

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0012a.7
(1015 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
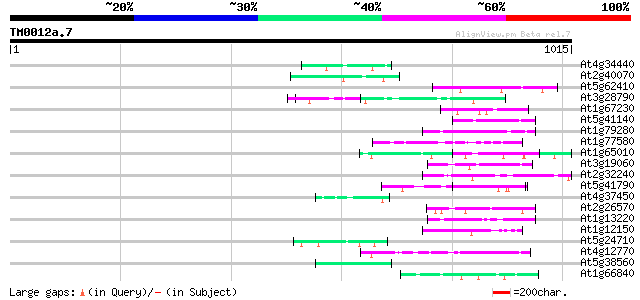
Score E
Sequences producing significant alignments: (bits) Value
At4g34440 serine/threonine protein kinase - like 50 5e-06
At2g40070 En/Spm-like transposon protein 50 5e-06
At5g62410 chromosomal protein - like 48 2e-05
At3g28790 unknown protein 48 3e-05
At1g67230 unknown protein 48 3e-05
At5g41140 putative protein 47 5e-05
At1g79280 hypothetical protein 47 5e-05
At1g77580 unknown protein 47 5e-05
At1g65010 hypothetical protein 47 5e-05
At3g19060 hypothetical protein 47 7e-05
At2g32240 putative myosin heavy chain 47 7e-05
At5g41790 myosin heavy chain-like protein 46 9e-05
At4g37450 arabinogalactan protein AGP18 46 9e-05
At2g26570 unknown protein 46 9e-05
At1g13220 putative nuclear matrix constituent protein 46 9e-05
At1g12150 hypothetical protein 46 9e-05
At5g24710 unknown protein 45 2e-04
At4g12770 auxilin-like protein 45 2e-04
At5g38560 putative protein 45 2e-04
At1g66840 hypothetical protein 45 2e-04
>At4g34440 serine/threonine protein kinase - like
Length = 670
Score = 50.4 bits (119), Expect = 5e-06
Identities = 43/181 (23%), Positives = 65/181 (35%), Gaps = 24/181 (13%)
Query: 528 AENQAESTKAPKRRRLTRTSSGAGTSNPGAQPTTAAAPKGKNVA------EASIAAATEP 581
A++ +S+ AP+ T S+G SN + PT ++P +++ AS + P
Sbjct: 2 ADSPVDSSPAPETSNGTPPSNGTSPSNESSPPTPPSSPPPSSISAPPPDISASFSPPPAP 61
Query: 582 TAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPH 641
S P S ++ V A S N + A TP+ P
Sbjct: 62 PTQETSPPTSPSSSPPVVANPS-----PQTPENPSPPAPEGSTPVTPPAPPQTPSNQSPE 116
Query: 642 QEAPPSPPPTRDA-----------GSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGS 690
+ PPSP D GS PSPP G ++ + S I +P G
Sbjct: 117 RPTPPSPGANDDRNRTNGGNNNRDGSTPSPPSSGNRTSGDGGSPSPPRSI--SPPQNSGD 174
Query: 691 S 691
S
Sbjct: 175 S 175
>At2g40070 En/Spm-like transposon protein
Length = 510
Score = 50.4 bits (119), Expect = 5e-06
Identities = 50/214 (23%), Positives = 85/214 (39%), Gaps = 23/214 (10%)
Query: 508 RMARFSPKFNTEGDAKKR---GGAENQAESTKAPKRRRLTRTSSGAGTSNPGAQPTTAAA 564
R SP ++ A +R G +T + LT S + S P ++ T ++A
Sbjct: 131 RQQTSSPGLSSSSGASRRPSSSGGPGSRPATPTGRSSTLTANSKSSRPSTPTSRATVSSA 190
Query: 565 PKGKNV-AEASIAAATEPTAVPASTPASAGATAAVAAES------SVGATAASAGVNATK 617
+ + ++++A T+PT + ST S+ A++ S G+ S TK
Sbjct: 191 TRPSLTNSRSTVSATTKPTPMSRSTSLSSSRLTPTASKPTTSTARSAGSVTRSTPSTTTK 250
Query: 618 AA--ASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATT- 674
+A + S TP+ + TP S P+ PP++ +P + S A TT
Sbjct: 251 SAGPSRSTTPLSRSTARSSTPTS------RPTLPPSKTISRSSTPTRRPIASASAATTTA 304
Query: 675 ----SEAAQIEQAPAPEVGSSSYYNMLPNAIEPS 704
S+ APA + + S L A P+
Sbjct: 305 NPTISQIKPSSPAPAKPMPTPSKNPALSRAASPT 338
>At5g62410 chromosomal protein - like
Length = 1175
Score = 48.1 bits (113), Expect = 2e-05
Identities = 60/251 (23%), Positives = 117/251 (45%), Gaps = 26/251 (10%)
Query: 766 KHQEAAAAWEKRFDKLATQAGK-DKVYADKMIG-TAGIKIGELEDQLALMK-------EE 816
K +E AA ++RF +L+T + +K + + G ++G + LEDQL K E
Sbjct: 354 KSEEGAADLKQRFQELSTTLEECEKEHQGVLAGKSSGDEEKCLEDQLRDAKIAVGTAGTE 413
Query: 817 ADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKALAEQEKKSAES 876
+L + C+KE ++ + +++ E I E+EL ++E VKKAL +
Sbjct: 414 LKQLKTKIEHCEKELKERKSQLMSKLEEAIEVENELGARKNDVEHVKKALESIPYNEGQM 473
Query: 877 LALAK---SDMEAV------MQATSEEIKKATETHAEALATKD-AEIASQLAKIKSLEDE 926
AL K +++E V ++ S ++ T+++ + D +++ +AK+ ++D
Sbjct: 474 EALEKDRGAELEVVQRLEDKVRGLSAQLANFQFTYSDPVRNFDRSKVKGVVAKLIKVKDR 533
Query: 927 LATEKAKAIEAREQAADIALDNRERGFYLAKDQAQH----LYPNFDFSAMGVMKEI--TT 980
++ A + A + D+ +D+ + G L ++ A + P + V + T
Sbjct: 534 -SSMTALEVTAGGKLYDVVVDSEDTGKQLLQNGALRRRVTIIPLNKIQSYVVQPRVQQAT 592
Query: 981 AGLVGPDDPPL 991
A LVG D+ L
Sbjct: 593 ARLVGKDNAEL 603
Score = 36.2 bits (82), Expect = 0.097
Identities = 52/243 (21%), Positives = 100/243 (40%), Gaps = 61/243 (25%)
Query: 758 ERLKAESAKHQEAAAAWEKRFDKLA--------TQAGKDKVYADKMIGTAGIKIGELEDQ 809
E+L+ E +++ + A D+L QA K + A +G K+G+++ +
Sbjct: 208 EKLRKEKSQYMQWANG-NAELDRLRRFCIAFEYVQAEKIRDNAVLGVGEMKAKLGKIDAE 266
Query: 810 LALMKEEADELDASLRACKKEKEQA----------EKDSIA------------RGEALIA 847
+EE E + ++A + KE + + DS+A + + L+
Sbjct: 267 TEKTQEEIQEFEKQIKALTQAKEASMGGEVKTLSEKVDSLAQEMTRESSKLNNKEDTLLG 326
Query: 848 KESELAVLCAELELVKKALAEQE---KKSAESLALAKSDMEAVMQATSEEIKKATETHAE 904
++ + + +E +KK++ E+ KKS E A D++ Q S +++ + H
Sbjct: 327 EKENVEKIVHSIEDLKKSVKERAAAVKKSEEGAA----DLKQRFQELSTTLEECEKEHQG 382
Query: 905 ALATK--------------DAEIASQLA---------KIKSLEDELATEKAKAIEAREQA 941
LA K DA+IA A KI+ E EL K++ + E+A
Sbjct: 383 VLAGKSSGDEEKCLEDQLRDAKIAVGTAGTELKQLKTKIEHCEKELKERKSQLMSKLEEA 442
Query: 942 ADI 944
++
Sbjct: 443 IEV 445
Score = 31.2 bits (69), Expect = 3.1
Identities = 32/173 (18%), Positives = 72/173 (41%), Gaps = 6/173 (3%)
Query: 758 ERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEA 817
E L+ ++ +E A++ FD ++ K + G ++ +LE + +K +
Sbjct: 748 EELEEAKSQIKEKELAYKNCFDAVSKLENSIKDHDKNREG----RLKDLEKNIKTIKAQM 803
Query: 818 DELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESL 877
L++ + EKE+ + A + + ES L L ++ + + EQ K ++L
Sbjct: 804 QAASKDLKSHENEKEKLVMEEEAMKQEQSSLESHLTSLETQISTLTSEVDEQRAK-VDAL 862
Query: 878 ALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATE 930
+ A ++ ++K+ +T T + +L+ +K +L E
Sbjct: 863 QKIHDESLAELKLIHAKMKEC-DTQISGFVTDQEKCLQKLSDMKLERKKLENE 914
>At3g28790 unknown protein
Length = 608
Score = 47.8 bits (112), Expect = 3e-05
Identities = 83/395 (21%), Positives = 146/395 (36%), Gaps = 41/395 (10%)
Query: 518 TEGDAKKRGGAENQAESTKAPKRRRLTRTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAA 577
TE +K A+ + S + T S +G S G+ T + P ++
Sbjct: 239 TEAGSKSSSSAKTKEVSGGSSGNTYKDTTGSSSGASPSGSPTPTPSTPTPSTPTPSTPTP 298
Query: 578 ATEPTAVPA-STPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETP 636
+T + P STPA + T A S G+ +AS + + S G K ET
Sbjct: 299 STPTPSTPTPSTPAPS--TPAAGKTSEKGSESASMKKESNSKSESESAASGSVSKTKETN 356
Query: 637 KSPPHQ-------EAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVG 689
K + SP + PS G+ S G+A+ S A + + G
Sbjct: 357 KGSSGDTYKDTTGTSSGSPSGSPSGSPTPSTSTDGKASSKGSASASAGA----SASASAG 412
Query: 690 SSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEK 749
+S+ +E A ++ K S + +E F EK
Sbjct: 413 ASA----------SAEESAASQKKESNSKSSSSSSSTTSVKEVETQTSSEVNSFISNLEK 462
Query: 750 FDAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQ 809
N E ++ E K +A+A KL+T K+ V + +A KI E
Sbjct: 463 KYTGNSEL-KVFFEKLKTSMSASA------KLSTSNAKELVTG---MRSAASKIAEAMMF 512
Query: 810 LALMKEEADELDASLRACKKEKEQAEKD------SIARGEALIA-KESELAVLCAELELV 862
++ +++E S+ +C++E Q+ K+ I G+ + + +++EL + E V
Sbjct: 513 VSSRFSKSEETKTSMASCQQEVMQSLKELQDINSQIVSGKTVTSTQQTELKQTITKWEQV 572
Query: 863 KKALAEQEKKSAESLALAKSDMEAVMQATSEEIKK 897
E S+ S + + S + Q +++ K
Sbjct: 573 TTQFVETAASSSSSSSSSSSSSSSSSQGSAKMAMK 607
Score = 43.5 bits (101), Expect = 6e-04
Identities = 36/140 (25%), Positives = 61/140 (42%), Gaps = 17/140 (12%)
Query: 503 KARERRMARFSPKFNTEGDAKKRGGAENQAESTKAPKR-------RRLTRTSSGAGTSNP 555
K E+ S K + ++ A TK + + T TSSG+ + +P
Sbjct: 320 KTSEKGSESASMKKESNSKSESESAASGSVSKTKETNKGSSGDTYKDTTGTSSGSPSGSP 379
Query: 556 GAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNA 615
PT + + GK ++ S +A+ A ASA A A+ +AE S A+ +
Sbjct: 380 SGSPTPSTSTDGKASSKGSASAS-------AGASASASAGASASAEES---AASQKKESN 429
Query: 616 TKAAASSDTPIGEKEKENET 635
+K+++SS + KE E +T
Sbjct: 430 SKSSSSSSSTTSVKEVETQT 449
Score = 42.4 bits (98), Expect = 0.001
Identities = 80/398 (20%), Positives = 137/398 (34%), Gaps = 61/398 (15%)
Query: 546 TSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVG 605
+S +SN +Q T++ + G SI T S +S+ T V+ SS
Sbjct: 204 SSDNESSSNTKSQGTSSKS--GSESTAGSIETNTGSKTEAGSKSSSSAKTKEVSGGSSGN 261
Query: 606 ATAASAGVNATKAAASSDTPIGEKEKENETPKSP-PHQEAPPSPPPTRDAGSMPSPPHQG 664
+ G ++ + + S TP TP +P P P +P P+ S P+P
Sbjct: 262 TYKDTTGSSSGASPSGSPTP----TPSTPTPSTPTPSTPTPSTPTPSTPTPSTPAP---- 313
Query: 665 EKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSRG 724
S P A TSE + E S S SE +G E S G
Sbjct: 314 --STPAAGKTSEKGSESASMKKESNSKS----------ESESAASGSVSKTKETNKGSSG 361
Query: 725 LNDTKEETLACLLRAGCIFAHTFEKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLATQ 784
DT ++T T + + + S D A+
Sbjct: 362 --DTYKDTTG-----------TSSGSPSGSPSGSPTPSTST-------------DGKASS 395
Query: 785 AGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEA 844
G A + E+ A K+E++ +S + ++ E + + +
Sbjct: 396 KGSASASAGASASASAGASASAEESAASQKKESNSKSSSSSSSTTSVKEVETQTSSEVNS 455
Query: 845 LIAK-------ESELAVLCAELELVKKALAEQEKKSAESLAL----AKSDMEAVMQATSE 893
I+ SEL V +L+ A A+ +A+ L A S + M S
Sbjct: 456 FISNLEKKYTGNSELKVFFEKLKTSMSASAKLSTSNAKELVTGMRSAASKIAEAMMFVSS 515
Query: 894 EIKKATETHAEALATKDAEIASQLAKIKSLEDELATEK 931
K+ ET ++A+ E+ L +++ + ++ + K
Sbjct: 516 RFSKSEETKT-SMASCQQEVMQSLKELQDINSQIVSGK 552
>At1g67230 unknown protein
Length = 1132
Score = 47.8 bits (112), Expect = 3e-05
Identities = 50/188 (26%), Positives = 80/188 (41%), Gaps = 32/188 (17%)
Query: 779 DKLATQAGKDKVYADKMIGTAGIKIGELED-------QLALMKEEADELDASLRACKKEK 831
DK+ Q GK+ A K I A + + +LED LAL ++E D L S+ +E
Sbjct: 264 DKIIKQKGKELEEAQKKIDAANLAVKKLEDDVSSRIKDLALREQETDVLKKSIETKAREL 323
Query: 832 EQAEKDSIARGEALIAK-----ESELAVLCAELEL------------VKKALAEQEKKSA 874
+ ++ AR + + + +++L E EL +K +AE EK+ A
Sbjct: 324 QALQEKLEAREKMAVQQLVDEHQAKLDSTQREFELEMEQKRKSIDDSLKSKVAEVEKREA 383
Query: 875 ESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAE---IASQLAKIKSLEDELATEK 931
E ME + + + + E H E D I+ + +KS E L TEK
Sbjct: 384 E-----WKHMEEKVAKREQALDRKLEKHKEKENDFDLRLKGISGREKALKSEEKALETEK 438
Query: 932 AKAIEARE 939
K +E +E
Sbjct: 439 KKLLEDKE 446
Score = 42.0 bits (97), Expect = 0.002
Identities = 50/252 (19%), Positives = 121/252 (47%), Gaps = 21/252 (8%)
Query: 715 VIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFDAANVEAERLKAESAKHQEAAAAW 774
V + +++ + D E+ + + G +K DAAN+ ++L+ + + + A
Sbjct: 246 VAKSQMIVKQREDRANESDKIIKQKGKELEEAQKKIDAANLAVKKLEDDVSSRIKDLALR 305
Query: 775 EKRFD----KLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLRACKKE 830
E+ D + T+A + + +K+ + + +L D E +LD++ R + E
Sbjct: 306 EQETDVLKKSIETKARELQALQEKLEAREKMAVQQLVD------EHQAKLDSTQREFELE 359
Query: 831 KEQAEKDSIARGEALIAKESELAVLCAELELVKKALAEQEK---KSAESLALAKSDMEAV 887
EQ K + ++L +K +E+ AE + +++ +A++E+ + E ++D +
Sbjct: 360 MEQKRK---SIDDSLKSKVAEVEKREAEWKHMEEKVAKREQALDRKLEKHKEKENDFDLR 416
Query: 888 MQATSEEIKKATETHAEALATKDAEIASQ---LAKIKSLEDELATE-KAKAIEAREQAAD 943
++ S +KA ++ +AL T+ ++ + +K+L ++++ E +A+ E ++ +
Sbjct: 417 LKGISGR-EKALKSEEKALETEKKKLLEDKEIILNLKALVEKVSGENQAQLSEINKEKDE 475
Query: 944 IALDNRERGFYL 955
+ + ER YL
Sbjct: 476 LRVTEEERSEYL 487
Score = 35.8 bits (81), Expect = 0.13
Identities = 32/146 (21%), Positives = 68/146 (45%), Gaps = 10/146 (6%)
Query: 802 KIGELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEAL--IAKESE-----LAV 854
+I + Q L+++EA++L A + +KE E+ ++ G L I + E + +
Sbjct: 496 QIEKCRSQQELLQKEAEDLKAQRESFEKEWEELDERKAKIGNELKNITDQKEKLERHIHL 555
Query: 855 LCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIA 914
L+ K+A E ++ E+L +AK+ M+ + K E+ L +I
Sbjct: 556 EEERLKKEKQAANENMERELETLEVAKASFAETMEYERSMLSKKAESERSQLL---HDIE 612
Query: 915 SQLAKIKSLEDELATEKAKAIEAREQ 940
+ K++S + EK + ++A+++
Sbjct: 613 MRKRKLESDMQTILEEKERELQAKKK 638
>At5g41140 putative protein
Length = 983
Score = 47.0 bits (110), Expect = 5e-05
Identities = 39/156 (25%), Positives = 73/156 (46%), Gaps = 10/156 (6%)
Query: 802 KIGELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELEL 861
K+ EL + L +E + A L K++KE D + ++ E+ +L +LE
Sbjct: 693 KLNELSGKTDLKTKEMKRMSADLEYQKRQKEDVNADLT---HEITRRKDEIEILRLDLEE 749
Query: 862 VKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKA---TETHAEALATKDAEIAS--- 915
+K+ E E +E L + EAV+ A +++ A + +L+ ++EI +
Sbjct: 750 TRKSSMETEASLSEELQRIIDEKEAVITALKSQLETAIAPCDNLKHSLSNNESEIENLRK 809
Query: 916 QLAKIKSLEDELATEKAKAIEAREQAADIALDNRER 951
Q+ +++S E E E+ +E RE +AD +R
Sbjct: 810 QVVQVRS-ELEKKEEEMANLENREASADNITKTEQR 844
Score = 33.5 bits (75), Expect = 0.63
Identities = 35/155 (22%), Positives = 73/155 (46%), Gaps = 9/155 (5%)
Query: 802 KIGELEDQLALMKEEADE----LDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCA 857
+I ELE Q+ M+EE ++ + + A + K + E+ +I EAL + A +
Sbjct: 572 RIKELETQIKGMEEELEKQAQIFEGDIEAVTRAKVEQEQRAIEAEEALRKTRWKNASVAG 631
Query: 858 ELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQL 917
+++ K ++EQ S+ A K M+A+ + + E++ E L + E+
Sbjct: 632 KIQDEFKRISEQ--MSSTLAANEKVTMKAMTE--TRELRMQKRQLEELLMNANDELRVNR 687
Query: 918 AKIKSLEDELATE-KAKAIEAREQAADIALDNRER 951
+ ++ +EL+ + K E + +AD+ R++
Sbjct: 688 VEYEAKLNELSGKTDLKTKEMKRMSADLEYQKRQK 722
>At1g79280 hypothetical protein
Length = 2111
Score = 47.0 bits (110), Expect = 5e-05
Identities = 50/207 (24%), Positives = 86/207 (41%), Gaps = 13/207 (6%)
Query: 748 EKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 807
EK A + AE K + Q + EK T KD +K + A ++
Sbjct: 1389 EKLKAKDAHAEDCKKVLLEKQNKISLLEKEL----TNCKKDLSEREKRLDDAQQAQATMQ 1444
Query: 808 DQLALMKEEADE---LDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKK 864
+ K+E ++ + +L K++ E+ EKD +++ +AK+ E A A
Sbjct: 1445 SEFNKQKQELEKNKKIHYTLNMTKRKYEK-EKDELSKQNQSLAKQLEEAKEEAGKRTTTD 1503
Query: 865 ALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLE 924
A+ EQ K E ++ + +E++K TE L KD E+ + ++ KS+E
Sbjct: 1504 AVVEQSVKEREEKEKRIQILDKYVHQLKDEVRKKTED----LKKKDEELTKERSERKSVE 1559
Query: 925 DELATEKAKAIEAREQAADIALDNRER 951
E+ K I+ + D L ER
Sbjct: 1560 KEVGDSLTK-IKKEKTKVDEELAKLER 1585
Score = 38.9 bits (89), Expect = 0.015
Identities = 47/175 (26%), Positives = 80/175 (44%), Gaps = 17/175 (9%)
Query: 799 AGIKIGE-LEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCA 857
AG +IG + +L KEE ++L + + K Q + SIA+ K+ E A
Sbjct: 955 AGFRIGSAMSIELRTAKEEIEKLRGEVESSKSHMLQYK--SIAQVNETALKQMESAHENF 1012
Query: 858 ELELVKKALAEQEKKSAESLALAK--SDMEAVMQATSEEIKKATETHAEALATKDAEIAS 915
LE K+ Q AE ++L + S++E SE++ A +AL + AEIAS
Sbjct: 1013 RLEAEKR----QRSLEAELVSLRERVSELENDCIQKSEQLATAAAGKEDALLSASAEIAS 1068
Query: 916 -------QLAKIKSLEDELATEKAKAIEAREQAADIALDNRERGFYLAKDQAQHL 963
+ ++I+++ +++T K +E + +A N ER L + Q L
Sbjct: 1069 LREENLVKKSQIEAMNIQMSTLK-NDLETEHEKWRVAQRNYERQVILLSETIQEL 1122
Score = 34.7 bits (78), Expect = 0.28
Identities = 37/157 (23%), Positives = 69/157 (43%), Gaps = 10/157 (6%)
Query: 799 AGIKIGELEDQLALMKEEADELDASLRACKKEKEQA----EKDSIARGEALIAKESELAV 854
A + ++ + + E D + A A EQ E+ ++ + + ES+ A
Sbjct: 18 ASVVAERADEYIRKIYAELDSVRAKADAASITAEQTCSLLEQKYLSLSQDFSSLESQNAK 77
Query: 855 LCAELE--LVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAE 912
L ++ + L + A ++ +K ++ K M E+ K+ E L KDAE
Sbjct: 78 LQSDFDDRLAELAQSQAQKHQLHLQSIEKDGEVERMSTEMSELHKSKRQLMELLEQKDAE 137
Query: 913 IASQLAKIKSLEDELA--TEKAKAIEAR--EQAADIA 945
I+ + + IKS D++ T+ + EAR E A++A
Sbjct: 138 ISEKNSTIKSYLDKIVKLTDTSSEKEARLAEATAELA 174
Score = 31.6 bits (70), Expect = 2.4
Identities = 44/195 (22%), Positives = 92/195 (46%), Gaps = 25/195 (12%)
Query: 748 EKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLA-TQAGKDKVYADKMIGTAGIKIGEL 806
+K+ + + + L++++AK Q + ++ R +LA +QA K +++ + E
Sbjct: 59 QKYLSLSQDFSSLESQNAKLQ---SDFDDRLAELAQSQAQKHQLHLQSI---------EK 106
Query: 807 EDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKAL 866
+ ++ M E EL S R + EQ + + + K S + ++ +
Sbjct: 107 DGEVERMSTEMSELHKSKRQLMELLEQKDAE-------ISEKNSTIKSYLDKIVKLTDTS 159
Query: 867 AEQEKKSAESLA-LAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLED 925
+E+E + AE+ A LA+S +A+ S+E K+ TE HA+ L + A+++
Sbjct: 160 SEKEARLAEATAELARS--QAMCSRLSQE-KELTERHAKWLDEELTAKVDSYAELRRRHS 216
Query: 926 ELATE-KAKAIEARE 939
+L +E AK ++ +
Sbjct: 217 DLESEMSAKLVDVEK 231
>At1g77580 unknown protein
Length = 779
Score = 47.0 bits (110), Expect = 5e-05
Identities = 63/275 (22%), Positives = 122/275 (43%), Gaps = 43/275 (15%)
Query: 656 SMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLAGLNRDV 715
++P P KS P + T E + A E+ +L + I+ E L L
Sbjct: 313 ALPETPDGNGKSGPESVTEEVVVPSENSLASEI------EVLTSRIKELEEKLEKLEA-- 364
Query: 716 IEKEVLSRGLNDTKEETLACLLRAGCIFAHTFE---KFDAANVEAERLKAESAKHQEAAA 772
EK L + +EE + + + + + T E K + E E LK+E ++E A
Sbjct: 365 -EKHELENEVKCNREEAVVHIENSEVLTSRTKELEEKLEKLEAEKEELKSEVKCNREKAV 423
Query: 773 AWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLRACKKEKE 832
+ + + A+ + T+ K ELE+QL ++ E EL++ ++ C +E
Sbjct: 424 VHVE-----------NSLAAEIEVLTSRTK--ELEEQLEKLEAEKVELESEVK-CNRE-- 467
Query: 833 QAEKDSIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATS 892
EA+ E+ LA E+E++ + + E+K E L + K ++++ ++ +
Sbjct: 468 ----------EAVAQVENSLA---TEIEVLTCRIKQLEEK-LEKLEVEKDELKSEVKC-N 512
Query: 893 EEIKKATETHAEALATKDAEIASQLAKIKSLEDEL 927
E++ EA+A + E+ ++L K++ + EL
Sbjct: 513 REVESTLRFELEAIACEKMELENKLEKLEVEKAEL 547
Score = 38.5 bits (88), Expect = 0.020
Identities = 50/223 (22%), Positives = 97/223 (43%), Gaps = 43/223 (19%)
Query: 728 TKEETLACLLRAGCIFAHTFEKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGK 787
T+ E L C ++ EK + VE + LK+E ++E + + +A
Sbjct: 479 TEIEVLTCRIK------QLEEKLEKLEVEKDELKSEVKCNREVESTLRFELEAIA----- 527
Query: 788 DKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIA 847
+KM ELE++L ++ E EL S K + E+++ L
Sbjct: 528 ----CEKM---------ELENKLEKLEVEKAELQISFDIIKDKYEESQV-------CLQE 567
Query: 848 KESELAVLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALA 907
E++L + E++LV + AE E ++ A AK+ A +++ E+++K E A
Sbjct: 568 IETKLGEIQTEMKLVNELKAEVESQTIAMEADAKTK-SAKIESLEEDMRK------ERFA 620
Query: 908 TKDAEIASQLAKIKSLEDELATEKAKAIEAREQAADIALDNRE 950
+ K ++LE+E++ K +I++ + I ++ E
Sbjct: 621 FDELR-----RKCEALEEEISLHKENSIKSENKEPKIKQEDIE 658
Score = 38.1 bits (87), Expect = 0.025
Identities = 53/190 (27%), Positives = 85/190 (43%), Gaps = 26/190 (13%)
Query: 748 EKFDAA--NVEA--ERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKI 803
EK AA NV A + +K +EA A WEK A + V + + A K
Sbjct: 67 EKLSAALANVSAKDDLVKQHVKVAEEAVAGWEK--------AENEVVELKEKLEAADDKN 118
Query: 804 GELEDQLALMKEEADELDASLRACKKEKEQAEKDS-IARGEALIAKESELAVLCAELELV 862
LED+++ + E LR + E+EQ +D+ I R + L + + L E ++
Sbjct: 119 RVLEDRVSHLDGALKECVRQLRQARDEQEQRIQDAVIERTQELQSSRTSL-----ENQIF 173
Query: 863 KKALAEQE-KKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQ----- 916
+ A +E + AES+A + + A EE++ T L+T+ AE AS+
Sbjct: 174 ETATKSEELSQMAESVAKENVMLRHELLARCEELE--IRTIERDLSTQAAETASKQQLDS 231
Query: 917 LAKIKSLEDE 926
+ K+ LE E
Sbjct: 232 IKKVAKLEAE 241
>At1g65010 hypothetical protein
Length = 1318
Score = 47.0 bits (110), Expect = 5e-05
Identities = 49/168 (29%), Positives = 78/168 (46%), Gaps = 21/168 (12%)
Query: 801 IKIGELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELE 860
+K EL+ QL ++E+ + D + KK+K +A D KESE V A E
Sbjct: 51 VKGTELQTQLNQIQEDLKKADEQIELLKKDKAKAIDD---------LKESEKLVEEAN-E 100
Query: 861 LVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKI 920
+K+ALA Q K++ ES + K + QA E ++K T L + ++ A ++ +
Sbjct: 101 KLKEALAAQ-KRAEESFEVEKFRAVELEQAGLEAVQKKDVTSKNELESIRSQHALDISAL 159
Query: 921 KSLEDEL----------ATEKAKAIEAREQAADIALDNRERGFYLAKD 958
S +EL A K KA+ E+A IA + E+ LA +
Sbjct: 160 LSTTEELQRVKHELSMTADAKNKALSHAEEATKIAEIHAEKAEILASE 207
Score = 45.1 bits (105), Expect = 2e-04
Identities = 107/440 (24%), Positives = 169/440 (38%), Gaps = 71/440 (16%)
Query: 634 ETPKSPPHQEAPPSPPPTR---------DAGSMPSPPHQGEKSCPGAATTSEAAQIEQAP 684
ETP+S P SPPP R D+ S SP S + T +++ +
Sbjct: 2 ETPRSKP------SPPPPRLSKLSASKSDSNSA-SPVPNTRLSLDRSPPTKVHSRLVKGT 54
Query: 685 APEVGSSSYYNMLPNAIEPSEFLLAGLNR---DVIEKEVLSRGLNDTKEETLACLLRAGC 741
+ + L A E E L + D+ E E L N+ +E LA RA
Sbjct: 55 ELQTQLNQIQEDLKKADEQIELLKKDKAKAIDDLKESEKLVEEANEKLKEALAAQKRAEE 114
Query: 742 IFAHTFEKFDAANVEAERLKA------------ESAKHQEAAAAWEKRFDKLATQAGKDK 789
F EKF A +E L+A ES + Q A Q K +
Sbjct: 115 SFE--VEKFRAVELEQAGLEAVQKKDVTSKNELESIRSQHALDISALLSTTEELQRVKHE 172
Query: 790 VYADKMIGTAGIKIGELEDQLALMKEEADELDAS----LRACKKEKEQAEKDSIARGEAL 845
+ + E ++A + E E+ AS L+A KE EK++I E +
Sbjct: 173 LSMTADAKNKALSHAEEATKIAEIHAEKAEILASELGRLKALLGSKE--EKEAIEGNEIV 230
Query: 846 IAKESELAVLCAELELVKKALAEQEKKSAESLA-LAKSDMEAVMQATS------------ 892
+SE+ +L ELE K ++ E K E L K D+EA A S
Sbjct: 231 SKLKSEIELLRGELE--KVSILESSLKEQEGLVEQLKVDLEAAKMAESCTNSSVEEWKNK 288
Query: 893 -EEIKKATE--THAEALATKDAE-IASQLAKIKSLEDELAT------EKAKAIEAREQAA 942
E++K E +++ A++ E + QLA++ + E + EK + +E +A
Sbjct: 289 VHELEKEVEESNRSKSSASESMESVMKQLAELNHVLHETKSDNAAQKEKIELLEKTIEAQ 348
Query: 943 DIALDNRERGFYLAKDQAQHLYPNFDF--SAMGVMKEITTAGL-----VGPDDPPLIDQN 995
L+ R +AK++A L + S + + +E T L + L+DQ
Sbjct: 349 RTDLEEYGRQVCIAKEEASKLENLVESIKSELEISQEEKTRALDNEKAATSNIQNLLDQR 408
Query: 996 LWTATEEEEEEEEQEKENNE 1015
+ E E + E+EK +
Sbjct: 409 TELSIELERCKVEEEKSKKD 428
Score = 32.3 bits (72), Expect = 1.4
Identities = 30/118 (25%), Positives = 52/118 (43%), Gaps = 15/118 (12%)
Query: 815 EEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKALAEQEKKSA 874
++ EL L CK E+E+++KD + AL +E + A L + ++ L E +
Sbjct: 406 DQRTELSIELERCKVEEEKSKKDMESLTLALQEASTESSEAKATLLVCQEELKNCESQ-V 464
Query: 875 ESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKA 932
+SL LA K+ E + + L EI S + + S+++E KA
Sbjct: 465 DSLKLAS--------------KETNEKYEKMLEDARNEIDSLKSTVDSIQNEFENSKA 508
>At3g19060 hypothetical protein
Length = 1647
Score = 46.6 bits (109), Expect = 7e-05
Identities = 52/201 (25%), Positives = 90/201 (43%), Gaps = 27/201 (13%)
Query: 756 EAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKE 815
E LK E K + A E R+ + A K YAD E E+++ L++
Sbjct: 1388 EVLNLKEEFGKMKSEAKEMEARYIEAQQIAESRKTYAD-----------EREEEVKLLEG 1436
Query: 816 EADELDASLRACKKE----KEQAEKDSIARGEALIAKESELAVLCAELELVKKALAEQEK 871
+EL+ ++ + + K++AE+ + R E E EL + ++E + A E ++
Sbjct: 1437 SVEELEYTINVLENKVNVVKDEAERQRLQREEL----EMELHTIRQQMESARNADEEMKR 1492
Query: 872 KSAE---SLALAKSDMEAVMQATSE---EIKKATETHAEALATKDAEIASQLAKIKSLED 925
E LA AK +EA+ + T++ EI + +E +E +A+ + + K K L
Sbjct: 1493 ILDEKHMDLAQAKKHIEALERNTADQKTEITQLSEHISELNLHAEAQASEYMHKFKEL-- 1550
Query: 926 ELATEKAKAIEAREQAADIAL 946
E E+ K QA D +L
Sbjct: 1551 EAMAEQVKPEIHVSQAIDSSL 1571
Score = 37.4 bits (85), Expect = 0.043
Identities = 43/222 (19%), Positives = 90/222 (40%), Gaps = 23/222 (10%)
Query: 748 EKFDAAN----VEAERLKAESAKHQEAAAAWEKRFDKLAT----------QAGKDKVYAD 793
EK A N EAE L AE +E +K + + Q +
Sbjct: 1313 EKLSAENKDIRAEAEDLLAEKCSLEEEMIQTKKVSESMEMELFNLRNALGQLNDTVAFTQ 1372
Query: 794 KMIGTAGIKIGELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELA 853
+ + A + L+D++ +KEE ++ + + + +A++ + +R +E E+
Sbjct: 1373 RKLNDAIDERDNLQDEVLNLKEEFGKMKSEAKEMEARYIEAQQIAESRKTYADEREEEVK 1432
Query: 854 VLCAELELVKKALAEQEKK------SAESLALAKSDMEAVMQATSEEIKKATETHAE--- 904
+L +E ++ + E K AE L + ++E + ++++ A E
Sbjct: 1433 LLEGSVEELEYTINVLENKVNVVKDEAERQRLQREELEMELHTIRQQMESARNADEEMKR 1492
Query: 905 ALATKDAEIASQLAKIKSLEDELATEKAKAIEAREQAADIAL 946
L K ++A I++LE A +K + + E +++ L
Sbjct: 1493 ILDEKHMDLAQAKKHIEALERNTADQKTEITQLSEHISELNL 1534
Score = 33.1 bits (74), Expect = 0.82
Identities = 42/190 (22%), Positives = 83/190 (43%), Gaps = 25/190 (13%)
Query: 790 VYADKMIGTAGIKIGELED------QLALMKEEADELDASL-------RACKKEKEQAEK 836
V+ + + T +K ELED L + +E+ E+ +L R C+++ K
Sbjct: 1261 VHVEALEKTLALKTFELEDAVSHAQMLEVRLQESKEITRNLEVDTEKARKCQEKLSAENK 1320
Query: 837 DSIARGEALIAKESELAVLCAELELVK-KALAEQEKKSAESLALAKSDMEAVMQATSEEI 895
D A E L+A++ L E E+++ K ++E + +L A + + T ++
Sbjct: 1321 DIRAEAEDLLAEKCSL-----EEEMIQTKKVSESMEMELFNLRNALGQLNDTVAFTQRKL 1375
Query: 896 KKATETHAEALATKDAEIASQLAKIKSLEDELATEKAKAIEARE--QAADIALDNRERGF 953
A + + L + + + K+KS E+ +A+ IEA++ ++ D RE
Sbjct: 1376 NDAID-ERDNLQDEVLNLKEEFGKMKSEAKEM---EARYIEAQQIAESRKTYADEREEEV 1431
Query: 954 YLAKDQAQHL 963
L + + L
Sbjct: 1432 KLLEGSVEEL 1441
Score = 32.7 bits (73), Expect = 1.1
Identities = 44/179 (24%), Positives = 72/179 (39%), Gaps = 20/179 (11%)
Query: 755 VEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMK 814
VE LK E + + + + LA Q M K+ LE ++A MK
Sbjct: 604 VEMANLKGEKEELKASEKRSLSNLNDLAAQICNLNTVMSNMEEQYEHKMETLEHEIAKMK 663
Query: 815 EEADELDASLRACKKEKEQAE----KDSIARGEALIAKESELAVLCAELELVKKALAEQE 870
EAD+ K+ E+A+ + I E +IA E ++ L +Q+
Sbjct: 664 IEADQEYVENLCILKKFEEAQGTIREADITVNELVIANEK-----------MRFDLEKQK 712
Query: 871 KKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELAT 929
K+ + K+ +E + + S +K+ E LA + S L I +L +ELAT
Sbjct: 713 KRGISLVGEKKALVEKLQELESINVKE-----NEKLAYLEKLFESSLMGIGNLVEELAT 766
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 46.6 bits (109), Expect = 7e-05
Identities = 73/278 (26%), Positives = 124/278 (44%), Gaps = 40/278 (14%)
Query: 748 EKFDAANVEAERLKAESAKHQEA---AAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIG 804
E FDA +E E + + + +E +A ++F++L Q+ +AD + K
Sbjct: 184 EAFDALGIELESSRKKLIELEEGLKRSAEEAQKFEELHKQSAS---HAD----SESQKAL 236
Query: 805 ELEDQLALMKEEADELDASLRACKKE-KEQAEKDSIARG--EALIAKESELAVLCAELEL 861
E + L KE A E++ + + ++E KE EK S AL + ELA + EL L
Sbjct: 237 EFSELLKSTKESAKEMEEKMASLQQEIKELNEKMSENEKVEAALKSSAGELAAVQEELAL 296
Query: 862 VKKALAEQEKKSAESLALAKSDMEAVMQATSE-EIKKATETHAEALATKDAEIASQLAKI 920
K L E E+K + + AL + + T E E KKA+E + +L+ +
Sbjct: 297 SKSRLLETEQKVSSTEAL-------IDELTQELEQKKASE----------SRFKEELSVL 339
Query: 921 KSLEDELATEKAKAIEAREQAADIALDNRERGFY--LAKDQAQHL-YPNFDFSAMGVMKE 977
+ L+ + +AK E + +A + +E+ L+KDQ + L N + + KE
Sbjct: 340 QDLDAQTKGLQAKLSEQEGINSKLAEELKEKELLESLSKDQEEKLRTANEKLAEVLKEKE 399
Query: 978 ITTAGLVGPDDPPLIDQNLWTATEEEEEEEEQEKENNE 1015
A + + N+ T TE E EE+ K ++E
Sbjct: 400 ALEANVAE------VTSNVATVTEVCNELEEKLKTSDE 431
Score = 44.7 bits (104), Expect = 3e-04
Identities = 65/279 (23%), Positives = 116/279 (41%), Gaps = 22/279 (7%)
Query: 748 EKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 807
E+F + + EA L + + ++E ++LA +GK +K+ T G ++ E
Sbjct: 813 EEFTSRDSEASSLTEKLRDLEGKIKSYE---EQLAEASGKSSSLKEKLEQTLG-RLAAAE 868
Query: 808 DQLALMKEEADEL-DASLRACKKEKEQAE-----KDSIARGEALIAKES-ELAVLCAELE 860
+K+E D+ + SL++ + + AE K I E LI S E LE
Sbjct: 869 SVNEKLKQEFDQAQEKSLQSSSESELLAETNNQLKIKIQELEGLIGSGSVEKETALKRLE 928
Query: 861 LVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKI 920
+ ++E +S++ + K+ + EE KK + T+ E+ L+K+
Sbjct: 929 EAIERFNQKETESSDLVEKLKTHENQI-----EEYKKLAHEASGVADTRKVELEDALSKL 983
Query: 921 KSLEDELATEKAKAIEAREQAADIALDNRERGFYLAKDQAQHLYPNFDFSAMGVMKEITT 980
K+LE + AK +++ D+A N + LA ++ SA+ KE T
Sbjct: 984 KNLESTIEELGAKCQGLEKESGDLAEVNLKLNLELANHGSEANELQTKLSALEAEKEQTA 1043
Query: 981 AGLVGPDDPPLIDQNLWTATEEEEEEEEQ----EKENNE 1015
L I+ T E E+ + Q +ENN+
Sbjct: 1044 NELEA--SKTTIEDLTKQLTSEGEKLQSQISSHTEENNQ 1080
Score = 42.0 bits (97), Expect = 0.002
Identities = 42/201 (20%), Positives = 95/201 (46%), Gaps = 13/201 (6%)
Query: 754 NVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALM 813
N+E +E+ + Q +A E ++ A + K + + + +L+ Q++
Sbjct: 1015 NLELANHGSEANELQTKLSALEAEKEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSH 1074
Query: 814 KEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELA-VLCAELELVKKALAEQEKK 872
EE ++++A ++ K+E + IA+ E + ES A L +E+E ++ AE+
Sbjct: 1075 TEENNQVNAMFQSTKEELQSV----IAKLEEQLTVESSKADTLVSEIEKLRAVAAEKSVL 1130
Query: 873 SAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSL--EDELATE 930
+ ++E + ++K+ E A A + K AE+ S+L + + + E ++ E
Sbjct: 1131 ESHF-----EELEKTLSEVKAQLKENVENAATA-SVKVAELTSKLQEHEHIAGERDVLNE 1184
Query: 931 KAKAIEAREQAADIALDNRER 951
+ ++ QAA ++D +++
Sbjct: 1185 QVLQLQKELQAAQSSIDEQKQ 1205
Score = 40.8 bits (94), Expect = 0.004
Identities = 43/207 (20%), Positives = 84/207 (39%), Gaps = 44/207 (21%)
Query: 754 NVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALM 813
N+E E + S++ E A + K + T A + ++ + +K + E +L +
Sbjct: 475 NLELEDVVRSSSQAAEEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKEL 534
Query: 814 KEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKALAEQEKKS 873
E++ EL ++ ++EK+QA + E ++K+
Sbjct: 535 SEKSSELQTAIEVAEEEKKQATTQ----------------------------MQEYKQKA 566
Query: 874 AE-SLALAKS---------DMEAVMQATSEEIKKATETHAEALATKDAEIASQL------ 917
+E L+L +S D+ +Q +E +A TH ++ + +SQ
Sbjct: 567 SELELSLTQSSARNSELEEDLRIALQKGAEHEDRANTTHQRSIELEGLCQSSQSKHEDAE 626
Query: 918 AKIKSLEDELATEKAKAIEAREQAADI 944
++K LE L TEK + E EQ + +
Sbjct: 627 GRLKDLELLLQTEKYRIQELEEQVSSL 653
Score = 40.0 bits (92), Expect = 0.007
Identities = 40/171 (23%), Positives = 75/171 (43%), Gaps = 27/171 (15%)
Query: 801 IKIGELEDQLALMKEEADELDASLRACKKEKEQAEKD------SIARGEALIAK-ESELA 853
+K LE L + E EL +L A EK++ E I+ E L+ +EL
Sbjct: 683 VKSSSLEAALNIATENEKELTENLNAVTSEKKKLEATVDEYSVKISESENLLESIRNELN 742
Query: 854 VLCAELELVK---KALAEQEKKSAESLALAKS-----------------DMEAVMQATSE 893
V +LE ++ KA QE + E L A+ ++EA+ Q+ S
Sbjct: 743 VTQGKLESIENDLKAAGLQESEVMEKLKSAEESLEQKGREIDEATTKRMELEALHQSLSI 802
Query: 894 EIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKAKAIEAREQAADI 944
+ + + E ++D+E +S K++ LE ++ + + + EA +++ +
Sbjct: 803 DSEHRLQKAMEEFTSRDSEASSLTEKLRDLEGKIKSYEEQLAEASGKSSSL 853
Score = 37.4 bits (85), Expect = 0.043
Identities = 40/167 (23%), Positives = 69/167 (40%), Gaps = 19/167 (11%)
Query: 806 LEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKA 865
+E+Q +++ + L +++ ++ E + L ESE L EL K+
Sbjct: 70 VEEQKEVIERSSSGSQRELHESQEKAKELELELERVAGELKRYESENTHLKDELLSAKEK 129
Query: 866 LAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLED 925
L E EKK D+E V + E+I + E H+ L + + + S AK K L +
Sbjct: 130 LEETEKKHG--------DLEVVQKKQQEKIVEGEERHSSQLKSLEDALQSHDAKDKELTE 181
Query: 926 ----------ELATEKAKAIEAREQAADIALDNRERGFYLAKDQAQH 962
EL + + K IE E+ + + ++ L K A H
Sbjct: 182 VKEAFDALGIELESSRKKLIEL-EEGLKRSAEEAQKFEELHKQSASH 227
Score = 37.0 bits (84), Expect = 0.057
Identities = 58/257 (22%), Positives = 103/257 (39%), Gaps = 12/257 (4%)
Query: 756 EAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKE 815
E++ E E A KR++ T + + A + + K G+LE +E
Sbjct: 90 ESQEKAKELELELERVAGELKRYESENTHLKDELLSAKEKLEETEKKHGDLEVVQKKQQE 149
Query: 816 EADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKALAEQEKKSAE 875
+ E + + K E A + A+ + L + L ELE +K L E E+
Sbjct: 150 KIVEGEERHSSQLKSLEDALQSHDAKDKELTEVKEAFDALGIELESSRKKLIELEE---- 205
Query: 876 SLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKAKAI 935
L +S EA EE+ K + +HA++ + K E + L K E+ EK ++
Sbjct: 206 --GLKRSAEEA---QKFEELHKQSASHADSESQKALEFSELLKSTKESAKEM-EEKMASL 259
Query: 936 EAREQAADIALDNRERGFYLAKDQAQHLYPNFDFSAMGVMKEITTAGLVGPDDPPLIDQN 995
+ + + + E+ K A L + A+ + + T V + LID+
Sbjct: 260 QQEIKELNEKMSENEKVEAALKSSAGELAAVQEELALSKSRLLETEQKVSSTE-ALIDE- 317
Query: 996 LWTATEEEEEEEEQEKE 1012
L E+++ E + KE
Sbjct: 318 LTQELEQKKASESRFKE 334
Score = 34.7 bits (78), Expect = 0.28
Identities = 74/373 (19%), Positives = 140/373 (36%), Gaps = 67/373 (17%)
Query: 614 NATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAAT 673
N KAA ++ + EK K E +E + + ++ HQ + +
Sbjct: 753 NDLKAAGLQESEVMEKLKSAEESLEQKGREIDEATTKRMELEAL----HQ-------SLS 801
Query: 674 TSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSRGLNDTKEETL 733
++++A S + L + E + + E S L + E+TL
Sbjct: 802 IDSEHRLQKAMEEFTSRDSEASSLTEKLRDLEGKIKSYEEQLAEASGKSSSLKEKLEQTL 861
Query: 734 ACLLRAGCIFAHTFEKFDAANVEAERLKAES-----------AKHQE-----AAAAWEK- 776
L A + ++FD A ++ + +ES K QE + + EK
Sbjct: 862 GRLAAAESVNEKLKQEFDQAQEKSLQSSSESELLAETNNQLKIKIQELEGLIGSGSVEKE 921
Query: 777 -----------RFDKLATQAG--------------KDKVYADKMIGTAGIKIGELEDQLA 811
RF++ T++ + K A + G A + ELED L+
Sbjct: 922 TALKRLEEAIERFNQKETESSDLVEKLKTHENQIEEYKKLAHEASGVADTRKVELEDALS 981
Query: 812 LMK---EEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKALAE 868
+K +EL A + +KE + ++ L SE EL+ AL
Sbjct: 982 KLKNLESTIEELGAKCQGLEKESGDLAEVNLKLNLELANHGSE----ANELQTKLSALEA 1037
Query: 869 QEKKSAESLALAKS---DMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKS--- 922
+++++A L +K+ D+ + + E+++ +H E +A S +++S
Sbjct: 1038 EKEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSHTEENNQVNAMFQSTKEELQSVIA 1097
Query: 923 -LEDELATEKAKA 934
LE++L E +KA
Sbjct: 1098 KLEEQLTVESSKA 1110
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 46.2 bits (108), Expect = 9e-05
Identities = 32/148 (21%), Positives = 77/148 (51%), Gaps = 16/148 (10%)
Query: 802 KIGELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELEL 861
++ ELE QL L+++ +L ASL A ++EK+ + + L +S++ L EL
Sbjct: 486 RLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILEITDELKQAQSKVQELVTELAE 545
Query: 862 VKKALAEQEKKSAESLALAKS----------DMEAVMQATSEEIKK------ATETHAEA 905
K L ++E + + + + ++ ++EA +++ E++K+ ++E +
Sbjct: 546 SKDTLTQKENELSSFVEVHEAHKRDSSSQVKELEARVESAEEQVKELNQNLNSSEEEKKI 605
Query: 906 LATKDAEIASQLAKIKSLEDELATEKAK 933
L+ + +E++ ++ + +S EL++E +
Sbjct: 606 LSQQISEMSIKIKRAESTIQELSSESER 633
Score = 44.3 bits (103), Expect = 4e-04
Identities = 56/284 (19%), Positives = 121/284 (41%), Gaps = 45/284 (15%)
Query: 674 TSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLA----GLNRDVIEKEVLSRGLNDTK 729
+ ++ +E E SSS L IE SE L+A LN EK++LS+ + +
Sbjct: 48 SEHSSLVELHKTHERESSSQVKELEAHIESSEKLVADFTQSLNNAEEEKKLLSQKIAELS 107
Query: 730 EETLACLLRAGCIFAHTFEKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDK 789
E A + L +ES + +E+ + E+ L +
Sbjct: 108 NE------------------IQEAQNTMQELMSESGQLKESHSVKERELFSL-------R 142
Query: 790 VYADKMIGTAGIKIGELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKE 849
+ + + ELE QL K++ +L ASL+A ++E + ++ L +
Sbjct: 143 DIHEIHQRDSSTRASELEAQLESSKQQVSDLSASLKAAEEENKAISSKNVETMNKLEQTQ 202
Query: 850 SELAVLCAELELVKKALAEQEKKSAESLALAKS----------DMEAVMQATSEEIKKAT 899
+ + L AEL +K + E+E + + + + ++ ++E ++++ + + +
Sbjct: 203 NTIQELMAELGKLKDSHREKESELSSLVEVHETHQRDSSIHVKELEEQVESSKKLVAELN 262
Query: 900 ET------HAEALATKDAEIASQLAKIKSLEDELATEKAKAIEA 937
+T + L+ K AE+++++ + ++ EL +E + E+
Sbjct: 263 QTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSESGQLKES 306
Score = 37.4 bits (85), Expect = 0.043
Identities = 49/219 (22%), Positives = 87/219 (39%), Gaps = 17/219 (7%)
Query: 755 VEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKV-YADKMIGTAGIKIGELEDQLALM 813
++ L+ E+ Q + E R K DK+ A I I L+++L +
Sbjct: 937 IKGRELELETLGKQRSELDEELRTKKEENVQMHDKINVASSEIMALTELINNLKNELDSL 996
Query: 814 KEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKALAEQEK-- 871
+ + E +A L K+EK + +AL+ +E+ L E + + + E E
Sbjct: 997 QVQKSETEAELEREKQEKSELSNQITDVQKALVEQEAAYNTLEEEHKQINELFKETEATL 1056
Query: 872 -------KSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLE 924
K A+ L + +T ++ E+ L K EI + + KI ++E
Sbjct: 1057 NKVTVDYKEAQRLLEERGKEVTSRDSTIGVHEETMESLRNELEMKGDEIETLMEKISNIE 1116
Query: 925 DELATEKAKAIEAREQAADIALDNRERGFYLAKDQAQHL 963
+L K + EQ L +E F K++A+HL
Sbjct: 1117 VKLRLSNQK-LRVTEQ----VLTEKEEAF--RKEEAKHL 1148
Score = 33.1 bits (74), Expect = 0.82
Identities = 68/299 (22%), Positives = 122/299 (40%), Gaps = 55/299 (18%)
Query: 690 SSSYYNMLPNAIEPSEFLLAGLNRDVI----EKEVLSRGL----NDTKE--ETLACLLRA 739
SS + L +E S+ L+A LN+ + EK+VLS+ + N+ KE T+ L+
Sbjct: 240 SSIHVKELEEQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSE 299
Query: 740 GCIFAHTF-----EKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLA--TQAGKDKVYA 792
+ + F ++ + S + E A E +++ T KD
Sbjct: 300 SGQLKESHSVKDRDLFSLRDIHETHQRESSTRVSELEAQLESSEQRISDLTVDLKDAEEE 359
Query: 793 DKMIGTAGIKI-GELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESE 851
+K I + ++I +LE +KE DEL L+ KEKE + + +A
Sbjct: 360 NKAISSKNLEIMDKLEQAQNTIKELMDEL-GELKDRHKEKESELSSLVKSADQQVAD--- 415
Query: 852 LAVLCAELELVKKAL--AEQEKKSAESLALAKSDMEAVMQAT-------SEEIKKA---- 898
+K++L AE+EKK L S+ Q T SE++K++
Sbjct: 416 ----------MKQSLDNAEEEKKMLSQRILDISNEIQEAQKTIQEHMSESEQLKESHGVK 465
Query: 899 ----------TETHAEALATKDAEIASQLAKIKSLEDELATEKAKAIEAREQAADIALD 947
ETH +T+ +E+ +QL ++ +L+ A E ++ + + L+
Sbjct: 466 ERELTGLRDIHETHQRESSTRLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILE 524
Score = 31.6 bits (70), Expect = 2.4
Identities = 53/260 (20%), Positives = 102/260 (38%), Gaps = 21/260 (8%)
Query: 707 LLAGLNRDVIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFDAANVEAERLKAESAK 766
L A L+ ++KE + + + EE + R ++ + + + L+ + K
Sbjct: 838 LRAELDSMSVQKEEVEKQMVCKSEEASVKIKRLDDEVNGLRQQVASLDSQRAELEIQLEK 897
Query: 767 HQEAAAAWEKRFDKLATQA-GKDKVYADKMIGTAGI--KIGELEDQLALMKEEADELDAS 823
E + + + L + K KV+ + G+ KI E +L + ++ ELD
Sbjct: 898 KSEEISEYLSQITNLKEEIINKVKVHESILEEINGLSEKIKGRELELETLGKQRSELDEE 957
Query: 824 LRACKKEKEQA-EKDSIARGEALIAKE------SELAVLCAELELVKKALAEQEKKSAE- 875
LR K+E Q +K ++A E + E +EL L + + L ++++ +E
Sbjct: 958 LRTKKEENVQMHDKINVASSEIMALTELINNLKNELDSLQVQKSETEAELEREKQEKSEL 1017
Query: 876 -----SLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATE 930
+ A + EA EE K+ E E AT + + LE E
Sbjct: 1018 SNQITDVQKALVEQEAAYNTLEEEHKQINELFKETEATLNKVTVDYKEAQRLLE-----E 1072
Query: 931 KAKAIEAREQAADIALDNRE 950
+ K + +R+ + + E
Sbjct: 1073 RGKEVTSRDSTIGVHEETME 1092
>At4g37450 arabinogalactan protein AGP18
Length = 209
Score = 46.2 bits (108), Expect = 9e-05
Identities = 37/138 (26%), Positives = 55/138 (39%), Gaps = 17/138 (12%)
Query: 553 SNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAG 612
S+P PTT +AP + A T PT PA TP +A A++ V + S + S
Sbjct: 26 SSPTKSPTTPSAP---TTSPTKSPAVTSPTTAPAKTP-TASASSPVESPKSPAPVSES-- 79
Query: 613 VNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAA 672
+ TP+ E P +PP P P D+ P + P +
Sbjct: 80 -------SPPPTPVPESSPPVPAPMVSSPVSSPPVPAPVADSPPAPVAAPVADVPAPAPS 132
Query: 673 ----TTSEAAQIEQAPAP 686
TT ++ + + APAP
Sbjct: 133 KHKKTTKKSKKHQAAPAP 150
Score = 35.8 bits (81), Expect = 0.13
Identities = 33/131 (25%), Positives = 48/131 (36%), Gaps = 10/131 (7%)
Query: 550 AGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVP-ASTPASAGATAAVAAESSVGATA 608
A S+P P + A + + ++ P P S+P S+ A A+S A
Sbjct: 61 ASASSPVESPKSPAPVSESSPPPTPVPESSPPVPAPMVSSPVSSPPVPAPVADSPPAPVA 120
Query: 609 ASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPP-----PTRDAGSMPSPPHQ 663
A A + + +K P P PP+PP P DA S P P
Sbjct: 121 APVADVPAPAPSKHKKTTKKSKKHQAAPAPAPELLGPPAPPTESPGPNSDAFS-PGPSAD 179
Query: 664 GEKSCPGAATT 674
+ GAA+T
Sbjct: 180 DQS---GAAST 187
>At2g26570 unknown protein
Length = 807
Score = 46.2 bits (108), Expect = 9e-05
Identities = 54/219 (24%), Positives = 95/219 (42%), Gaps = 31/219 (14%)
Query: 755 VEAERLKAESAKHQE-----AAAAWEKRFDK----LATQAGKDKVYADKMIGTAGIKIGE 805
+E E+ S K +E A A+ E D+ +A+ K+K +KM+ E
Sbjct: 497 LEKEKSTLASIKQREGMASIAVASIEAEIDRTRSEIASVQSKEKDAREKMV--------E 548
Query: 806 LEDQLALMKEEADELDA-------SLRACKKEKEQAEKDSIARGEALIAKESELAVLCAE 858
L QL EEADE + LR K+E EQA+ + L A + E+ A
Sbjct: 549 LPKQLQQAAEEADEAKSLAEVAREELRKAKEEAEQAKAGASTMESRLFAAQKEIEAAKAS 608
Query: 859 LELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLA 918
L A+ E +S +L +D + + EE + ++ EA +A +A+ ++
Sbjct: 609 ERLALAAIKALE-ESESTLKANDTDSPRSVTLSLEEYYELSKRAHEAEELANARVAAAVS 667
Query: 919 KIKSLED------ELATEKAKAIEAREQAADIALDNRER 951
+I+ ++ E E + ++AR++A A + E+
Sbjct: 668 RIEEAKETEMRSLEKLEEVNRDMDARKKALKEATEKAEK 706
Score = 34.7 bits (78), Expect = 0.28
Identities = 48/188 (25%), Positives = 86/188 (45%), Gaps = 29/188 (15%)
Query: 748 EKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 807
++ + NV E+ AE + + A+++ + +K + K + G A I + +E
Sbjct: 467 KELEEVNVNIEKAAAEVSCLKLASSSLQLELEKEKSTLASIK----QREGMASIAVASIE 522
Query: 808 DQLALMKEEADELDASLRACKKEKEQAEKD-SIARGEALIAKESELAVLCAEL--ELVKK 864
++ + E + + KEK+ EK + + A+E++ A AE+ E ++K
Sbjct: 523 AEIDRTRSEIASVQS------KEKDAREKMVELPKQLQQAAEEADEAKSLAEVAREELRK 576
Query: 865 ALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLE 924
A K+ AE S ME+ + A +EI+ A +E LA LA IK+LE
Sbjct: 577 A-----KEEAEQAKAGASTMESRLFAAQKEIEAAKA--SERLA---------LAAIKALE 620
Query: 925 DELATEKA 932
+ +T KA
Sbjct: 621 ESESTLKA 628
>At1g13220 putative nuclear matrix constituent protein
Length = 1128
Score = 46.2 bits (108), Expect = 9e-05
Identities = 44/198 (22%), Positives = 85/198 (42%), Gaps = 22/198 (11%)
Query: 756 EAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKM--IGTAGIKIGELEDQLALM 813
E E + K +E WEK+ GK++ ++ + K+ E+E +L L
Sbjct: 250 ERESYEGTFQKQREYLNEWEKKLQ------GKEESITEQKRNLNQREEKVNEIEKKLKLK 303
Query: 814 KEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKALAEQEKKS 873
++E +E + + + ++ E+D R E L KE E L ++ L+ K E E ++
Sbjct: 304 EKELEEWNRKVDLSMSKSKETEEDITKRLEELTTKEKEAHTL--QITLLAK---ENELRA 358
Query: 874 AESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKAK 933
E +A+ EI+K + E L +K E + +I+ D+ K +
Sbjct: 359 FEEKLIARE---------GTEIQKLIDDQKEVLGSKMLEFELECEEIRKSLDKELQRKIE 409
Query: 934 AIEAREQAADIALDNRER 951
+E ++ D + + E+
Sbjct: 410 ELERQKVEIDHSEEKLEK 427
Score = 31.6 bits (70), Expect = 2.4
Identities = 46/230 (20%), Positives = 94/230 (40%), Gaps = 33/230 (14%)
Query: 805 ELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKK 864
+LE +L +KE + A + EK+Q D E+L + E+ + AE+ K+
Sbjct: 445 DLEAKLKTIKEREKIIQAEEKRLSLEKQQLLSDK----ESLEDLQQEIEKIRAEM-TKKE 499
Query: 865 ALAEQEKKSAESLALAKSDMEAVMQATSE---EIKKATETHAEALATKDAEIASQLAKIK 921
+ E+E KS E + K + E ++ SE +I+K + H E L+ + + + + +
Sbjct: 500 EMIEEECKSLE---IKKEEREEYLRLQSELKSQIEK-SRVHEEFLSKEVENLKQEKERFE 555
Query: 922 S----LEDELATEKAKAIEAREQAADIALDNRERGFYLAKD-----------------QA 960
L+++ A + I E+ G L K+ Q
Sbjct: 556 KEWEILDEKQAVYNKERIRISEEKEKFERFQLLEGERLKKEESALRVQIMQELDDIRLQR 615
Query: 961 QHLYPNFDFSAMGVMKEITTAGLVGPDDPPLIDQNLWTATEEEEEEEEQE 1010
+ N + + +++ DD ++ +NL +E +E++E++
Sbjct: 616 ESFEANMEHERSALQEKVKLEQSKVIDDLEMMRRNLEIELQERKEQDEKD 665
Score = 30.0 bits (66), Expect = 6.9
Identities = 44/225 (19%), Positives = 92/225 (40%), Gaps = 27/225 (12%)
Query: 748 EKFDAANVEAERLKAESAKHQEAAAAWEKRF----DKLATQAGKDKVYAD---KMIGTAG 800
EK A + E + E K E + + +L +Q K +V+ + K +
Sbjct: 489 EKIRAEMTKKEEMIEEECKSLEIKKEEREEYLRLQSELKSQIEKSRVHEEFLSKEVENLK 548
Query: 801 IKIGELEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLC---- 856
+ E + ++ E+ + +EKE+ E+ + GE L +ES L V
Sbjct: 549 QEKERFEKEWEILDEKQAVYNKERIRISEEKEKFERFQLLEGERLKKEESALRVQIMQEL 608
Query: 857 -----------AELELVKKALAE----QEKKSAESLALAKSDMEAVMQATSEEIKK-ATE 900
A +E + AL E ++ K + L + + ++E +Q E+ +K +
Sbjct: 609 DDIRLQRESFEANMEHERSALQEKVKLEQSKVIDDLEMMRRNLEIELQERKEQDEKDLLD 668
Query: 901 THAEALATKDAEIASQLAKIKSLEDELATEKAKAIEAREQAADIA 945
A+ + AE++ + ++L E+ +K ++++ +IA
Sbjct: 669 RMAQFEDKRMAELSDINHQKQALNREMEEMMSKRSALQKESEEIA 713
>At1g12150 hypothetical protein
Length = 548
Score = 46.2 bits (108), Expect = 9e-05
Identities = 51/194 (26%), Positives = 92/194 (47%), Gaps = 33/194 (17%)
Query: 748 EKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 807
E +++ E++K E+ + + AA ++ + L + + A++ + I E+E
Sbjct: 362 EALKQESLKLEQMKTEAIEARNEAANMNRKIESLKKETEAAMIAAEEAEKRLELVIREVE 421
Query: 808 DQLALMKEEADELD--ASLRACKKEKEQA----------EKDSIARG--EALIAKESELA 853
+ + ++ +E+ + + KK+ E++ E +S+ RG E A E +LA
Sbjct: 422 EAKSAEEKVREEMKMISQKQESKKQDEESSGSKIKITIQEFESLKRGAGETEAAIEKKLA 481
Query: 854 VLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEI 913
+ AELE + K AE + K +EA ++A EE+K+ATE LA K AE
Sbjct: 482 TIAAELEEINKRRAEADNK-----------LEANLKAI-EEMKQATE-----LAQKSAES 524
Query: 914 ASQLAKIKSLEDEL 927
A A + +E EL
Sbjct: 525 AE--AAKRMVESEL 536
Score = 33.9 bits (76), Expect = 0.48
Identities = 41/207 (19%), Positives = 89/207 (42%), Gaps = 18/207 (8%)
Query: 748 EKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 807
E++ + VE + K + K +++ ++ D AT A A + + K+ EL
Sbjct: 152 EQYISTTVELDAAKQQLNKIRQS---FDSAMDFKAT-ALNQAAEAQRALQVNSAKVNELS 207
Query: 808 DQLALMKEEADELDASLRACKKEKEQA----EKDSIARGEALIAKESELAVLCAELELVK 863
+++ MK+ +L L A + +E A EKD + +E+E +L +++
Sbjct: 208 KEISDMKDAIHQL--KLAAAQNLQEHANIVKEKDDLRECYRTAVEEAEKKLL-----VLR 260
Query: 864 KALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSL 923
K E E + + +L + + ++ EE+KKA E+ + E+ +++
Sbjct: 261 K---EYEPELSRTLEAKLLETTSEIEVLREEMKKAHESEMNTVKIITNELNEATMRLQEA 317
Query: 924 EDELATEKAKAIEAREQAADIALDNRE 950
D+ + ++ R + D+ + E
Sbjct: 318 ADDECSLRSLVNSLRMELEDLRREREE 344
>At5g24710 unknown protein
Length = 1003
Score = 45.4 bits (106), Expect = 2e-04
Identities = 52/197 (26%), Positives = 78/197 (39%), Gaps = 29/197 (14%)
Query: 514 PKFNTEGDAKKRG-------GAENQAESTKAPKRRRLTRTSSGAGTSNPGA--------- 557
PK + DA ++ G + S APK+ LT+ ++ + P A
Sbjct: 785 PKLTSLSDASRKPPIEILPPGMSSIFASITAPKKPLLTQKTAQPEVAKPLALEEPTKPLA 844
Query: 558 --QPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNA 615
P ++ AP+ ++ E + AA + A + A TAAVA E+ TAA A V+
Sbjct: 845 IEAPPSSEAPQTESAPETAAAAESPAPETAAVAESPAPGTAAVA-EAPASETAA-APVDG 902
Query: 616 TKAAASSDTPIGEKE----KENETPKSPPHQEAPPSPPPTRDAGSMP-----SPPHQGEK 666
S+ P EKE +E P S P+ E S T + P +PP
Sbjct: 903 PVTETVSEPPPVEKEETSLEEKSDPSSTPNTETATSTENTSQTTTTPESVTTAPPEPITT 962
Query: 667 SCPGAATTSEAAQIEQA 683
+ P TT+ E A
Sbjct: 963 APPETVTTTAVKPTENA 979
>At4g12770 auxilin-like protein
Length = 909
Score = 45.4 bits (106), Expect = 2e-04
Identities = 68/317 (21%), Positives = 131/317 (40%), Gaps = 42/317 (13%)
Query: 636 PKSPPHQEAPPSPPPTR------DAGSMPSPPHQGEKSCPGAATTSE--AAQIEQAPAPE 687
P S P PP P PTR + S+P+ + G A+ + A+Q+++
Sbjct: 366 PTSAPPPTRPPPPRPTRPIKKKVNEPSIPTSAYHSHVPSSGRASVNSPTASQMDELDDFS 425
Query: 688 VG-SSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSRG-LNDTKEETLACLLRAGCIFAH 745
+G + + N P +PS +G + DV S + D + +A F H
Sbjct: 426 IGRNQTAANGYP---DPS----SGEDSDVFSTAAASAAAMKDAMD-------KAEAKFRH 471
Query: 746 TFEKFDAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGE 805
E+ + +++A R + H E + E+ + + +++ + E
Sbjct: 472 AKERREKESLKASR--SREGDHTENYDSRERELREKQVRLDRERAERE----------AE 519
Query: 806 LEDQLALMKEEADELDASLRACKKEKEQAEKDSIARGEALIAKESELAVLCAELELVKKA 865
+E A E +E + + ++E+E+ A+E A+++
Sbjct: 520 MEKTQA---REREEREREQKRIERERERLLARQAVERATREARERAATEAHAKVQRAAVG 576
Query: 866 LAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLED 925
++ AE A+ ++ EA +A + +KA + AEA +AE+ + AK+++ +
Sbjct: 577 KVTDARERAERAAVQRAHAEARERAAAGAREKAEKAAAEARERANAEVREKEAKVRA--E 634
Query: 926 ELATEKAKAIEAREQAA 942
A E+A A EAR +AA
Sbjct: 635 RAAVERA-AAEARGRAA 650
Score = 29.6 bits (65), Expect = 9.1
Identities = 54/241 (22%), Positives = 84/241 (34%), Gaps = 32/241 (13%)
Query: 503 KARERRMARFSPKFNTEGDAKKRGGAENQAESTKAP------KRRRLTRTSSGAGTSNPG 556
+ RER +AR + + T +A++R E A+ +A R R R + +
Sbjct: 540 RERERLLARQAVERATR-EARERAATEAHAKVQRAAVGKVTDARERAERAAVQRAHAEAR 598
Query: 557 AQPTTAAAPKG-KNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNA 615
+ A K K AEA A E A A A AAE+ G AA A
Sbjct: 599 ERAAAGAREKAEKAAAEARERANAEVREKEAKVRAERAAVERAAAEAR-GRAAAQA---K 654
Query: 616 TKAAASSDTPIGEKEKENETPKSPPHQEAPPSPP------------PTRDAGSMPSPPHQ 663
K ++ + P S P Q P P +R + +PS P +
Sbjct: 655 AKQQQENNNDLDSFFNSVSRPSSVPRQRTNPPDPFQDSWNKGGSFESSRPSSRVPSGPTE 714
Query: 664 GEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSR 723
+ A + + S + LPN + + L+ L RD + ++R
Sbjct: 715 NLRKASSATNIVD-------DLSSIFGGSAISELPNLV-GFKMLMEKLKRDDVPGWNVTR 766
Query: 724 G 724
G
Sbjct: 767 G 767
>At5g38560 putative protein
Length = 681
Score = 45.1 bits (105), Expect = 2e-04
Identities = 34/141 (24%), Positives = 49/141 (34%), Gaps = 2/141 (1%)
Query: 553 SNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAG 612
+ P A P P ++++ P V + P+S+ + S A+S
Sbjct: 30 TTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPSSSPPPSPPVITSPPPTVASSPP 89
Query: 613 VNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSP--PPTRDAGSMPSPPHQGEKSCPG 670
A+ TP +T PP +A PSP P T + PSP GE P
Sbjct: 90 PPVVIASPPPSTPATTPPAPPQTVSPPPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPP 149
Query: 671 AATTSEAAQIEQAPAPEVGSS 691
T S P P +S
Sbjct: 150 GETPSPPKPSPSTPTPTTTTS 170
Score = 40.8 bits (94), Expect = 0.004
Identities = 32/143 (22%), Positives = 54/143 (37%), Gaps = 9/143 (6%)
Query: 547 SSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGA 606
SS +S+P P +P ++A++ P V AS P S AT A +V
Sbjct: 62 SSPPPSSSPPPSPPVITSPP------PTVASSPPPPVVIASPPPSTPATTPPAPPQTVSP 115
Query: 607 TAA-SAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPS--PPHQ 663
A + ++ P ETP P +PP P P+ + + PP
Sbjct: 116 PPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTSPPPPP 175
Query: 664 GEKSCPGAATTSEAAQIEQAPAP 686
+ P ++ ++ + + P P
Sbjct: 176 ATSASPPSSNPTDPSTLAPPPTP 198
Score = 37.0 bits (84), Expect = 0.057
Identities = 22/71 (30%), Positives = 32/71 (44%)
Query: 621 SSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQI 680
SS T + + TP +PP PPSPP + SPP S P +++ + +
Sbjct: 17 SSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPSSSPPPSPPV 76
Query: 681 EQAPAPEVGSS 691
+P P V SS
Sbjct: 77 ITSPPPTVASS 87
Score = 33.1 bits (74), Expect = 0.82
Identities = 38/165 (23%), Positives = 53/165 (32%), Gaps = 16/165 (9%)
Query: 543 LTRTSSGAGTSNPG---AQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVA 599
L+ SS + T+ P QPTT +AP T P + P S P ++
Sbjct: 10 LSPPSSNSSTTAPPPLQTQPTTPSAPP----------PVTPPPSPPQSPPPVVSSSPPPP 59
Query: 600 AESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPS 659
SS S+ + +S P + P P + PP P
Sbjct: 60 VVSS---PPPSSSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPP 116
Query: 660 PPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPS 704
PP S P TT+ + +P E S P PS
Sbjct: 117 PPPDASPSPPAPTTTNPPPKPSPSPPGETPSPPGETPSPPKPSPS 161
Score = 31.2 bits (69), Expect = 3.1
Identities = 28/130 (21%), Positives = 42/130 (31%), Gaps = 9/130 (6%)
Query: 581 PTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPP 640
P + + +++ TA ++ +A V + S P+ SPP
Sbjct: 6 PLPILSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPP 65
Query: 641 HQEAPP-------SPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSY 693
+PP SPPPT S P PP P T+ A + P +S
Sbjct: 66 PSSSPPPSPPVITSPPPT--VASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDASP 123
Query: 694 YNMLPNAIEP 703
P P
Sbjct: 124 SPPAPTTTNP 133
Score = 30.0 bits (66), Expect = 6.9
Identities = 22/98 (22%), Positives = 34/98 (34%), Gaps = 1/98 (1%)
Query: 553 SNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAG 612
+ P A P T + P + A S A T P +P+ G T + E+ + +
Sbjct: 104 TTPPAPPQTVSPPPPPD-ASPSPPAPTTTNPPPKPSPSPPGETPSPPGETPSPPKPSPST 162
Query: 613 VNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPP 650
T + P + P P PP+P P
Sbjct: 163 PTPTTTTSPPPPPATSASPPSSNPTDPSTLAPPPTPLP 200
>At1g66840 hypothetical protein
Length = 607
Score = 45.1 bits (105), Expect = 2e-04
Identities = 67/289 (23%), Positives = 106/289 (36%), Gaps = 50/289 (17%)
Query: 707 LLAGLNRDVIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFDAANVEAERLKAESAK 766
L+ + R V E +SR N E L + E +A E + AE
Sbjct: 242 LVKEMERKVQRNESMSRSKNRAFERGKDNL----SVLKEVTEATEAKKAELASINAELFC 297
Query: 767 HQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEE---------- 816
K FD A K+ + DKMI + + L +L + K++
Sbjct: 298 LVNTMDTLRKEFD----HAKKETAWLDKMIQKDDVMLERLNTKLLIAKDQLEAVSKAEER 353
Query: 817 ----ADELDASLRACKKEKEQAEKDSIA-RGEALIA----KESELAVLCAELELVKKALA 867
AD L S K ++E A+K+ + R EA I +++E E EL+ K
Sbjct: 354 ISYLADNLTTSFEKLKSDREAAKKEELKLREEARIINNEIQKTETGFDGKEKELLSKLDE 413
Query: 868 EQEKKSAESLALAKSDMEAVMQATSE-------------------EIKKATETHAEALAT 908
++ K AESLAL K +E +++ T E E HAE A
Sbjct: 414 LEKAKHAESLALEK--LETMVEKTMETREMESRRNSTITISRFEYEYLSGKACHAEETAE 471
Query: 909 KDAEIASQLAKIKSLEDELATEKAKAIEAREQAADIALDNRERGFYLAK 957
K E A +A +++L+ K + + L+ F + +
Sbjct: 472 KKVEAA--MAWVEALKASTKAIMIKTESLKRVSGKTMLEEERESFRMQR 518
Score = 41.6 bits (96), Expect = 0.002
Identities = 47/213 (22%), Positives = 94/213 (44%), Gaps = 38/213 (17%)
Query: 787 KDKVYADKMIGTAGIKIGELEDQLALMKEEADELDA-----------SLRACKKEKEQAE 835
++KV A+K + ++ E L +K E D + +L+ CK+ +EQ E
Sbjct: 133 REKVVAEKEVMELESRMEENLKLLESLKLEVDVANEEHVLVEVAKIEALKECKEVEEQRE 192
Query: 836 KDSIARGEALIAK-----------------ESELAVLCAELELVK---KALAEQEKKSAE 875
K+ E+L + E+ELA ++E+++ K + E E+K
Sbjct: 193 KERKEVSESLHKRKKRIREMIREIERSKNFENELAETLLDIEMLETQLKLVKEMERKVQR 252
Query: 876 SLALAKSDMEAVMQ-----ATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDEL--A 928
+ ++++S A + + +E+ +ATE LA+ +AE+ + + +L E A
Sbjct: 253 NESMSRSKNRAFERGKDNLSVLKEVTEATEAKKAELASINAELFCLVNTMDTLRKEFDHA 312
Query: 929 TEKAKAIEAREQAADIALDNRERGFYLAKDQAQ 961
++ ++ Q D+ L+ +AKDQ +
Sbjct: 313 KKETAWLDKMIQKDDVMLERLNTKLLIAKDQLE 345
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.129 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,226,879
Number of Sequences: 26719
Number of extensions: 1078198
Number of successful extensions: 8904
Number of sequences better than 10.0: 459
Number of HSP's better than 10.0 without gapping: 104
Number of HSP's successfully gapped in prelim test: 364
Number of HSP's that attempted gapping in prelim test: 6758
Number of HSP's gapped (non-prelim): 1494
length of query: 1015
length of database: 11,318,596
effective HSP length: 109
effective length of query: 906
effective length of database: 8,406,225
effective search space: 7616039850
effective search space used: 7616039850
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0012a.7


