTYILDE 295
R+ ++ V DE A +A +N + +T ++ + RAS +I E
Sbjct: 275 DRFKLVVTNITVPDE---AAIVALRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIELE 331
Query: 296 EDDAFKRKRAKLEKGDTSPKRVKKDRSGEDKGDGKQQRSGEGKSVFKLTKEQLYPRRDDY 355
E+ ++ K V V K+ + D
Sbjct: 332 EEKLILARKHNSTKTPACKDAV----------------------VIKVGPD------DSN 363
Query: 356 EQRRPWQSKSHRQREETDMVMNTDVSDMLRGASDANLVDEPEA-PKYHPRDANPKKWCEF 414
E R+ R+ T +++T+ D D P A P Y CE+
Sbjct: 364 EPRQHLDRNPSAGRKPTSFLVSTETPDAKPWNKYIRDADSPAAGPMY----------CEY 413
Query: 415 HRSAGHDTDDCWTLHREIDKLIRAGYQ-GNRQGQWRNSGDQNKAHKREEERA------DT 467
H+S H T++C R + L+ A Y+ G + NK +R E A T
Sbjct: 414 HKSRAHSTENC----RFLQGLLMAKYKSGGITIECDRPPINNKNQRRNETTARQYLNDQT 469
Query: 468 KGKKKQESAAIATKGADDTFA--QHSGPPVGT-------INTIAGGFGGGGDTRAARKRH 518
K E I + ADD A Q +G + ++ I GG D+ + K++
Sbjct: 470 KPPTPAEQGIITS--ADDPAAKRQRNGKAIAAEPVVVRQVHVIMGGLQNCSDSVRSIKQY 527
Query: 519 VRAVNSVHEVAFGFV-----HPD----ITISMADFEGIKPHKDDPIVVQLRMNSFNVRRV 569
+ V VA+ +P+ I+ + D EG+ +DP+VV+L ++ V RV
Sbjct: 528 RKKAEMV--VAWPSSTSTTRNPNQSAPISFTDVDLEGLDTPHNDPLVVELIISDSRVTRV 585
Query: 570 LLDQGSSADIIYGDAFDKLGLTDNGLTPYAGTLVGFAGEQVMVRGYIDLDTIFGEDECAR 629
L+D GSS D+I+ D + +TD + P + L GF G+ VM G I L G
Sbjct: 586 LIDTGSSVDLIFKDVLTAMNITDRQIKPVSKPLAGFDGDFVMTIGTIKLPIFVG----GL 641
Query: 630 VLKVRYLVLQVVASYNVIIGRNTLNRLCAVISTAHLAVKYPLSSG 674
+ V+++V+ A YNVI+G ++++ A+ ST H VK+P +G
Sbjct: 642 IAWVKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVKFPTHNG 686
>At3g32290 hypothetical protein
Length = 612
Score = 102 bits (255), Expect = 6e-22
Identities = 122/501 (24%), Positives = 203/501 (40%), Gaps = 69/501 (13%)
Query: 123 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAASDAVKCRMFPSTFKS 182
PF+ + + I D K L ++ ++G SDPK HL K II S
Sbjct: 164 PFTHMISNAIISDPGK-LRIEYFNGSSDPKGHL-----KSFII----------------S 201
Query: 183 TAMAWFTTLPR---GSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMA 239
A A F R S+ +F++ S+ FL Q+S + + DL+++ QQ E L++++A
Sbjct: 202 VARAKFRQEERDAGNSVDSFQELSTLFLKQYSVLIDLGTSDADLWSLSQQPNEPLRDFLA 261
Query: 240 RYSAASVKVEDEEPRACALAFKNGL-LPGGLNSKLTRKPARSMGEIRARASTYILDEEDD 298
++ + KVE A A K L +L ++ + RAS Y+ EE+
Sbjct: 262 KFRSTLAKVEGINDVAALSALKKALWYKSEFRKELNLSKPLTIRDALHRASDYVSHEEEM 321
Query: 299 AFKRKRAKLEKGDTSPKRVKKDRSGEDKGDGKQQRS---GEGKSVFKLTKEQLYPRRDDY 355
KR + K +P+ K S + G Q + EG++ + R +
Sbjct: 322 ELVAKRHEPSK--QTPRIDKSQHSAPNHKKGAQGWTFVHHEGRNFSGAHNYKADTPRGEA 379
Query: 356 EQRRPWQSKSHRQREETDMVMNTDVSDMLRGASDANLVDEPEAPKYHPRDANPKKWCEFH 415
+ R R RE D+P N +++CE H
Sbjct: 380 ARGRGRGRGRGRGRESYTWTK-----------------DQPAG--------NEQEYCELH 414
Query: 416 RSAGHDTDDCWTLHREIDKLIRAGYQGNRQGQWRNSGDQNKAHKREEERADTKGKKKQES 475
+S GH T C +L ++ AG G + + +A K + E+ + +Q +
Sbjct: 415 KSYGHHTSRCRSLGAKLVAKFLAGEIGGGL-----TIEDLEAEKGKTEQVNAVANPEQAA 469
Query: 476 AAI----ATKGADDTFAQHSGPPV--GTINTIAGGFGGGGDTRAARKRHVRAVNSVHEVA 529
A +G + A P G I TI G DT + K + R ++ A
Sbjct: 470 PAANPERPKRGRGNREADDDEPEAARGRIFTILGDSAFCQDTAPSIKAYQRKADANRNWA 529
Query: 530 FGFVHP--DITISMADFEGIKPHKDDPIVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDK 587
F P ++T D G+ +DP+V+ L + FNV RVL+D GS+ DII+ +
Sbjct: 530 RPFNGPNDEVTFHERDTNGLDRPHNDPLVITLTIGDFNVERVLVDTGSTLDIIFLTTLRE 589
Query: 588 LGLTDNGLTPYAGTLVGFAGE 608
+ + + P ++GF+G+
Sbjct: 590 MKIDMTQIVPTPRPVLGFSGK 610
>At4g04050 putative transposon protein
Length = 681
Score = 98.6 bits (244), Expect = 1e-20
Identities = 81/302 (26%), Positives = 141/302 (45%), Gaps = 36/302 (11%)
Query: 394 DEPEAPKYHPRDANPKKWCEFHRSAGHDTDDCWTLHREIDKLIRAGYQGNRQGQWRNSGD 453
+E EA + + + K+C++H+ G+ T++C R + KLI AG G + +
Sbjct: 368 EENEAAENESPELDLGKYCKYHKKRGYSTEEC----RAVKKLIAAG--GKTKKGSNPKVE 421
Query: 454 QNKAHKREEERADTKGKKKQESAAIATKGADD----------------TFAQHSGPPVGT 497
++EEE+ + K+++ I T + +H P
Sbjct: 422 TPPPDEQEEEQTPKQKKRERTPEGITTPPPAPHKKNMRFPLRLLKFCRSATEHLSPK--R 479
Query: 498 INTIAGGFGGGGDTRAARKRHVRAVNSVHEVAFGFVHPDITISMADFEGI---KPHKDDP 554
I+ I GG D+ + K H R +S + + P+ I+ + E KPH DD
Sbjct: 480 IDFIMGGSQLCNDSINSIKTHQRKADSYTKGKSPMMGPNHQITFWESETTDLNKPH-DDA 538
Query: 555 IVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTDNGLTPYAGTLVGFAGEQVMVRG 614
+V+++ + ++ + +++D GSS D+++ DAF ++G D+ L L GFAG+ G
Sbjct: 539 LVIRIDVGNYELSHIMIDTGSSVDVLFYDAFKRMGHLDSELQGRKTPLTGFAGDTTFSLG 598
Query: 615 YIDLDTIFGEDECARVLK--VRYLVLQVVASYNVIIGRNTLNRLCAVISTAHLAVKYPLS 672
I L TI AR ++ +LV+ A +N I+GR L+ + AV ST H +K+P
Sbjct: 599 TIQLPTI------ARGVRRLTSFLVVNKKAPFNAILGRPWLHAMKAVPSTYHQCIKFPSD 652
Query: 673 SG 674
G
Sbjct: 653 KG 654
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 93.2 bits (230), Expect = 5e-19
Identities = 81/335 (24%), Positives = 142/335 (42%), Gaps = 34/335 (10%)
Query: 377 NTDVSDMLRGASDANLVDEPEAPKYHPRDANPK----KWCEFHRSAGHDTDDCWTLHR-- 430
NTD L S A D +HP +A + ++C+FH+ +GH T C L
Sbjct: 153 NTDPQQFLSSFSVATQRDH-----FHPSEAEVENRLDQYCDFHKRSGHSTAACRHLQSIL 207
Query: 431 -------EIDKLIRAGYQGNRQGQWRNSGDQNKA------HKREEERADTKGKKKQESAA 477
+I+ R N R D N H ++ A ++++
Sbjct: 208 LNKYKKGDIEVQHRQYKSHNNTYAARGGRDGNNVFHRLGPHTGRQQEAPPANEEERHPDM 267
Query: 478 IATKGADDTFAQHSGPPVGT--INTIAGGFGGGGDTRAARKRHVR--AVNSVHEVAFGFV 533
K + QH+ PV +N I GG D+ + K +++ A A +
Sbjct: 268 EPPKKNRENDQQHNDAPVPRRRVNMIMGGLTACRDSFRSIKEYIKSGAATLWSSPATKEM 327
Query: 534 HPDITISMADFEGIKPHKDDPIVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTDN 593
P +T + D G+ +DP+V++L + V R+L+D GSS ++++ D K+ + D
Sbjct: 328 TP-LTFTSEDLFGVDLPHNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDR 386
Query: 594 GLTPYAGTLVGFAGEQVMVRGYIDLDTIFGEDECARVLKVRYLVLQVVASYNVIIGRNTL 653
+ P L GF G +M G I L G +++V+ YN+I+G +
Sbjct: 387 HIKPSVRPLTGFDGNTMMTNGTIKLPIYLG----GAATWHKFVVVDKPTIYNIILGTPWI 442
Query: 654 NRLCAVISTAHLAVKYPLSSGKVGKLKVDQKMARE 688
+ + A+ S+ H +K P S G + ++ +Q +A +
Sbjct: 443 HDMQAIPSSYHQCIKIPTSIG-IETIRGNQNLAHD 476
Score = 32.7 bits (73), Expect = 0.73
Identities = 38/162 (23%), Positives = 66/162 (40%), Gaps = 17/162 (10%)
Query: 2 ETRRRKQHHSPMRQRISPPRRPHRVDLDSPVRTAGVQSPSHPPSPPPPPSPSQVGSREHS 61
+T R QH + ++P D +P+ V++ +P P VG
Sbjct: 16 DTPDRIQHAA-----VAPTGGEQGKDATNPIEVNPVENEPTLNTPTPT-----VGLEPGV 65
Query: 62 PGNSPAPEQQPAVTQEQWRHLMRSI-GNIQQRNEHLQAQLDFYRREQRDDGSREADSVAE 120
G P P + LMR+I IQ+ +QA + Q + E + + E
Sbjct: 66 QGMEGRPALTPPARPDDPGALMRNILAQIQELRSTIQAMSQ--KVHQATSSTPELEQILE 123
Query: 121 FR---PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFN 159
PF+ + +V++ + K + L SY G++DP+ L F+
Sbjct: 124 KNQKTPFAVHISNVSVRHDNK-IKLKSYEGNTDPQQFLSSFS 164
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 79.0 bits (193), Expect = 9e-15
Identities = 58/214 (27%), Positives = 104/214 (48%), Gaps = 9/214 (4%)
Query: 123 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVII----AASDAVKCRMFPS 178
PF+ + S+ I D+ K L L+SY+G DPK +L F + A D C++F
Sbjct: 144 PFTPQITSLRIRDSRK-LNLESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSE 202
Query: 179 TFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYM 238
A+ WFT L GSISNF + S FL Q+S ++ ++ DL+N+ Q E+L+ ++
Sbjct: 203 NLCGQALMWFTQLEPGSISNFNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAFI 262
Query: 239 ARYSAASVKVEDEEPRACALAFKNGL-LPGGLNSKLTRKPARSMGEIRARASTYI-LDEE 296
++ K+ ++ A + GL L ++ + RA+ ++ +++E
Sbjct: 263 TKFKYVLSKLSRISQQSALSALRKGLWYDSRFKEDLILHKLDTIQDALFRANNWMEVEDE 322
Query: 297 DDAFKRKRAKLEKGDTSPKRVKKDRSGEDKGDGK 330
++F ++ + + T P KK E++G K
Sbjct: 323 KESFAKRDKQAKPAVTFPP--KKFEPRENQGPRK 354
Score = 42.4 bits (98), Expect = 0.001
Identities = 35/144 (24%), Positives = 64/144 (44%), Gaps = 13/144 (9%)
Query: 461 EEERADTKGKKKQESAAIATKGADDTFAQHSGP-PVGTI---NTIAGGFGGGG------- 509
E+E+ + KQ A+ ++ GP G + N + F G G
Sbjct: 320 EDEKESFAKRDKQAKPAVTFPPKKFEPRENQGPRKFGLLPFNNNVGKQFQGKGRSNTWIR 379
Query: 510 DTRAARKRHVRAVNSVHEVA--FGFVHPDITISMADFEGIKPHKDDPIVVQLRMNSFNVR 567
D + K+H R V+ ++ F I + + ++ DD +VV L + +F V
Sbjct: 380 DESSVIKKHSRNVHLKLSLSEEVDFQSTSILFDEKETQHLERSHDDALVVTLDVANFEVS 439
Query: 568 RVLLDQGSSADIIYGDAFDKLGLT 591
R+L+D GSS D+I+ +++G++
Sbjct: 440 RILIDTGSSVDLIFLSTLERMGIS 463
>At2g13330 F14O4.9
Length = 889
Score = 78.6 bits (192), Expect = 1e-14
Identities = 49/145 (33%), Positives = 81/145 (55%), Gaps = 10/145 (6%)
Query: 548 KPHKDDPIVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTDNGLTPYAGTLVGFAG 607
KPH DD +V+++ + ++ + +++D GSS D+++ DAF + G D+ L L GFAG
Sbjct: 59 KPH-DDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSKLQGRKTPLTGFAG 117
Query: 608 EQVMVRGYIDLDTIFGEDECARVLK--VRYLVLQVVASYNVIIGRNTLNRLCAVISTAHL 665
+ G I L TI AR ++ +LV+ A +N I+GR L+ + AV ST H
Sbjct: 118 DTTFSIGTIQLPTI------ARGVRQLTNFLVVDKKAPFNAILGRPWLHVMKAVPSTYHQ 171
Query: 666 AVKYPLSSGKVGKLKVDQKMARECY 690
+K+P G + + Q+ +R+CY
Sbjct: 172 CIKFPSYKG-IAVVYGSQRSSRKCY 195
>At1g34080 putative protein
Length = 811
Score = 78.6 bits (192), Expect = 1e-14
Identities = 94/372 (25%), Positives = 142/372 (37%), Gaps = 57/372 (15%)
Query: 367 RQREETDMVMNTDVSDMLRGA-SDANLVDEPEAPKYHPRDAN-----------PKKWCEF 414
R RE + + D L G + P P + P+ N P +CE
Sbjct: 310 RFRESLTVNRPESIQDALHGELPKTSPKASPSKPPFSPKGKNGAFSSNKWVRDPDAYCEI 369
Query: 415 HRSAGHDTDDCWTLHREIDKLIRAGYQGNRQGQWRNSGDQNKAHKREEERADTKGKKKQE 474
H+ GH T DC L R + +G E+ + K+K +
Sbjct: 370 HKMNGHATKDCKALGRLLAAKYVSGEIPYIDVTTIELAQTAYDVLDSEKNSPPNKKQKSD 429
Query: 475 SAAIATKGADDTFAQHSGPPVGTINTIAGGFGGGGDTRAARKRHVRAVNSVHEVAFGFVH 534
A +A KG P + I GG D+ + K H V V
Sbjct: 430 PALVAPKG-----------PKKMVKVIMGGSKLFQDSISVIKHHEGKV----------VT 468
Query: 535 PDITISMADFEGIKPHKDDPIVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTDNG 594
PD KP DD +V+ L + + V+R+L+D GSS D+I+ D ++G++
Sbjct: 469 PD-----------KP-DDDALVISLDVGNCEVQRILIDTGSSVDLIFLDTLVRMGISKKD 516
Query: 595 LTPYAGTLVGFAGEQVMVRGYIDLDTIFGEDECARVLKVRYLVLQVVASYNVIIGRNTLN 654
+ LV F E M G I L V + + V A+YN+I+G L
Sbjct: 517 IKGAPSPLVSFTIETSMSLGTITLPV----TAQGVVKMIEFTVFDRPAAYNIILGTPWLY 572
Query: 655 RLCAVISTAHLAVKYPLSS-------GKVGKLKVDQKMARECYNNCLNLYGKKSALVGHR 707
+ AV ST H VK+PL + KVG V++ + E + + L G + G
Sbjct: 573 EMKAVPSTYHQCVKFPLPTRAAPNPFKKVGNDLVERVVINEEHPDQLVGIGANLEMTGIS 632
Query: 708 CYEIEASDENLD 719
E+ + D N+D
Sbjct: 633 T-EVISHDLNVD 643
Score = 45.4 bits (106), Expect = 1e-04
Identities = 37/142 (26%), Positives = 56/142 (39%), Gaps = 29/142 (20%)
Query: 123 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAASDAVKCRMFPSTFKS 182
PF+ + + I + + L Y+G DPK++L F +IA +
Sbjct: 193 PFTLRISKLRIRE-FQDFKLPGYNGKGDPKEYLTSFQ----VIAGRE------------- 234
Query: 183 TAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYS 242
GSI NF+ S+ F+ Q+ VT L+N+ Q E L+ Y +
Sbjct: 235 -----------GSIDNFKQLSTAFIKQYEYFIKSDVTEAQLWNLSQAADEPLRTYNTAFK 283
Query: 243 AASVKVEDEEPRACALAFKNGL 264
V+V A A KNGL
Sbjct: 284 EIMVQVPGLSDSAAQSALKNGL 305
>At4g20490 putative protein
Length = 853
Score = 75.9 bits (185), Expect = 8e-14
Identities = 72/303 (23%), Positives = 130/303 (42%), Gaps = 27/303 (8%)
Query: 419 GHDTDDCWTLHREIDKL----IRAGYQGNRQGQWRNSG-----------DQNKAHKR--E 461
GH T++C L + K + A Y R SG +Q+ H+ E
Sbjct: 326 GHSTEECKHLLNILGKYKSGEVEAVYHPKNGKNKRKSGSKQSQAGPTHEEQHVDHRLLLE 385
Query: 462 EERADTKGKKKQESAAIATKGADDTFAQHSGPPVGTINTIAGGFGGGGDTRAARKRHVRA 521
++R + + E + + P G I+ I G D+ +A K+ R
Sbjct: 386 DDRVEEARVPENELPGPPKRWRGQQPQRAPDHPRGRISMIMAGLSDDEDSVSALKKRARQ 445
Query: 522 VNSVHEV--AFGFVHPDITISMADFEGIKPHKDDPIVVQLRMNSFNVRRVLLDQGSSADI 579
V SV A I + D GI+ +D +V++L M F+V R+L+D GSS ++
Sbjct: 446 VCSVRVAPEAEPLSQEPIIFTSEDASGIQHPHNDALVIKLVMEDFDVERILIDTGSSVNL 505
Query: 580 IYGDAFDKLGLTDNGLTPYAGTLVGFAGEQVMVRGYIDLDTIFGEDECARVLKVRYLVLQ 639
++ K+G++++ + P + G+ GE M G I L G K +++V+
Sbjct: 506 MFLKTLFKMGISESKIMPKIRPMTGYDGEAKMSIGEIKLQVQVG----GITQKTKFVVID 561
Query: 640 VVASYNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKVGKLKVDQKMARECYNNCLNLYGK 699
YN I+G + + A+ ST ++ P G L+ ++ A+E N + + G+
Sbjct: 562 SEPIYNAILGSPWIYSMKAIPSTNLHTIRQP--EGLTDMLRDREEAAKE--GNLIAIKGR 617
Query: 700 KSA 702
+++
Sbjct: 618 RAS 620
Score = 46.6 bits (109), Expect = 5e-05
Identities = 21/69 (30%), Positives = 37/69 (53%)
Query: 173 CRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGE 232
C++F A+ F+ L SI NF S+ FL Q+ + +DL+++ Q+ GE
Sbjct: 222 CQLFVEGLTGNALTCFSRLEANSIDNFTQLSTAFLKQYRVFIQPGASSSDLWSMTQENGE 281
Query: 233 SLKEYMARY 241
+L +Y+ R+
Sbjct: 282 TLNDYLGRF 290
>At4g03770 hypothetical protein
Length = 464
Score = 74.3 bits (181), Expect = 2e-13
Identities = 57/212 (26%), Positives = 100/212 (46%), Gaps = 13/212 (6%)
Query: 123 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAAS-------DAVKCRM 175
PF+ + + I D K L ++ ++G SDPK HL F ++ +A + DA C +
Sbjct: 218 PFTHMISNAIISDPGK-LRIEYFNGSSDPKGHLKLF---IISVARAKFRPEERDAGLCHL 273
Query: 176 FPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLK 235
F K A+ WFT L S+ +F++ S+ FL Q+S + + DL+++ QQ E L+
Sbjct: 274 FVEHLKGPALNWFTRLKGNSVDSFQELSTLFLKQYSVLIDPSTSDADLWSLSQQPNEPLR 333
Query: 236 EYMARYSAASVKVEDEEPRACALAFKNGL-LPGGLNSK-LTRKPARSMGEIRARASTYIL 293
+++A++ + KVE A K L L L++K L R+ + A
Sbjct: 334 DFLAKFRSTLAKVEGINDLAALSTLKKALCLRARLSAKFLAREIGGGLTIKDLEAEKGKT 393
Query: 294 DEEDDAFKRKRAKLEKGDTSPKRVKKDRSGED 325
++ + ++A PKR + +R +D
Sbjct: 394 EQVNAVANPEQAAPAANSEGPKRGRGNREADD 425
>At3g31520 hypothetical protein
Length = 528
Score = 70.1 bits (170), Expect = 4e-12
Identities = 45/149 (30%), Positives = 76/149 (50%), Gaps = 11/149 (7%)
Query: 123 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAAS-------DAVKCRM 175
PF+ + + I D K L ++ ++G SDPK HL F ++ +A + DA C +
Sbjct: 226 PFTHRISNAIISDPGK-LRIEYFNGSSDPKRHLKSF---IISVARTKFRPEERDAGLCHL 281
Query: 176 FPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLK 235
F K A+ WF+ L S+ +F++ S+ FL Q+S + + DL+++ QQ E L+
Sbjct: 282 FVEHLKGPALCWFSRLEGNSMDSFQELSTLFLKQYSVLIDLGTSDADLWSLSQQPNEPLR 341
Query: 236 EYMARYSAASVKVEDEEPRACALAFKNGL 264
+++A+ + KVE A A K L
Sbjct: 342 DFLAKLRSTLAKVEGINDVAALSALKKAL 370
>At1g37180 hypothetical protein
Length = 661
Score = 69.3 bits (168), Expect = 7e-12
Identities = 56/240 (23%), Positives = 95/240 (39%), Gaps = 25/240 (10%)
Query: 48 PPPSPSQVGSREHSPGNSPAPEQQPAVTQEQWRHLM--------RSIGNIQQRNEHLQAQ 99
PPPS + +P S +P AV+ E + I + +Q + ++
Sbjct: 203 PPPSVEVGTTSTQNPTTSSSPAGFAAVSNETIARTLVPLDPAIRLPISDPRQEPSAVNSE 262
Query: 100 LDFYRRE---------QRDDGSREADSVAEFR---PFSEDVESVAIPDNMKTLVLDSYSG 147
L + + Q + E + V E PF+ + + I + + L Y+G
Sbjct: 263 LQSLQDQIWAMNAKVHQATTSAPEVEKVIEATRRTPFTPRISKLRIRE-FRDFKLPVYNG 321
Query: 148 DSDPKDHLLYFNTKMVIIAAS----DAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFS 203
DP +HL F + + DA C++F A+ WFT L GSI NF+ S
Sbjct: 322 KGDPNEHLTSFQVIVRRVPLEPYEEDAGLCKLFSENLSGPALTWFTQLEEGSIDNFKQLS 381
Query: 204 SKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASVKVEDEEPRACALAFKNG 263
+ F+ Q+ +T L+N+ Q + L+ Y+ + V++ A KNG
Sbjct: 382 TAFIKQYGYFIKSDITEAHLWNLSQSADDPLRTYIIVFKEIMVQIPSLSDSTAQFALKNG 441
Score = 34.7 bits (78), Expect = 0.19
Identities = 34/141 (24%), Positives = 56/141 (39%), Gaps = 32/141 (22%)
Query: 396 PEAPKYHPRDAN----PKKW-------CEFHRSAGHDTDDCWTLHREIDKLIRAGYQGNR 444
P P + P+ N KW C+ H+ GH T DC + + +L+ A Y
Sbjct: 458 PSKPPFSPKGKNGAFPSNKWVQDLNAYCDIHKMNGHATKDC----KALGRLLAAKYASGE 513
Query: 445 QGQW-----RNSGDQNKAHKREEERADTKGKKKQESAAIATKGADDTFAQHSGPPVGTIN 499
++ + N+ + EE +K K+K++ A + KG P +
Sbjct: 514 ISEFDITPIELAQTANETLESEEASPPSK-KQKRDPALVIPKG-----------PKKMVK 561
Query: 500 TIAGGFGGGGDTRAARKRHVR 520
I GG D+ +A K+H R
Sbjct: 562 VIMGGSKLFRDSISAIKQHER 582
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 66.2 bits (160), Expect = 6e-11
Identities = 49/174 (28%), Positives = 77/174 (44%), Gaps = 18/174 (10%)
Query: 80 RHLMRSIGNIQQRNEHLQAQLDFYRRE--QRDDGSREADSVAEFR---PFSEDVESVAIP 134
RH +R + + ++LQ Q+ + Q + E + V E PF+ + + I
Sbjct: 109 RHRLRILSPLNSELQNLQDQIWAMNAKVHQATTSAPEVEKVIEATRRTPFTPRISKLRIR 168
Query: 135 DNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAA--------SDAVKCRMFPSTFKSTAMA 186
+ + L Y+G D K+HL F +IA DA C++F A+
Sbjct: 169 E-FRDFKLPVYNGKGDLKEHLTSFQ----VIAGRVPLEPHEEDAGLCKLFSENLFGLALT 223
Query: 187 WFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMAR 240
WFT L GSI NF+ S+ F+ Q+ N +T L+N Q E L+ Y+ R
Sbjct: 224 WFTQLEEGSIDNFKQLSTAFIKQYEYFINSDITEAHLWNFSQSADEPLRTYIYR 277
Score = 34.3 bits (77), Expect = 0.25
Identities = 19/47 (40%), Positives = 25/47 (52%)
Query: 630 VLKVRYLVLQVVASYNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKV 676
V V + V A+YNVI+G L + V ST H VK+P GK+
Sbjct: 366 VKMVEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPTPVGKM 412
>At2g12870 putative retroelement pol polyprotein
Length = 411
Score = 65.5 bits (158), Expect = 1e-10
Identities = 53/191 (27%), Positives = 82/191 (42%), Gaps = 14/191 (7%)
Query: 123 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAASDAVK--------CR 174
PF+ + S I D K L +D ++G S PK HL F +I AA D K C
Sbjct: 61 PFTRKISSAIISDPGK-LRIDYFNGSSYPKGHLKSF----IIYAARDKFKPEERDAGLCH 115
Query: 175 MFPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESL 234
+F K + F L + +F + S+ FL QFS + +DL+++ QQ E L
Sbjct: 116 LFVEHLKGPVLVLFLRLEGNFVDSFEELSTLFLKQFSVLIDPGTLDSDLWSLSQQPNEPL 175
Query: 235 KEYMARYSAASVKVEDEEPRACALAFKNGL-LPGGLNSKLTRKPARSMGEIRARASTYIL 293
+ + ++ + VE A A K L +L +++ + RAS Y+
Sbjct: 176 MDLLTKFRSTLASVEGIIDVAALSALKEALWYKSEFRKELNLSKPQTICDALHRASDYVS 235
Query: 294 DEEDDAFKRKR 304
EE+ KR
Sbjct: 236 HEEEMELLSKR 246
Score = 38.1 bits (87), Expect = 0.017
Identities = 28/101 (27%), Positives = 47/101 (45%), Gaps = 6/101 (5%)
Query: 481 KGADDTFAQHSGPPVGTINTIAGGFGGGGDTRAARKRHVRAVNSVHEVAFGFVHPD--IT 538
+G + + S G I TI G DT + ++A +S + F PD +T
Sbjct: 307 RGNREAYDDESEAARGRIFTILGDSVFCHDTTTS----IKAYHSNRNLTRPFNGPDDKVT 362
Query: 539 ISMADFEGIKPHKDDPIVVQLRMNSFNVRRVLLDQGSSADI 579
++ + +DP+V+ L + +FNV RVL+D GS +
Sbjct: 363 FHESNINCLDQPHNDPLVITLTIGAFNVERVLVDTGSDISL 403
>At1g41780 hypothetical protein
Length = 246
Score = 65.5 bits (158), Expect = 1e-10
Identities = 37/123 (30%), Positives = 63/123 (51%), Gaps = 5/123 (4%)
Query: 123 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIA----ASDAVKCRMFPS 178
PF+ + + I D K L ++ +SG SDPK HL F + SDA C +F
Sbjct: 125 PFTRKISNAIISDPEK-LRIEYFSGSSDPKGHLKSFKISVARAKFKPEESDAGLCHLFVE 183
Query: 179 TFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYM 238
K + WF+ L S+ +F++ S+ FL Q+S + + DL+++ QQ E L +++
Sbjct: 184 HLKGPVLDWFSRLEGNSVDSFQELSTLFLKQYSVLIDPGTSDVDLWSLSQQPNEPLSDFL 243
Query: 239 ARY 241
++
Sbjct: 244 TKF 246
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 64.3 bits (155), Expect = 2e-10
Identities = 133/633 (21%), Positives = 221/633 (34%), Gaps = 123/633 (19%)
Query: 50 PSPSQVGS-REHSPGNSPAPEQQPAVTQEQWRHLMRSIGNIQQRNEHLQAQL-DFYRREQ 107
P+ VGS EH+ N+P + +P V R G + + L+ L D +
Sbjct: 158 PASIVVGSIDEHTRENAPNRDNRPNVDTRARRTEDDDTGARDRELQQLRTTLKDTSSKMH 217
Query: 108 RDDGSR-EADSVAEFR---PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMV 163
R S E D V E PF+ + V I ++ + L +Y G DP+ L + +
Sbjct: 218 RAVSSTPEVDKVLEETQRSPFTSCISDVRIR-HISKIKLANYEGLVDPRPFLTSVSIAIG 276
Query: 164 IIAASD----AVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVT 219
SD A C++F A+ WF+ L SI + ++ FL + + +
Sbjct: 277 RAHFSDEDRDAGSCQLFVKHLSGAALTWFSRLEANSIDSVHALTTSFLKNYGVFMEKGAS 336
Query: 220 INDLYNIRQQEGESLKEYMARYSAASVKVEDEEPRACALAFKNGLLPGGLNSKLTR-KPA 278
DL+ + Q ESL+ ++ R+ V + A A A +N L G + T KP
Sbjct: 337 NVDLWTMAQTAKESLRSFIGRFKEIVTSVATPDDAAIA-ALRNALWHGDSSRCTTLCKPF 395
Query: 279 RSMGEIRARASTYILDEEDDAFKRKRAKLEKGDTSPKRVKKDRSGEDKGDGKQQRSGEGK 338
G +A + + K ++G + R D K+++ +G
Sbjct: 396 HRQGSAKAPIA--------------KEKPKEGH------HESRQHYDADYAKEEKLKKGT 435
Query: 339 SVFKLTKEQLYPRRDDYEQRRPWQSKSHRQREETDMVMNTDVSDMLRGASDANLVDEPEA 398
S + PR D +PW + + D
Sbjct: 436 SYYV---GDTAPRSD-----KPWNKWNRSADAKGD------------------------- 462
Query: 399 PKYHPRDANPKKWCEFHRSAGHDTDDCWTLHREIDKLIRAG--------------YQGNR 444
+K+ EFH+ GH TD+C L + + G + N
Sbjct: 463 ----------QKYYEFHKIPGHSTDECRQLQTLLLTKFKKGDLDIEPDRRRTGTNDRDNT 512
Query: 445 QGQWRNSGDQNKAHKREEERADTKGKK--KQESAAIATKGADDTFAQHSGPPVG--TINT 500
+ N+ N+ KR ++ D + ++ + A K D Q P N
Sbjct: 513 HRRVENTDRDNQDDKRRQDETDRRAERIHDDDRRAEPHKRNHDAPRQFEDEPASKRRRNM 572
Query: 501 IAGGFGGGGDTRAARKRHVRAVNSVH-----EVAFGFVHPDITISMADFEGIKPHKDDPI 555
I GG D+ + K + R + + IT S AD + +DP+
Sbjct: 573 IMGGLTACRDSVRSIKSYRRQGEKQRAWIEKNTSAVECNEPITFSEADI-ALPGSHNDPL 631
Query: 556 VVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTDNGLTPYAGTLVGFAGEQVMVRGY 615
VV+L++ + ++ +D S D + L GF G+ +M G
Sbjct: 632 VVELKIGENVLLQLEIDVRSFQDKVQ-------------------PLTGFDGDTIMTVGT 672
Query: 616 IDLDTIFGEDECARVLKVRYLVLQVVASYNVII 648
I L G V + V+ YNVI+
Sbjct: 673 ITLPIYVG----GTVSWFEFAVVDKPIIYNVIL 701
>At4g03800
Length = 637
Score = 63.9 bits (154), Expect = 3e-10
Identities = 67/262 (25%), Positives = 114/262 (42%), Gaps = 37/262 (14%)
Query: 416 RSAGHDTDDCWTLHREIDKLIRAGYQGNRQGQWRNSGDQNKAHKREEERADTKGKKKQES 475
R H T DC L R + +L +G N D K + + ++T+ K+
Sbjct: 77 RVTEHLTKDCIVLKRYLAELWASGDLSKF-----NIDDFVKQYHEARDNSETQNSKRPRL 131
Query: 476 AAIATKGADDTFAQHSGPPVGTINTIAGGFGGGGDTRAARKRH---VRAVNSVHEVAFGF 532
T + G IN I GG ++ +A K+H V +S+ E +
Sbjct: 132 ----------TNEEEPRSSKGKINVILGGSKLCHNSVSAIKKHRPNVLLKSSLSEEV-NY 180
Query: 533 VHPDITISMADFEGIKPHKDDPIVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTD 592
++ + I+ DD +++ L + +F + R+L+D GSS D+I+
Sbjct: 181 QCSSVSFDEEETRHIERPHDDALIITLDVANFKISRILVDTGSSVDLIF----------- 229
Query: 593 NGLTPYAGTLVGFAGEQVMVRGYIDLDTIFGEDECARVLKVRYLVLQVVASYNVIIGRNT 652
L P LV F E M G I L + G + +++ V ++V A+YN+I+G
Sbjct: 230 --LGP-PSPLVAFTSESAMSLGTIKLPVLAG--KMSKI--VDFVVFDKPATYNIILGTPW 282
Query: 653 LNRLCAVISTAHLAVKYPLSSG 674
+ ++ AV ST H +K+P S+G
Sbjct: 283 IFQMKAVPSTYHQWLKFPTSNG 304
>At3g30655 hypothetical protein, 3' partial
Length = 660
Score = 50.8 bits (120), Expect = 3e-06
Identities = 33/103 (32%), Positives = 48/103 (46%), Gaps = 6/103 (5%)
Query: 150 DPKDHL-----LYFNTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSS 204
DP DHL LY TK+ ++ D K R+FP + A W TLP+ SI+ + D
Sbjct: 61 DPLDHLDEFERLYGLTKINGVS-EDGFKLRLFPFSLGDKAHLWEKTLPQNSITAWDDCKK 119
Query: 205 KFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASVK 247
FL +F +N N++ Q++ ES E R+ K
Sbjct: 120 AFLAKFFSNSRTARLRNEISGFTQKQNESFGEAWERFKGYQTK 162
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 50.4 bits (119), Expect = 3e-06
Identities = 30/107 (28%), Positives = 46/107 (42%), Gaps = 4/107 (3%)
Query: 150 DPKDHLLYFNTKMVII----AASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 205
DP DHL F+ + + D K R+FP + A W +LP+GSI+++ D
Sbjct: 61 DPLDHLDEFDRLCSLTKINRVSEDGFKLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKA 120
Query: 206 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASVKVEDEE 252
FL +F +N ND+ Q E+ E R+ + E
Sbjct: 121 FLAKFFSNSRTARLRNDISGFTQTNNETFYEAWERFKGYQTQCPHHE 167
>At2g07480 F9A16.15
Length = 1012
Score = 50.1 bits (118), Expect = 4e-06
Identities = 50/214 (23%), Positives = 83/214 (38%), Gaps = 15/214 (7%)
Query: 150 DPKDHLLYFNTKMVII----AASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 205
DP DHL F+ + + ++ K R+FP + A W TLP SI + D
Sbjct: 56 DPLDHLDNFDRLCSLTKINGVSEESFKLRLFPFSLGDKAHLWEKTLPVKSIDTWDDCKKA 115
Query: 206 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASVKVEDEE---PRACALAFK- 261
FL +F +N + +++ Q+ ES E R+ + + E P+ L
Sbjct: 116 FLAKFFSNSRKARLRSEISGFNQKNSESFSEAWERFKGYTTQCPHHESLPPQYSILRCLP 175
Query: 262 --NGLLPGGLNSKLTRKPARSMGEIRARASTYILDEEDDAFKRKRAKLEKGDTSPKRVKK 319
LL N + K E+ + + +D + R + D K +K
Sbjct: 176 KIRMLLDTASNGNILNKDVAEGWELVENLAQSHRNYNEDYDRTNRGSSDSEDKHKKEIKT 235
Query: 320 DRSGEDKGDGKQQRSGEGKSVFKLTKEQLYPRRD 353
DK QQR +V+ +T+E+L +D
Sbjct: 236 LNDRIDKLVLAQQR-----NVYYITEEELTQLQD 264
>At4g08050
Length = 1428
Score = 49.7 bits (117), Expect = 6e-06
Identities = 30/96 (31%), Positives = 43/96 (44%), Gaps = 4/96 (4%)
Query: 150 DPKDHLLYF----NTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 205
DP DHL F N + + D K R+FP + A W L SI+ + D+
Sbjct: 61 DPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLSHDSITTWDDYKKA 120
Query: 206 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARY 241
FL +F +N N++Y Q+ GES E R+
Sbjct: 121 FLSKFFSNARTARLRNEIYGFSQKTGESFCEAWERF 156
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,340,843
Number of Sequences: 26719
Number of extensions: 837867
Number of successful extensions: 8437
Number of sequences better than 10.0: 281
Number of HSP's better than 10.0 without gapping: 139
Number of HSP's successfully gapped in prelim test: 152
Number of HSP's that attempted gapping in prelim test: 5362
Number of HSP's gapped (non-prelim): 1393
length of query: 719
length of database: 11,318,596
effective HSP length: 106
effective length of query: 613
effective length of database: 8,486,382
effective search space: 5202152166
effective search space used: 5202152166
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0012a.8


