

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0012a.3
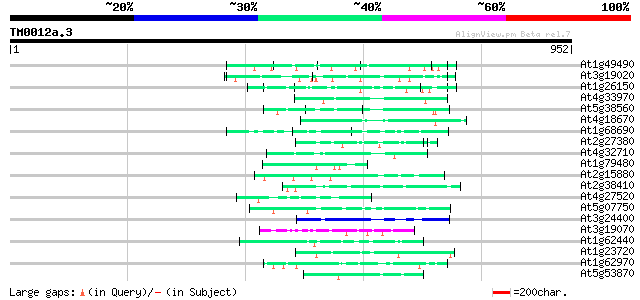
(952 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g49490 hypothetical protein 71 3e-12
At3g19020 hypothetical protein 63 9e-10
At1g26150 Pto kinase interactor, putative 62 2e-09
At4g33970 extensin-like protein 57 4e-08
At5g38560 putative protein 53 9e-07
At4g18670 extensin-like protein 53 9e-07
At1g68690 protein kinase, putative 52 2e-06
At2g27380 hypothetical protein 50 5e-06
At4g32710 putative protein kinase 50 6e-06
At1g79480 hypothetical protein 49 1e-05
At2g15880 unknown protein 48 2e-05
At2g38410 unknown protein 47 4e-05
At4g27520 early nodulin-like 2 predicted GPI-anchored protein 47 5e-05
At5g07750 putative protein 47 7e-05
At3g24400 protein kinase, putative 46 1e-04
At3g19070 hypothetical protein 46 1e-04
At1g62440 putative extensin-like protein (gnl|PID|e1310400 46 1e-04
At1g23720 unknown protein 46 1e-04
At1g62970 unknown protein 45 1e-04
At5g53870 predicted GPI-anchored protein 45 2e-04
>At1g49490 hypothetical protein
Length = 847
Score = 71.2 bits (173), Expect = 3e-12
Identities = 94/398 (23%), Positives = 139/398 (34%), Gaps = 59/398 (14%)
Query: 368 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDS----EDGPPQKKK 423
P PE PSP + + + E E +K KP +DS + P + K
Sbjct: 425 PKTPEQPSPKPQPPKHESPKPEEPENKHELPKQKESPKPQPSKPEDSPKPEQPKPEESPK 484
Query: 424 KKVRIVVKPTR-VEPAAEVV---------------RRTEPPARVTRSSAHSSKPALTSDD 467
+ + +PT+ V P E RR+ PP +V + +P + S
Sbjct: 485 PEQPQIPEPTKPVSPPNEAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVPPPQPPMPSPS 544
Query: 468 DLNLFDALPISALLQHS------SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPL 521
+ + P HS S+P P S P + PP SP P P P+
Sbjct: 545 PPSPIYSPPPPV---HSPPPPVYSSPPPPHVYSPPPPVASPPPPSPPPPVHSPPPP--PV 599
Query: 522 WNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAA 581
++ P S P S + P P S P P P P P + + P + P
Sbjct: 600 FS--PPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQ 657
Query: 582 RTT-------------DTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALE 628
T ++D S +PV P V S+PS SE + E A
Sbjct: 658 APTPSSETTQVPTPSSESDQSQILSPVQAPTPVQSSTPS--SEPTQVPTPSSSESYQAPN 715
Query: 629 EYYLTCPSPRRYPGPRPE--RLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQ---PD 683
+ P+P + P E ++ P +P Q P+++ H +P+Q P
Sbjct: 716 LSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAPTPILEPVHAPTPNSKPVQSPTPS 775
Query: 684 PEPEQSVSNQSSVRSPHPL------VETSDPHLGTSEP 715
EP S V +P P V +S P TS P
Sbjct: 776 SEPVSSPEQSEEVEAPEPTPVNPSSVPSSSPSTDTSIP 813
Score = 48.9 bits (115), Expect = 1e-05
Identities = 65/320 (20%), Positives = 102/320 (31%), Gaps = 59/320 (18%)
Query: 448 PARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPPHSP 507
P R + +P + D S+ + P P P+ QP + + P P
Sbjct: 398 PPRTSEPKPSKPEPVMPKPSD---------SSKPETPKTPEQPSPKPQPPKHESPKPEEP 448
Query: 508 RSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRT 567
+ P E+P + QPS+ ++ P +P E P+P Q + T
Sbjct: 449 ENKHELPKQKESP-----KPQPSKPED---------SPKPEQPKPEESPKPEQPQIPEPT 494
Query: 568 SDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSAL 627
S P + P P + D+SP N R +
Sbjct: 495 KPVSPPNEAQGPT-----------------PDDPYDASPVKNR---------RSPPPPKV 528
Query: 628 EEYYLTCPSPRRYPGPRPERLVDPDEPILANP--IQEADPLVQQAHPVPDQQEPIQPDPE 685
E+ + P P P + P P+ + P + + P P P P P P
Sbjct: 529 EDTRVPPPQPPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPP 588
Query: 686 PEQSVSNQSSVRSPHPLVETSDPHLGTSEP----NAPMINIGSPQ----GASEAHSSNHP 737
P V SP P V + P P ++P + SP H++N P
Sbjct: 589 PPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQP 648
Query: 738 ASPAPNLSIIPYTHLQPTSL 757
AP + P + T +
Sbjct: 649 PMGAPTPTQAPTPSSETTQV 668
Score = 46.6 bits (109), Expect = 7e-05
Identities = 63/265 (23%), Positives = 88/265 (32%), Gaps = 60/265 (22%)
Query: 523 NLLQNQPSRSDEPTSLLTI--PYD--------------PLSSEPIIHEQPEPNQTEP-QP 565
N LQN+PS+ L + P D S P +P+P++ EP P
Sbjct: 355 NCLQNRPSQKPAKQCLPVVSRPVDCSKDKCSGGSNGGSSPSPNPPRTSEPKPSKPEPVMP 414
Query: 566 RTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVS 625
+ SD S P P + +P P N E + K E K + S
Sbjct: 415 KPSDSSKPETPKTPEQPSPKPQPPKHESPKP-------EEPENKHE-LPKQKESPKPQPS 466
Query: 626 ALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQ-------- 677
E+ P P + P+PE P++P + P + P + P PD
Sbjct: 467 KPED----SPKPEQ---PKPEESPKPEQPQIPEPTKPVSPPNEAQGPTPDDPYDASPVKN 519
Query: 678 -----------------EPIQPDPEPEQSV-SNQSSVRSPHPLVETSDPHLGTSEPNAPM 719
+P P P P + S V SP P V +S P P P+
Sbjct: 520 RRSPPPPKVEDTRVPPPQPPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPV 579
Query: 720 INIGSPQGASEAHSSNHPA--SPAP 742
+ P HS P SP P
Sbjct: 580 ASPPPPSPPPPVHSPPPPPVFSPPP 604
Score = 44.7 bits (104), Expect = 3e-04
Identities = 51/227 (22%), Positives = 74/227 (32%), Gaps = 29/227 (12%)
Query: 368 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVR 427
PP P F PP + + + ++ P + T+ + D S+ P
Sbjct: 632 PPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSP-------- 683
Query: 428 IVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNP 487
V PT V+ + T+ P T SS+ S + S P+ A S
Sbjct: 684 -VQAPTPVQSSTPSSEPTQVP---TPSSSESYQAPNLSPVQA----PTPVQAPTTSSETS 735
Query: 488 LTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLS 547
P P S+ + + P P+P P+ P+ S EP S P
Sbjct: 736 QVPTPSSESNQSPSQAPTPILEPVHAPTPNSKPV-----QSPTPSSEPVS------SPEQ 784
Query: 548 SEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTP 594
SE + E PEP P S + S P D D P
Sbjct: 785 SEEV--EAPEPTPVNPSSVPSSSPSTDTSIPPPENNDDDDDGDFVLP 829
>At3g19020 hypothetical protein
Length = 951
Score = 62.8 bits (151), Expect = 9e-10
Identities = 100/416 (24%), Positives = 140/416 (33%), Gaps = 54/416 (12%)
Query: 368 PPAPEF---PSPPKRKAQRKMVLDESSEESDVPLVKKTKR-KPDDGDDDDSEDGPPQKKK 423
PP PE P PPK++ + EES P K + KP++ PP+++
Sbjct: 522 PPKPEESPKPQPPKQETPKP-------EESPKPQPPKQETPKPEESPKPQ----PPKQET 570
Query: 424 KKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQH 483
K KP P E +TE P P L S + +DA PI
Sbjct: 571 PKPEESPKPQ--PPKQEQPPKTEAPKM--------GSPPLESPVPNDPYDASPIKKRRPQ 620
Query: 484 SSNPLTP-----IPESQPAEQTTSPP--HSPRSSFFQPSPT----EAPLWNLLQNQPSRS 532
+P T P+S P PP HSP F P P P+++ P S
Sbjct: 621 PPSPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVYS--PPPPVHS 678
Query: 533 DEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAF 592
P + + P S P +H P P + P P HS P P F
Sbjct: 679 PPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPV---HSPPPPVHSPPPPVQSPPPPPVF 735
Query: 593 TP------VSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPE 646
+P S P V S P + V + + P P P P P
Sbjct: 736 SPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPP-PV 794
Query: 647 RLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETS 706
P PI + P P + P+P P+ P P S S + S P ++
Sbjct: 795 HSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSPMANAPTPSSSESGEISTPVQAPTPDSE 854
Query: 707 DPHLGTSEPNAPMINIG---SPQGASEAHS---SNHPASPAPNLSIIPYTHLQPTS 756
D + ++P+ SP E + S+ P AP S + P+S
Sbjct: 855 DIEAPSDSNHSPVFKSSPAPSPDSEPEVEAPVPSSEPEVEAPKQSEATPSSSPPSS 910
Score = 59.7 bits (143), Expect = 8e-09
Identities = 94/424 (22%), Positives = 135/424 (31%), Gaps = 65/424 (15%)
Query: 365 SRLPPAPEFPSP--PKRKA----QRKMVLDESSEES---DVPLVKKTKRKPDDGDDDDSE 415
S P P+ P P PK+++ Q K + +ES + P ++ K KP+ + S+
Sbjct: 449 SHEPSNPKEPKPESPKQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSK 508
Query: 416 DGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDAL 475
PP+ ++ P P E + +PP + T S KP +
Sbjct: 509 QEPPKPEE-------SPKPEPPKPEESPKPQPPKQETPKPEESPKPQPPKQE-----TPK 556
Query: 476 PISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSS--------FFQPSPTE----APLWN 523
P + TP PE P Q P++ P P + +P+
Sbjct: 557 PEESPKPQPPKQETPKPEESPKPQPPKQEQPPKTEAPKMGSPPLESPVPNDPYDASPIKK 616
Query: 524 LLQNQPSRSDEPTSLLTIPYDP------------------LSSEPIIHEQPEPNQTEPQP 565
PS S E T T P P S P +H P P + P P
Sbjct: 617 RRPQPPSPSTEETK-TTSPQSPPVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVYSPPPP 675
Query: 566 RTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVS 625
S P S P + + PV P V S P V
Sbjct: 676 VHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVQSPPPPPV- 734
Query: 626 ALEEYYLTCPSPRRYPGPRPERLVDPDEPILANP----IQEADPLVQQAHPVPDQQEPIQ 681
+ P P P P + P P+ + P P+ PV P+
Sbjct: 735 ------FSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVH 788
Query: 682 PDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNA--PMINIGSPQGASEAHSSNHPAS 739
P P S S + SP P V + P T P A PM N +P + S +
Sbjct: 789 SPPPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSPMANAPTPSSSESGEISTPVQA 848
Query: 740 PAPN 743
P P+
Sbjct: 849 PTPD 852
Score = 59.3 bits (142), Expect = 1e-08
Identities = 64/243 (26%), Positives = 90/243 (36%), Gaps = 34/243 (13%)
Query: 514 PSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAP 573
P PT P ++ EP + P S+P E P+P Q P+P T H P
Sbjct: 404 PKPTPTP----------KAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSHE-P 452
Query: 574 RASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNS---ESIRKFMEVRKEKVSALEEY 630
P + +S P P + SP + E + E K++ S E
Sbjct: 453 SNPKEPKPESPKQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEP- 511
Query: 631 YLTCPSPRRYP---GPRPERLVDPDEPILANPIQEADPLVQ---QAHPVPDQ----QEPI 680
P P P P+PE P P P E P Q Q P P++ Q P
Sbjct: 512 ----PKPEESPKPEPPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPK 567
Query: 681 QPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASP 740
Q P+PE+S Q + P +T P +G+ +P+ N P AS P P
Sbjct: 568 QETPKPEESPKPQPPKQEQPP--KTEAPKMGSPPLESPVPN--DPYDASPI-KKRRPQPP 622
Query: 741 APN 743
+P+
Sbjct: 623 SPS 625
Score = 58.5 bits (140), Expect = 2e-08
Identities = 81/379 (21%), Positives = 131/379 (34%), Gaps = 52/379 (13%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRI 428
P P+ P P K + + + P ++ KP+ + S P+ + K +
Sbjct: 408 PTPKAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSHEPSNPKEPKPESPK-QE 466
Query: 429 VVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPL 488
K + +P E ++ P + KP + Q SS
Sbjct: 467 SPKTEQPKPKPESPKQESPKQEAPKPEQPKPKP----------------ESPKQESSKQE 510
Query: 489 TPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSS 548
P PE P + P SP+ QP E P +P S +P
Sbjct: 511 PPKPEESPKPEPPKPEESPKP---QPPKQETP-------KPEESPKP------------- 547
Query: 549 EPIIHEQPEPNQT-EPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPS 607
+P E P+P ++ +PQP + P S +P + T + P ++S
Sbjct: 548 QPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQEQPPKTEAPKMGSP--PLESPVP 605
Query: 608 NNSESIRKFMEVRKEKVS-ALEEYYLTCP-SPRRYPGPRPERLVDPDEPILANPIQEADP 665
N+ + R + S + EE T P SP + P P + P P+ + P P
Sbjct: 606 NDPYDASPIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVFSPP----PP 661
Query: 666 LVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEP--NAPMINIG 723
+ PV P+ P P S V SP P V + P + + P ++P +
Sbjct: 662 MHSPPPPVYSPPPPVHSPPPPPVH-SPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVH 720
Query: 724 SPQGASEAHSSNHPASPAP 742
SP ++ SP P
Sbjct: 721 SPPPPVQSPPPPPVFSPPP 739
Score = 40.4 bits (93), Expect = 0.005
Identities = 34/122 (27%), Positives = 51/122 (40%), Gaps = 11/122 (9%)
Query: 635 PSPRRYPGPR---PERLVDP---DEPILANPIQEADPLVQQAHPVPD--QQEPIQPDPEP 686
PSP+ P P+ P++ ++P +EP + P E P QQ P P+ EP P EP
Sbjct: 402 PSPKPTPTPKAPEPKKEINPPNLEEP--SKPKPEESPKPQQPSPKPETPSHEPSNPK-EP 458
Query: 687 EQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSI 746
+ Q S ++ P + P + + AP P+ S S+ P P S
Sbjct: 459 KPESPKQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPEESP 518
Query: 747 IP 748
P
Sbjct: 519 KP 520
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 62.0 bits (149), Expect = 2e-09
Identities = 79/298 (26%), Positives = 104/298 (34%), Gaps = 65/298 (21%)
Query: 484 SSNPLTPIPESQPA---EQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLT 540
+S PL +P QP+ + TSP P + +P E N P++S P
Sbjct: 19 ASPPLMALPPPQPSFPGDNATSPTREPTNG----NPPETT------NTPAQSSPP----- 63
Query: 541 IPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPIN 600
P PLSS P P P+ T P P T S P + P T+ P N
Sbjct: 64 -PETPLSSPPPEPSPPSPSLTGPPPTTIPVSPPPEPSPPPPLPTEAPP---------PAN 113
Query: 601 VVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPE---RLVDPDEPILA 657
V S P +S A +T PSP P P PE L PD P +
Sbjct: 114 PVSSPPPESSPP-------PPPPTEAPPTTPITSPSPPTNPPPPPESPPSLPAPDPP--S 164
Query: 658 NPIQEADPLVQQAH--------------PVPDQQEPIQPDPEPEQSVSNQSSVRSP---- 699
NP+ LV +H P P + P P E + + S SP
Sbjct: 165 NPL-PPPKLVPPSHSPPRHLPSPPASEIPPPPRHLPSPPASERPSTPPSDSEHPSPPPPG 223
Query: 700 HPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQPTSL 757
HP P G+ P SP S++ HP+ P+P +P P L
Sbjct: 224 HPKRREQPPPPGSKRPTP------SPPSPSDSKRPVHPSPPSPPEETLPPPKPSPDPL 275
Score = 54.3 bits (129), Expect = 3e-07
Identities = 80/321 (24%), Positives = 107/321 (32%), Gaps = 72/321 (22%)
Query: 432 PTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPI 491
P + P E + PP S + + P T +P+S + S P P
Sbjct: 57 PAQSSPPPETPLSSPPPEPSPPSPSLTGPPPTT----------IPVSPPPEPSPPPPLPT 106
Query: 492 PESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPI 551
PA +SPP P SS P PTEAP P S P T P P S P
Sbjct: 107 EAPPPANPVSSPP--PESSPPPPPPTEAP-----PTTPITSPSPP---TNPPPPPESPPS 156
Query: 552 IHEQPEPNQTEPQPRT--SDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNN 609
+ P+ P P+ HS PR P A +P P + S+P ++
Sbjct: 157 LPAPDPPSNPLPPPKLVPPSHSPPRHLPSPPASEIPPPPRHLPSP---PASERPSTPPSD 213
Query: 610 SESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQ 669
SE PSP P P+R P P P
Sbjct: 214 SEH----------------------PSPP--PPGHPKRREQPPPPGSKRPTPSP------ 243
Query: 670 AHPVP-DQQEPIQPDP--EPEQSVSNQSSVRSPHPLVETSDPHL-----GTSEPNAPMIN 721
P P D + P+ P P PE+++ P P +S P L S P+ P +
Sbjct: 244 --PSPSDSKRPVHPSPPSPPEETLPPPKPSPDPLPSNSSSPPTLLPPSSVVSPPSPPRKS 301
Query: 722 IGSPQGASEAHSSNHPASPAP 742
+ P S P +P P
Sbjct: 302 VSGPDNPS-------PNNPTP 315
Score = 52.4 bits (124), Expect = 1e-06
Identities = 82/305 (26%), Positives = 113/305 (36%), Gaps = 32/305 (10%)
Query: 404 RKPDDGDDDDSEDGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPAR---VTRSSAHSSK 460
R+P +G+ ++ + P Q + P EP+ T PP V+ S
Sbjct: 43 REPTNGNPPETTNTPAQSSPPPETPLSSPPP-EPSPPSPSLTGPPPTTIPVSPPPEPSPP 101
Query: 461 PALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAP 520
P L ++ A P+S+ SS P P E+ P TSP SP ++ P P E+P
Sbjct: 102 PPLPTEAPP---PANPVSSPPPESSPPPPPPTEAPPTTPITSP--SPPTN--PPPPPESP 154
Query: 521 LWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPA 580
+ PS P L+ P S P H P P +E P +P AS RP+
Sbjct: 155 PSLPAPDPPSNPLPPPKLV-----PPSHSPPRH-LPSPPASEIPPPPRHLPSPPASERPS 208
Query: 581 ARTTDTDSSTAFTP-------VSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLT 633
+D++ + P P +PS S S K V S EE T
Sbjct: 209 TPPSDSEHPSPPPPGHPKRREQPPPPGSKRPTPSPPSPSDSK-RPVHPSPPSPPEE---T 264
Query: 634 CPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPV--PDQQEPIQPDPEPEQSVS 691
P P+ P P P P P L P P V PD P P P + S S
Sbjct: 265 LPPPKPSPDPLPSNSSSP--PTLLPPSSVVSPPSPPRKSVSGPDNPSPNNPTPVTDNSSS 322
Query: 692 NQSSV 696
+ S+
Sbjct: 323 SGISI 327
Score = 32.7 bits (73), Expect = 1.00
Identities = 40/147 (27%), Positives = 49/147 (33%), Gaps = 27/147 (18%)
Query: 368 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVR 427
PPA E P PP+ S S P + P G E PP K
Sbjct: 186 PPASEIPPPPRHLPSP----PASERPSTPPSDSEHPSPPPPGHPKRREQPPPPGSK---- 237
Query: 428 IVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNP 487
+PT P+ +R P + S P T D LP +SS+P
Sbjct: 238 ---RPTPSPPSPSDSKRPVHP-------SPPSPPEETLPPPKPSPDPLP-----SNSSSP 282
Query: 488 LTPIPESQPAEQTTSPPHSPRSSFFQP 514
T +P S SPP PR S P
Sbjct: 283 PTLLPPS----SVVSPPSPPRKSVSGP 305
>At4g33970 extensin-like protein
Length = 699
Score = 57.4 bits (137), Expect = 4e-08
Identities = 70/283 (24%), Positives = 99/283 (34%), Gaps = 47/283 (16%)
Query: 484 SSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPS-----RSDEPTSL 538
SS P P P +P T+P H P P +P+ +PS EP+ +
Sbjct: 416 SSTPSKPSPVHKPTPVPTTPVHKPTPVPTTPVQKPSPVPTTPVQKPSPVPTTPVHEPSPV 475
Query: 539 LTIPYD---PLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASAR-PAARTTDTDSSTAFTP 594
L P D P+ S P+ ++P+P + PQP +P R P ++ ++P
Sbjct: 476 LATPVDKPSPVPSRPV--QKPQPPKESPQPDDPYDQSPVTKRRSPPPAPVNSPPPPVYSP 533
Query: 595 VSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEP 654
P V P +S P P P P P + P P
Sbjct: 534 PPPPPPVHSPPPPVHSPP----------------------PPPVYSPPPPPPPVHSPPPP 571
Query: 655 ILA--NPIQEADPLVQQAHP---VPDQQEPIQPDPEPEQSVSNQSSVRSPHPLV-----E 704
+ + P+ P V P P P+ P P S V SP P V
Sbjct: 572 VFSPPPPVYSPPPPVHSPPPPVHSPPPPAPVHSPPPPVHSPPPPPPVYSPPPPVFSPPPS 631
Query: 705 TSDPHLGTSEPNAPMIN---IGSPQGASEAHSSNHPA-SPAPN 743
S P + + P P IN + SP A PA +PAP+
Sbjct: 632 QSPPVVYSPPPRPPKINSPPVQSPPPAPVEKKETPPAHAPAPS 674
Score = 40.4 bits (93), Expect = 0.005
Identities = 52/252 (20%), Positives = 77/252 (29%), Gaps = 47/252 (18%)
Query: 436 EPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQ 495
EP+ + + P+ V +P S + +D P++ + S P P+
Sbjct: 471 EPSPVLATPVDKPSPVPSRPVQKPQPPKESPQPDDPYDQSPVT---KRRSPPPAPVNSPP 527
Query: 496 PAEQTTSPPHSPRSSFFQP--SPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIH 553
P + PP P S P SP P+++ P P + + P S P +H
Sbjct: 528 PPVYSPPPPPPPVHSPPPPVHSPPPPPVYSPPPPPPPVHSPPPPVFSPPPPVYSPPPPVH 587
Query: 554 EQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESI 613
P P + P P P + P PV P V S P + S
Sbjct: 588 SPPPPVHSPPPPAPVHSPPPPVHSPPPP-----------PPVYSPPPPVFSPPPSQS--- 633
Query: 614 RKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPV 673
P P PRP ++ P P+Q P +
Sbjct: 634 ---------------------PPVVYSPPPRPPKINSP-------PVQSPPPAPVEKKET 665
Query: 674 PDQQEPIQPDPE 685
P P D E
Sbjct: 666 PPAHAPAPSDDE 677
>At5g38560 putative protein
Length = 681
Score = 52.8 bits (125), Expect = 9e-07
Identities = 57/258 (22%), Positives = 84/258 (32%), Gaps = 51/258 (19%)
Query: 502 SPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQT 561
SPP S SS P P LQ QP+ P + P P S P++ P P
Sbjct: 11 SPPSS-NSSTTAPPP--------LQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVV 61
Query: 562 EPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRK 621
P P +S P+ + T + P+ + PS +
Sbjct: 62 SSPP-------PSSSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPAT---------- 104
Query: 622 EKVSALEEYYLTCPSPRRY--PGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEP 679
T P+P + P P P+ P P NP + P P P + P
Sbjct: 105 -----------TPPAPPQTVSPPPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPPGETP 153
Query: 680 IQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNA--------PMI----NIGSPQG 727
P P P ++ P P S P ++P+ P++ I P G
Sbjct: 154 SPPKPSPSTPTPTTTTSPPPPPATSASPPSSNPTDPSTLAPPPTPLPVVPREKPIAKPTG 213
Query: 728 ASEAHSSNHPASPAPNLS 745
+ + +N S +P S
Sbjct: 214 PASNNGNNTLPSSSPGKS 231
Score = 44.7 bits (104), Expect = 3e-04
Identities = 46/172 (26%), Positives = 61/172 (34%), Gaps = 16/172 (9%)
Query: 432 PTRVEPAAEVVRRTEPPARVTR----SSAHSSKPALTSDDDLNLFDALPISALLQHSSNP 487
P+ + VV + PP V+ SS S P +TS + P ++ S P
Sbjct: 42 PSPPQSPPPVVSSSPPPPVVSSPPPSSSPPPSPPVITSPPPT--VASSPPPPVVIASPPP 99
Query: 488 LTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLS 547
TP QT SPP P +S P+PT P S P P
Sbjct: 100 STPATTPPAPPQTVSPPPPPDASPSPPAPTTT------NPPPKPSPSPPGETPSPPGETP 153
Query: 548 SEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPI 599
S P +P P+ P TS P SA P + S+ A P P+
Sbjct: 154 SPP----KPSPSTPTPTTTTSPPPPPATSASPPSSNPTDPSTLAPPPTPLPV 201
>At4g18670 extensin-like protein
Length = 839
Score = 52.8 bits (125), Expect = 9e-07
Identities = 73/289 (25%), Positives = 99/289 (33%), Gaps = 47/289 (16%)
Query: 494 SQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIH 553
++P SPP +P PSP+ +P + P + P P + S P
Sbjct: 404 TRPPVVVPSPPTTPSPGGSPPSPSISPSPPITVPSPPTTPSPGGSPPSP-SIVPSPPSTT 462
Query: 554 EQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESI 613
P T P T S P + P + S T TP P SSP
Sbjct: 463 PSPGSPPTSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPTPGGSP----PSSP------- 511
Query: 614 RKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPV 673
T PSP G P + P PI P + P + P
Sbjct: 512 -------------------TTPSP---GGSPPSPSISPSPPITV-PSPPSTPTSPGSPPS 548
Query: 674 PDQQEPIQPDPEPEQSVSNQSSVRSP---HPLVETSDPHLGTSEPNAPMIN----IGSPQ 726
P P P P P + S + SP P + S P G S P++P I SP
Sbjct: 549 PSSPTPSSPIPSP-PTPSTPPTPISPGQNSPPIIPSPPFTGPSPPSSPSPPLPPVIPSPP 607
Query: 727 GASEAHSSNHPASPAPNLSIIPYTHLQPTSLSECINIFYHEASLRLRNV 775
SS P++P P + P+ HL LS + + H RLR++
Sbjct: 608 IVGPTPSSPPPSTPTPGTLLHPH-HLLLPQLSHLPHQYLHH---RLRHI 652
Score = 42.0 bits (97), Expect = 0.002
Identities = 91/411 (22%), Positives = 127/411 (30%), Gaps = 71/411 (17%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSED---GPPQKKKKK 425
P PSPP + S S V P G S PP
Sbjct: 406 PPVVVPSPPTTPSPGGSPPSPSISPSPPITVPSPPTTPSPGGSPPSPSIVPSPPSTTPSP 465
Query: 426 VRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSS 485
PT P PP+ T + S P+ + P S+ S
Sbjct: 466 GSPPTSPTTPTPGGS------PPSSPTTPTPGGSPPSSPTTPTPG---GSPPSSPTTPSP 516
Query: 486 NPLTPIPESQPAEQTT--SPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPY 543
P P P+ T SPP +P S PSP+ + + + P+ S PT P
Sbjct: 517 GGSPPSPSISPSPPITVPSPPSTPTSPGSPPSPSSPTPSSPIPSPPTPSTPPT-----PI 571
Query: 544 DP-LSSEPIIHEQPEPNQTEPQPRTSDH--------SAPRASARPAARTTDTDS-STAFT 593
P +S PII P P T P P +S S P P++ T + T
Sbjct: 572 SPGQNSPPII---PSPPFTGPSPPSSPSPPLPPVIPSPPIVGPTPSSPPPSTPTPGTLLH 628
Query: 594 PVSFPINVVDS-------------SPSNNSESIRKFMEVRKEKVS-----------ALEE 629
P + + PS + +R+ + + + L
Sbjct: 629 PHHLLLPQLSHLPHQYLHHRLRHILPSRHRHLLRRKHTIHRNHLHHNPRNLQFTALPLLP 688
Query: 630 YYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQS 689
YLTC P P P ++P P P P I P P P
Sbjct: 689 LYLTCRHRLNLASPPP-----PAPYYYSSPQPPPPPHYSLPPPTPTYHY-ISPPPPP--- 739
Query: 690 VSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASP 740
+ + SP P ++ P + S P P ++ P S AH S P+ P
Sbjct: 740 ----TPIHSPPP--QSHPPCIEYSPPPPPTVHYNPPPPPSPAHYSPPPSPP 784
Score = 41.6 bits (96), Expect = 0.002
Identities = 41/129 (31%), Positives = 49/129 (37%), Gaps = 21/129 (16%)
Query: 485 SNPLTPIPESQPAEQTTSPPH--SPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIP 542
S+P TP P P T+P SP SS PSP +P PS S P+ +T+P
Sbjct: 483 SSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPSPGGSP--------PSPSISPSPPITVP 534
Query: 543 YDPLSSEPIIHEQP----EPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFP 598
P S P P P + P P S P P + S FT S P
Sbjct: 535 SPP--STPTSPGSPPSPSSPTPSSPIPSPPTPSTPPTPISPGQNSPPIIPSPPFTGPSPP 592
Query: 599 INVVDSSPS 607
SSPS
Sbjct: 593 -----SSPS 596
>At1g68690 protein kinase, putative
Length = 708
Score = 52.0 bits (123), Expect = 2e-06
Identities = 69/269 (25%), Positives = 99/269 (36%), Gaps = 35/269 (13%)
Query: 481 LQHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLT 540
L ++++P TP P + P + PP+ R+ P T +P P + P L
Sbjct: 23 LNNATSPATPPPVTSPLPPSAPPPN--RAPPPPPPVTTSP-------PPVANGAPPPPLP 73
Query: 541 IPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPIN 600
P + S P P T P P+ S P +++ P A SS P S P
Sbjct: 74 KPPESSSPPPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPP-PPASVP-- 130
Query: 601 VVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPI 660
P S S + V ++ SP P RP + P P +P
Sbjct: 131 -----PPRPSPSPPILVRSPPPSVRPIQ-------SPPPPPSDRPTQSPPPPSP--PSPP 176
Query: 661 QEADPLVQQAHPVPDQQEPIQ--PDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAP 718
E Q+ P P + P Q P P P S++ S +SP P E + P PN+P
Sbjct: 177 SERPT---QSPPSPPSERPTQSPPPPSPPSPPSDRPS-QSPPPPPEDTKPQPPRRSPNSP 232
Query: 719 MINIGSPQGASE---AHSSNHPASPAPNL 744
SP + SN+P+ P L
Sbjct: 233 PPTFSSPPRSPPEILVPGSNNPSQNNPTL 261
Score = 45.8 bits (107), Expect = 1e-04
Identities = 55/213 (25%), Positives = 73/213 (33%), Gaps = 56/213 (26%)
Query: 368 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVR 427
PP P PSPP + + S S P P S PP
Sbjct: 82 PPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLP-------SSPPPPAS------ 128
Query: 428 IVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNP 487
V P R P+ ++ R+ PP+ S P SD + + +P
Sbjct: 129 --VPPPRPSPSPPILVRSPPPS----VRPIQSPPPPPSD---------------RPTQSP 167
Query: 488 LTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLS 547
P P S P+E+ T P SP S +P++S P S P P S
Sbjct: 168 PPPSPPSPPSERPTQSPPSPPS-----------------ERPTQSPPPPS----PPSPPS 206
Query: 548 SEPIIHEQPEPNQTEPQ-PRTSDHSAPRASARP 579
P P P T+PQ PR S +S P + P
Sbjct: 207 DRPSQSPPPPPEDTKPQPPRRSPNSPPPTFSSP 239
Score = 38.9 bits (89), Expect = 0.014
Identities = 36/134 (26%), Positives = 48/134 (34%), Gaps = 12/134 (8%)
Query: 635 PSPRRYPGPRPERLVD--------PDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEP 686
P P P P+ ++ P +P++ +P A P P+P P P P
Sbjct: 73 PKPPESSSPPPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPASVPPP 132
Query: 687 EQSVSNQSSVRSPHPLVE--TSDPHLGTSEP--NAPMINIGSPQGASEAHSSNHPASPAP 742
S S VRSP P V S P + P + P + SP S P S P
Sbjct: 133 RPSPSPPILVRSPPPSVRPIQSPPPPPSDRPTQSPPPPSPPSPPSERPTQSPPSPPSERP 192
Query: 743 NLSIIPYTHLQPTS 756
S P + P S
Sbjct: 193 TQSPPPPSPPSPPS 206
Score = 36.6 bits (83), Expect = 0.069
Identities = 39/168 (23%), Positives = 53/168 (31%), Gaps = 19/168 (11%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRI 428
P P P PP+ + + S S P + P PP
Sbjct: 68 PPPPLPKPPESSSPPPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPA 127
Query: 429 VVKPTRVEPAAEVVRRTEPPA-------------RVTRSSAHSSKPALTSDDDLNLFDAL 475
V P R P+ ++ R+ PP+ R T+S S P+ S+ +
Sbjct: 128 SVPPPRPSPSPPILVRSPPPSVRPIQSPPPPPSDRPTQSPPPPSPPSPPSERPTQSPPSP 187
Query: 476 PISALLQHSSNPLTPIPESQPAEQTTSP------PHSPRSSFFQPSPT 517
P Q P P P S Q+ P P PR S P PT
Sbjct: 188 PSERPTQSPPPPSPPSPPSDRPSQSPPPPPEDTKPQPPRRSPNSPPPT 235
>At2g27380 hypothetical protein
Length = 761
Score = 50.4 bits (119), Expect = 5e-06
Identities = 56/244 (22%), Positives = 82/244 (32%), Gaps = 32/244 (13%)
Query: 485 SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
S P+ P P +P T SPP P P+PT +P P + T Y
Sbjct: 533 SPPIKPPPVHKPPTPTYSPPIKPPPIHKPPTPTYSP--------PIKPPPVHKPPTPTYS 584
Query: 545 PLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDT---DSSTAFTPVSFPINV 601
P P +H+ P P + P H P + P + + T P+ P
Sbjct: 585 PPIKPPPVHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPP--P 642
Query: 602 VDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQ 661
V P+ K V+K P+P P +P + P P + P++
Sbjct: 643 VHKPPTPTYSPPIKPPPVQKP------------PTPTYSPPVKPPPVQLPPTPTYSPPVK 690
Query: 662 EADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMIN 721
P Q P P P++P P S P P+ P + P+ P
Sbjct: 691 ---PPPVQVPPTPTYSPPVKPPPVQVPPTPTYSPPIKPPPVQVPPTP----TTPSPPQGG 743
Query: 722 IGSP 725
G+P
Sbjct: 744 YGTP 747
Score = 50.4 bits (119), Expect = 5e-06
Identities = 53/218 (24%), Positives = 79/218 (35%), Gaps = 23/218 (10%)
Query: 485 SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
S P+ P P P T SPP P P+PT +P +P PT + + P
Sbjct: 147 SPPVKPPPVQMPPTPTYSPPIKPPPVHKPPTPTYSP-----PIKPPVHKPPTPIYSPPIK 201
Query: 545 PLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDS 604
P P +H+ P P + P H P + P + TP+ P +
Sbjct: 202 P----PPVHKPPTPIYSPPIKPPPVHKPPTPTYSPPVKPPPVHKPP--TPIYSP--PIKP 253
Query: 605 SPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEAD 664
P + + V+ V T P+P P +P + P P + P++
Sbjct: 254 PPVHKPPTPIYSPPVKPPPVQ-------TPPTPIYSPPVKPPPVHKPPTPTYSPPVK--S 304
Query: 665 PLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPL 702
P VQ+ P P PI+P P + S P P+
Sbjct: 305 PPVQKP-PTPTYSPPIKPPPVQKPPTPTYSPPIKPPPV 341
Score = 48.5 bits (114), Expect = 2e-05
Identities = 53/221 (23%), Positives = 76/221 (33%), Gaps = 29/221 (13%)
Query: 485 SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
S P+ P P +P T SPP P P+PT +P ++ P + PT Y
Sbjct: 483 SPPVQPPPVQKPPTPTYSPPVKPPPIQKPPTPTYSP---PIKPPPVKPPTPT------YS 533
Query: 545 PLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDT---DSSTAFTPVSFPINV 601
P P +H+ P P + P H P + P + + T P+ P
Sbjct: 534 PPIKPPPVHKPPTPTYSPPIKPPPIHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPP--P 591
Query: 602 VDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQ 661
V P+ K V K P+P P +P + P P + PI+
Sbjct: 592 VHKPPTPTYSPPIKPPPVHKP------------PTPTYSPPIKPPPVHKPPTPTYSPPIK 639
Query: 662 EADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPL 702
P P P PI+P P + S P P+
Sbjct: 640 ---PPPVHKPPTPTYSPPIKPPPVQKPPTPTYSPPVKPPPV 677
Score = 48.1 bits (113), Expect = 2e-05
Identities = 54/229 (23%), Positives = 83/229 (35%), Gaps = 18/229 (7%)
Query: 485 SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
S P+ P P +P T SPP P P+PT +P L +P PT + + P
Sbjct: 383 SPPVKPPPIQKPPTPTYSPPIKPPPLQKPPTPTYSPPIKLPPVKP-----PTPIYSPPVK 437
Query: 545 PLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSST-AFTPVSFPINVVD 603
P P +H+ P P + P H P + P + T ++P P V
Sbjct: 438 P----PPVHKPPTPIYSPPVKPPPVHKPPTPTYSPPIKPPPVKPPTPTYSPPVQP-PPVQ 492
Query: 604 SSPSNNSESIRKFMEVRKEKVSA----LEEYYLTCPSPRRYPGPRPERLVDPDEPILANP 659
P+ K ++K ++ + P+P P +P + P P + P
Sbjct: 493 KPPTPTYSPPVKPPPIQKPPTPTYSPPIKPPPVKPPTPTYSPPIKPPPVHKPPTPTYSPP 552
Query: 660 IQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDP 708
I+ P P P PI+P P + S P P+ + P
Sbjct: 553 IK---PPPIHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPPTP 598
Score = 43.1 bits (100), Expect = 7e-04
Identities = 55/231 (23%), Positives = 78/231 (32%), Gaps = 20/231 (8%)
Query: 485 SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
S P+ P P +P T SPP P P+PT +P P +P T Y
Sbjct: 79 SPPIYPPPIQKPPTPTYSPPIYPPPIQKPPTPTYSP-----PIYPPPIQKPP---TPTYS 130
Query: 545 PLSSEPIIHEQPEPNQTEP-------QPRTSDHSAPRASARPAARTTDTDSSTAFTPVSF 597
P P I + P P+ + P P T +S P T T S PV
Sbjct: 131 PPIYPPPIQKPPTPSYSPPVKPPPVQMPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPVHK 190
Query: 598 PINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILA 657
P + S P + + + + P+P P +P + P PI +
Sbjct: 191 PPTPIYSPPIKPPPVHKPPTPIYSPPIKPPPVH--KPPTPTYSPPVKPPPVHKPPTPIYS 248
Query: 658 NPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDP 708
PI+ P P P P++P P S P P+ + P
Sbjct: 249 PPIK---PPPVHKPPTPIYSPPVKPPPVQTPPTPIYSPPVKPPPVHKPPTP 296
Score = 42.4 bits (98), Expect = 0.001
Identities = 49/218 (22%), Positives = 75/218 (33%), Gaps = 30/218 (13%)
Query: 485 SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
S P+ P +P T SPP P P+PT +P +P PT + + P
Sbjct: 299 SPPVKSPPVQKPPTPTYSPPIKPPPVQKPPTPTYSP-----PIKPPPVKPPTPIYSPPVK 353
Query: 545 PLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTD---SSTAFTPVSFPINV 601
P P +H+ P P + P H P P + + T P+ P
Sbjct: 354 P----PPVHKPPTPIYSPPVKPPPVHKPPTPIYSPPVKPPPIQKPPTPTYSPPIKPP--P 407
Query: 602 VDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQ 661
+ P+ K V+ P+P P +P + P PI + P++
Sbjct: 408 LQKPPTPTYSPPIKLPPVKP-------------PTPIYSPPVKPPPVHKPPTPIYSPPVK 454
Query: 662 EADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSP 699
P P P PI+P P + + V+ P
Sbjct: 455 ---PPPVHKPPTPTYSPPIKPPPVKPPTPTYSPPVQPP 489
Score = 42.4 bits (98), Expect = 0.001
Identities = 50/220 (22%), Positives = 76/220 (33%), Gaps = 31/220 (14%)
Query: 485 SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
S P+ P P +P T SPP P P+PT +P P + T Y
Sbjct: 567 SPPIKPPPVHKPPTPTYSPPIKPPPVHKPPTPTYSP--------PIKPPPVHKPPTPTYS 618
Query: 545 PLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTD---SSTAFTPVSFPINV 601
P P +H+ P P + P H P + P + + T PV P
Sbjct: 619 PPIKPPPVHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPPVQKPPTPTYSPPVKPPPVQ 678
Query: 602 VDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQ 661
+ +P+ + ++V P+P P +P + P P + PI+
Sbjct: 679 LPPTPTYSPPVKPPPVQVP--------------PTPTYSPPVKPPPVQVPPTPTYSPPIK 724
Query: 662 EADPLVQQAHPVPDQQEPIQ---PDPEPEQSVSNQSSVRS 698
P Q P P P Q P P +S+ +R+
Sbjct: 725 ---PPPVQVPPTPTTPSPPQGGYGTPPPYAYLSHPIDIRN 761
>At4g32710 putative protein kinase
Length = 731
Score = 50.1 bits (118), Expect = 6e-06
Identities = 76/283 (26%), Positives = 97/283 (33%), Gaps = 46/283 (16%)
Query: 437 PAAEVVRRTEPPARVTRSSAH--SSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPES 494
PA V + PPA SS P L S L+ A P ++ +P +P ES
Sbjct: 22 PADSVPDTSSPPAPPLSPLPPPLSSPPPLPSPPPLSAPTASPPPLPVE---SPPSPPIES 78
Query: 495 QPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHE 554
P SPP P S PSP + PS S P L P S P
Sbjct: 79 PPPPLLESPPPPPLESPSPPSPH--------VSAPSGS-PPLPFLPAKPSPPPSSPPSET 129
Query: 555 QPEPNQTEPQPRT--SDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSES 612
P N P PR+ S+ + P +A P + S PIN +PS N+ S
Sbjct: 130 VPPGNTISPPPRSLPSESTPPVNTASPPPPSPPRRRSGPKPSFPPPINSSPPNPSPNTPS 189
Query: 613 IRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDP-----DEPILANPIQEADPLV 667
+ P P P+P P P +P P
Sbjct: 190 L-----------------------PETSPPPKPPLSTTPFPSSSTPPPKKSPAAVTLPFF 226
Query: 668 QQAHPVPD--QQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDP 708
A +PD PI P EP+ S + S +P PLV S P
Sbjct: 227 GPAGQLPDGTVAPPIGPVIEPKTSPAESISPGTPQPLVPKSLP 269
Score = 31.6 bits (70), Expect = 2.2
Identities = 31/117 (26%), Positives = 47/117 (39%), Gaps = 12/117 (10%)
Query: 645 PERLVDPDEPI--LANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPL 702
P+ P P+ L P+ PL P P P P P +S + P PL
Sbjct: 27 PDTSSPPAPPLSPLPPPLSSPPPLPS---PPPLSAPTASPPPLPVESPPSPPIESPPPPL 83
Query: 703 VETSDPHLGTSEPNAPMINIGSPQGAS-----EAHSSNHPASPAPNLSIIPYTHLQP 754
+E S P P+ P ++ +P G+ A S P+SP P+ ++ P + P
Sbjct: 84 LE-SPPPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPPSSP-PSETVPPGNTISP 138
>At1g79480 hypothetical protein
Length = 356
Score = 48.9 bits (115), Expect = 1e-05
Identities = 51/209 (24%), Positives = 77/209 (36%), Gaps = 43/209 (20%)
Query: 430 VKPTRVEPAAEVVRRTEPPARVTRSSA--HSSKPALTSDDDLNLFDALPISALLQHSSNP 487
+ P P++ + PP +T + SS P + + + P + +SNP
Sbjct: 63 INPPNTPPSSSYPGLSPPPGPITLPNPPDSSSNPNSNPNPPESSSNPNPPDSSSNPNSNP 122
Query: 488 LTPIPESQPAEQTTSP--PHSPRSSFFQPSPTEA-------------PLWNLLQNQPSRS 532
P+ P E +++P P S + P+P E+ P + N P S
Sbjct: 123 NPPVTVPNPPESSSNPNPPDSSSNPNSNPNPPESSSNPNPPVTVPNPPESSSNPNPPESS 182
Query: 533 DEPTSLLTIPYDPLSSEP----IIHEQPE----------PNQTEPQPRTSDHSAPRASAR 578
P +TIPY P SS P I+ PE P +EP P T S P
Sbjct: 183 SNPNPPITIPYPPESSSPNPPEIVPSPPESGYTPGPVLGPPYSEPGPSTPTGSIP----- 237
Query: 579 PAARTTDTDSSTAFTPVSFPINVVDSSPS 607
+ SS P+ +P + SPS
Sbjct: 238 -------SPSSGFLPPIVYPPPMAPPSPS 259
Score = 37.0 bits (84), Expect = 0.053
Identities = 32/126 (25%), Positives = 50/126 (39%), Gaps = 12/126 (9%)
Query: 635 PSPRRYPG--PRPERLVDPDEP-ILANPIQEADPLVQQAHPVPDQQE--------PIQPD 683
P YPG P P + P+ P +NP +P ++P P P
Sbjct: 69 PPSSSYPGLSPPPGPITLPNPPDSSSNPNSNPNPPESSSNPNPPDSSSNPNSNPNPPVTV 128
Query: 684 PEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPN 743
P P +S SN + S +P +S PN P+ P+ +S + ++P P
Sbjct: 129 PNPPESSSNPNPPDSSSNPNSNPNPPESSSNPNPPVTVPNPPESSSNPNPPESSSNPNPP 188
Query: 744 LSIIPY 749
++ IPY
Sbjct: 189 IT-IPY 193
Score = 36.6 bits (83), Expect = 0.069
Identities = 33/117 (28%), Positives = 50/117 (42%), Gaps = 13/117 (11%)
Query: 636 SPRRYPGPRPERLVDPDEP-ILANPIQEADPLVQQAHPVPDQQEPIQPD--------PEP 686
+P P P PE +P+ P +NP +P V +P P PD P P
Sbjct: 95 NPNSNPNP-PESSSNPNPPDSSSNPNSNPNPPVTVPNPPESSSNPNPPDSSSNPNSNPNP 153
Query: 687 EQSVSNQSS-VRSPHPLVETSDPHL--GTSEPNAPMINIGSPQGASEAHSSNHPASP 740
+S SN + V P+P +S+P+ +S PN P+ P+ +S P+ P
Sbjct: 154 PESSSNPNPPVTVPNPPESSSNPNPPESSSNPNPPITIPYPPESSSPNPPEIVPSPP 210
>At2g15880 unknown protein
Length = 727
Score = 48.1 bits (113), Expect = 2e-05
Identities = 75/364 (20%), Positives = 114/364 (30%), Gaps = 61/364 (16%)
Query: 416 DGPPQKKKKKVRIVVK------PTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDL 469
D P Q+ K+ +V+ + + ++ P+ V H +P S
Sbjct: 360 DRPKQRSAKECAVVISRPVDCSKDKCAGGSSQATPSKSPSPVPTRPVHKPQPPKESPQPN 419
Query: 470 NLFDALPISAL------------LQHSSNPLTPIPESQPAEQTTSPPHSPRSS------- 510
+ ++ P+ + HS P + P S P T SP H P+
Sbjct: 420 DPYNQSPVKFRRSPPPPQQPHHHVVHSPPPASSPPTSPPVHSTPSPVHKPQPPKESPQPN 479
Query: 511 --------FFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIP-----YDPLSSEPIIHEQPE 557
F+ SP P+ + P S P + + P Y P P+ P
Sbjct: 480 DPYDQSPVKFRRSPPPPPVHSPPPPSPIHSPPPPPVYSPPPPPPVYSPPPPPPVYSPPPP 539
Query: 558 PNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFM 617
P P P P S P + + PV P V S P S +
Sbjct: 540 PPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVYSPPPPPVHSPPPPV 599
Query: 618 EVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQ 677
V + P P Y P P + P P+ + P P+ PV
Sbjct: 600 HSPPPPVHS--------PPPPVYSPPPPPPVHSPPPPVFSPP----PPVHSPPPPVYSPP 647
Query: 678 EPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLG---TSEPNAPMINIGSPQGASEAHSS 734
P+ P P V+SP P S P L +S P +N P+ S+ +
Sbjct: 648 PPVYSPPPP--------PVKSPPPPPVYSPPLLPPKMSSPPTQTPVNSPPPRTPSQTVEA 699
Query: 735 NHPA 738
P+
Sbjct: 700 PPPS 703
>At2g38410 unknown protein
Length = 671
Score = 47.4 bits (111), Expect = 4e-05
Identities = 77/315 (24%), Positives = 112/315 (35%), Gaps = 46/315 (14%)
Query: 464 TSDDDL-----NLFDALPISALLQH----SSNPLTPIPESQPAEQTTSPP--HSPRSSFF 512
T DD+L +L D+L I L +H S +PL P S P SP+SS
Sbjct: 289 TGDDELLGRGLDLNDSLQI-LLAKHDAIASGSPLPVQASGSPLSVQASKPADSSPKSSEA 347
Query: 513 QPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSA 572
+ S + A S P IP + + I E+ E + E HS
Sbjct: 348 KDSSSIAG-----------SSSP-----IPATVSTGKSPIDEEYEEEEDEFAQLARRHSK 391
Query: 573 PRASARPAARTTDTDSSTAFTPVS--FPINVVDSSPSNNSESIRKFMEVRKEKVSALEEY 630
P PA+ TTD S + S + + D P N+ + +++ +
Sbjct: 392 P-----PASVTTDPTSLESHNAASNALALALPDPPPPVNTTKEQDMIDLLSITLCTPS-- 444
Query: 631 YLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSV 690
T P+P P P P D + I P D V P QQ+P QP + S
Sbjct: 445 --TPPAPSSQPSPPPPAGSDQNTHIYPQPQPRFDSYVA---PWAQQQQPQQPQAQQGYSQ 499
Query: 691 SNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYT 750
Q + + + S G S+ P G Q +A P++ N PY
Sbjct: 500 HQQHQQQQGYSQPQHSQQQQGYSQLQQPQPQQGYSQSQPQAQVQMQPSTRPQN----PYE 555
Query: 751 HLQPTSLSECINIFY 765
+ P S N +Y
Sbjct: 556 YPPPPWASTSANAYY 570
>At4g27520 early nodulin-like 2 predicted GPI-anchored protein
Length = 349
Score = 47.0 bits (110), Expect = 5e-05
Identities = 61/242 (25%), Positives = 94/242 (38%), Gaps = 51/242 (21%)
Query: 386 VLDESSEESDVPLVKKTKRKPDDGDDDDSED--GPPQ---------KKKKKVRIVVKPTR 434
VL+ + + D K ++ DDGD + S D GP KK +K+ +VV R
Sbjct: 73 VLEVNKADYDACNTKNPIKRVDDGDSEISLDRYGPFYFISGNEDNCKKGQKLNVVVISAR 132
Query: 435 VEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPES 494
+ A+ S H++ P ++ + P A SS+P++P +
Sbjct: 133 IPSTAQ--------------SPHAAAPGSSTPGSMTP----PGGAHSPKSSSPVSPT--T 172
Query: 495 QPAEQTTSP--PHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPII 552
P TT P HSP+SS T P + +S P S T P P
Sbjct: 173 SPPGSTTPPGGAHSPKSSSAVSPATSPP-----GSMAPKSGSPVSPTTSPPAP------- 220
Query: 553 HEQPEPNQTEP-QPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSE 611
P T P P ++ ++P A P + +T SS T + SSP +NS
Sbjct: 221 -----PKSTSPVSPSSAPMTSPPAPMAPKSSSTIPPSSAPMTSPPGSMAPKSSSPVSNSP 275
Query: 612 SI 613
++
Sbjct: 276 TV 277
>At5g07750 putative protein
Length = 1289
Score = 46.6 bits (109), Expect = 7e-05
Identities = 88/360 (24%), Positives = 119/360 (32%), Gaps = 49/360 (13%)
Query: 408 DGDDDDSEDG--PPQKKKKKVRIVVKPTRVEPAAEVVRRT-----EPPARVTRSSAHSSK 460
DG S DG P K K +R V + A + E P+ + +S H +
Sbjct: 565 DGKRATSPDGVIPKDAKTKYLRASVSSPDMRSRAPICSSPDSSPKETPSSLPPASPHQAP 624
Query: 461 P---ALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPP--------HSPRS 509
P +LTS+ L + +++ P P Q PP P S
Sbjct: 625 PPLPSLTSEAKTVLHSSQAVASPPPPPPPPPLPTYSHYQTSQLPPPPPPPPPFSSERPNS 684
Query: 510 SFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSD 569
P P P P S+ P S +P P P E+P P P
Sbjct: 685 GTVLPPPPPPP-------PPFSSERPNSGTVLPPPPPPPLPFSSERPNSGTVLPPP---- 733
Query: 570 HSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEE 629
S P S +A ST+ P S P P S+ +K S L+
Sbjct: 734 PSPPWKSVYASALAIPAICSTSQAPTSSP---TPPPPPPAYYSV-------GQKSSDLQT 783
Query: 630 YYLTCPSPRRYPGPRPERLVDPD-EPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQ 688
L PSP P P P V + E +L P P + P P P P +
Sbjct: 784 SQL--PSPPPPPPPPPFASVRRNSETLLPPPPPPPPPPFASVRRNSETLLPPPPPPPPWK 841
Query: 689 SVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIP 748
S+ +S H TS +S P P SP ++A+ P P P SI P
Sbjct: 842 SL--YASTFETHEACSTS-----SSPPPPPPPPPFSPLNTTKANDYILPPPPLPYTSIAP 894
>At3g24400 protein kinase, putative
Length = 694
Score = 45.8 bits (107), Expect = 1e-04
Identities = 66/263 (25%), Positives = 79/263 (29%), Gaps = 59/263 (22%)
Query: 487 PLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPL 546
PL P QP T PP + P P L L P PT++ IP
Sbjct: 17 PLPIPPPPQPLPVTPPPPPTALPPALPPPPPPTALPPALPPPP----PPTTVPPIPPSTP 72
Query: 547 SSEPIIHEQP-EPNQTEPQ-PRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDS 604
S P + P P+ T P P T + P P+ T S TP P +
Sbjct: 73 SPPPPLTPSPLPPSPTTPSPPLTPSPTTPSPPLTPSPPPAIT-PSPPLTPSPLPPSPTTP 131
Query: 605 SPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEAD 664
SP S SI PSP P P P + P P
Sbjct: 132 SPPPPSPSI---------------------PSPPLTPSPPPSSPLRPSSP---------- 160
Query: 665 PLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIG- 723
P P P P P P P T P +G+ P P G
Sbjct: 161 -------PPPSPATPSTPPRSP------------PPPSTPTPPPRVGSLSPPPPASPSGG 201
Query: 724 -SPQGASEAHSSNHPASPAPNLS 745
SP S S+ PA + LS
Sbjct: 202 RSPSTPSTTPGSSPPAQSSKELS 224
Score = 39.7 bits (91), Expect = 0.008
Identities = 28/99 (28%), Positives = 40/99 (40%), Gaps = 3/99 (3%)
Query: 486 NPLTPIPESQPAEQTTSPPHSPR-SSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
+P TP P P+ T SPP +P PSP P + L P+ P +IP
Sbjct: 86 SPTTPSPPLTPSPTTPSPPLTPSPPPAITPSPPLTP--SPLPPSPTTPSPPPPSPSIPSP 143
Query: 545 PLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAART 583
PL+ P P+ P + + PR+ P+ T
Sbjct: 144 PLTPSPPPSSPLRPSSPPPPSPATPSTPPRSPPPPSTPT 182
Score = 33.1 bits (74), Expect = 0.76
Identities = 30/116 (25%), Positives = 38/116 (31%), Gaps = 11/116 (9%)
Query: 641 PGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPH 700
P P L P P PI + P + P P P+ P P S
Sbjct: 48 PTALPPALPPPPPPTTVPPIPPSTP----SPPPPLTPSPLPPSPTTPSPPLTPSPTTPSP 103
Query: 701 PLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQPTS 756
PL + P + S P P SP S P P+P++ P T P S
Sbjct: 104 PLTPSPPPAITPSPPLTPSPLPPSPTTPS-------PPPPSPSIPSPPLTPSPPPS 152
>At3g19070 hypothetical protein
Length = 346
Score = 45.8 bits (107), Expect = 1e-04
Identities = 80/282 (28%), Positives = 118/282 (41%), Gaps = 52/282 (18%)
Query: 424 KKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQH 483
KK+ K E AA++++ RS+A ++ AL D N LP ++
Sbjct: 29 KKLDEETKEDEDEDAAKIIQ--------DRSTAREAEIALLMDLRQNF---LPPPSM--- 74
Query: 484 SSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPY 543
SS+ L P P+ T +P S S QP APL + ++ S S +P S T+
Sbjct: 75 SSSSLQP-----PSSSTLAPSSS---SSLQPP---APLIDFFRSSVSYSHQPPSSSTLA- 122
Query: 544 DPLSSEPIIHEQPEPNQTEPQPRTSDH----SAPRASARPAARTTDTDSSTAFTPVSFPI 599
SS P + + P + + QP S S+ S +P++ +T SS F P S P
Sbjct: 123 --TSSSPSL-QPPSMSSSSLQPPASLREFFTSSVSYSHQPSSSSTLATSS--FFPSSMPY 177
Query: 600 NV--VDSS---------PSNNSESIRKFMEVRKEKVSALEEYY----LTCPSPRRYPGPR 644
+V DSS PS+ S SI+ E K + L + + PSP P R
Sbjct: 178 SVRPPDSSDRPSLREFFPSSPSSSIQP-PESSSSKRARLSNIFPSPLSSSPSPFVNPFLR 236
Query: 645 PERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEP 686
P+ +P P NPI + P + P P+ P P P
Sbjct: 237 PQAQ-EPTIPNFINPIPQISPGLPALFPNPNPNPNPNPIPNP 277
>At1g62440 putative extensin-like protein (gnl|PID|e1310400
Length = 786
Score = 45.8 bits (107), Expect = 1e-04
Identities = 75/318 (23%), Positives = 101/318 (31%), Gaps = 31/318 (9%)
Query: 390 SSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPA 449
SS+E P + ++ S P K VR++ P + R T PP
Sbjct: 360 SSKECSSPASRSVDCSKFGCNNFFSPPPPSFKMSPTVRVLPPPPPSSKMSPTFRATPPPP 419
Query: 450 RVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPPHSPRS 509
S + + P S F A P + S + P P E SPP P S
Sbjct: 420 SSKMSPSFRATPPPPSSKMSPSFRATPPPPSSKMSPSVKAYPPPPPPPEYEPSPP--PPS 477
Query: 510 SFFQPS----PTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEP--IIHEQPEPNQTEP 563
S PS P PL + P S P + + P P S P I+ P P P
Sbjct: 478 SEMSPSVRAYPPPPPL-----SPPPPSPPPPYIYSSPPPPSPSPPPPYIYSSPPPVVNCP 532
Query: 564 QPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEK 623
P T P+ P+ R + S P SSP + + +
Sbjct: 533 -PTTQSPPPPKYEQTPSPREY-------YPSPSPPYYQYTSSPPPPT-----YYATQSPP 579
Query: 624 VSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPD 683
YY P P P P P P+ P++Q P P P+
Sbjct: 580 PPPPPTYYAVQSPPPPPPVYYPPVTASPPPP----PVYYT-PVIQSPPPPPVYYSPVTQS 634
Query: 684 PEPEQSVSNQSSVRSPHP 701
P P V +SP P
Sbjct: 635 PPPPPPVYYPPVTQSPPP 652
Score = 31.6 bits (70), Expect = 2.2
Identities = 33/126 (26%), Positives = 45/126 (35%), Gaps = 19/126 (15%)
Query: 635 PSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQS 694
PS R P P ++ P + P V +A+P P +P P P S + S
Sbjct: 425 PSFRATPPPPSSKMSPSFRATPPPPSSKMSPSV-KAYPPPPPPPEYEPSPPPPSSEMSPS 483
Query: 695 SVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQP 754
P P P L P+ P I +SS P SP+P PY + P
Sbjct: 484 VRAYPPP------PPLSPPPPSPPPPYI---------YSSPPPPSPSPP---PPYIYSSP 525
Query: 755 TSLSEC 760
+ C
Sbjct: 526 PPVVNC 531
>At1g23720 unknown protein
Length = 895
Score = 45.8 bits (107), Expect = 1e-04
Identities = 72/293 (24%), Positives = 102/293 (34%), Gaps = 34/293 (11%)
Query: 485 SNPLTPI--PESQPAEQTTSPPH---SPRSSFFQPSPTEA----PLWNLLQNQPSRSDEP 535
S+P P P +P ++ PP+ SP ++ PSP P + + P P
Sbjct: 591 SSPPPPYYSPAPKPVYKSPPPPYVYNSPPPPYYSPSPKPTYKSPPPPYVYSSPPPPYYSP 650
Query: 536 TSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRA---SARPAARTTDTDSSTAF 592
T T P P ++ P P P P+ + S P S+ P + T
Sbjct: 651 TPKPTYKSPP---PPYVYSSPPPPYYSPSPKPTYKSPPPPYVYSSPPPPYYSPAPKPTYK 707
Query: 593 TPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPE-RLVDP 651
+P P V SSP S + S Y + P P Y P P+ P
Sbjct: 708 SP---PPPYVYSSPPPPYYS----PSPKPTYKSPPPPYVYSSPPPPPYYSPSPKVEYKSP 760
Query: 652 DEPILAN----PIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSD 707
P + + P P V+ P P P P P S S + +SP P S
Sbjct: 761 PPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYS-SPPPPPYYSPSPKVEYKSPPPPYVYSS 819
Query: 708 PHLGTSEPNAPMINIGSPQGASEAHSSNHPA--SPAPNLSI----IPYTHLQP 754
P T +P + SP +S PA SP+P + PY + P
Sbjct: 820 PPPPTYYSPSPKVEYKSPPPPYVYNSPPPPAYYSPSPKIEYKSPPPPYVYSSP 872
Score = 39.7 bits (91), Expect = 0.008
Identities = 83/353 (23%), Positives = 115/353 (32%), Gaps = 60/353 (16%)
Query: 432 PTRVEPAAEVVRRTEPPARVTRSS-----AHSSKPALTSDDDLNLFDALPISALLQHSSN 486
P P+ + + ++ PP V S + S KPA S ++ P
Sbjct: 268 PPYYSPSPKPIYKSPPPPYVYNSPPPPYYSPSPKPAYKSPPPPYVYSFPP---------- 317
Query: 487 PLTPIPESQPAEQTTSPPH---SPRSSFFQPSPTEA---------------PLWNLLQNQ 528
P P +P ++ PP+ SP ++ PSP A P ++
Sbjct: 318 PPYYSPSPKPVYKSPPPPYVYNSPPPPYYSPSPKPAYKSPPPPYVYSSPPPPYYSPSPKP 377
Query: 529 PSRSDEPTSLLTIPYDPLSSE-----------PIIHEQPEPNQTEPQPRTSDHSAPRASA 577
+S P + + P P S P I+ P P P P+ S S P
Sbjct: 378 TYKSPPPPYVYSSPPPPYYSPSPKPVYKSPPPPYIYNSPPPPYYSPSPKPSYKSPPPPYV 437
Query: 578 RPAARTTDTDSSTAFTPVSFPINVVDSSP-----SNNSESIRKFMEVRKEKVSALEEYYL 632
+ S T S P V SSP S + + + K S YY
Sbjct: 438 YSSPPPPYYSPSPKLTYKSSPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYS 497
Query: 633 TCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSN 692
P P Y P P + + P P P V P P + P P P S S
Sbjct: 498 PSPKP-SYKSPPPPYVYNSPPP----PYYSPSPKVIYKSP-PHPHVCVCPPPPPCYSHSP 551
Query: 693 QSSVRS-PHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPA--SPAP 742
+ +S P P V S P P+ SP +SS P SPAP
Sbjct: 552 KIEYKSPPTPYVYHSPPPPPYYSPSPKPAYKSSP--PPYVYSSPPPPYYSPAP 602
>At1g62970 unknown protein
Length = 825
Score = 45.4 bits (106), Expect = 1e-04
Identities = 85/336 (25%), Positives = 130/336 (38%), Gaps = 55/336 (16%)
Query: 431 KPTRVEPAAEVVRRTE-------PPARVTRSSAHSSKPA----LTSDDDLNLFDALPISA 479
KPT P +E VR +E PAR + S+ SS+ A L S D + + P +
Sbjct: 307 KPT---PVSEPVRHSELVPWQYSEPARQYQLSSRSSEAAQLSLLPSVSDSS-HASQPTRS 362
Query: 480 LLQHS-------SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRS 532
H+ S P P P SQP + P S S +P P N S+S
Sbjct: 363 NQSHAVSKPQPVSKPHPPFPMSQPPPTSNPFPLSQPPSNSKPFPMSQSSQNSKPFPVSQS 422
Query: 533 DEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAF 592
+ + L + S+P+ Q N + P P S P ++++P + +S F
Sbjct: 423 SQKSKPLLVSQSSQRSKPLPVSQSLQN-SNPFP----VSQPSSNSKPFPVSQPQPASNPF 477
Query: 593 TPVSFP-INVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDP 651
PVS P N S S S + R F + S + +P +P
Sbjct: 478 -PVSQPRPNSQPFSMSQPSSTARPFPASQPPAAS------------KSFPISQPP---TT 521
Query: 652 DEPILANPIQEADPLVQQAHPVPDQQEPI-QPDPEPEQSVSNQ----SSVRSPHPLVETS 706
+P ++ P + P+ P + P+ QP P + + +Q SS SP P V S
Sbjct: 522 SKPFVSQPPNTSKPMPVSQPPTTSKPLPVSQPPPTFQSTCPSQPPAASSSLSPLPPVFNS 581
Query: 707 DPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAP 742
T +P ++ +P EA + P +PAP
Sbjct: 582 -----TQSFQSPPVST-TPSAVPEASTIPSPPAPAP 611
Score = 41.2 bits (95), Expect = 0.003
Identities = 59/229 (25%), Positives = 86/229 (36%), Gaps = 39/229 (17%)
Query: 455 SAHSSKPALTSDDDLNLFDALPISALLQHS--------SNPLTPIPESQPAEQTTSP--- 503
S+ SKP L S LP+S LQ+S S+ P P SQP + ++P
Sbjct: 422 SSQKSKPLLVSQSSQRS-KPLPVSQSLQNSNPFPVSQPSSNSKPFPVSQP-QPASNPFPV 479
Query: 504 ----PHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPN 559
P+S S QPS T P S+ + I P +S+P + + P +
Sbjct: 480 SQPRPNSQPFSMSQPSSTARPF------PASQPPAASKSFPISQPPTTSKPFVSQPPNTS 533
Query: 560 QTEP--QPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPI-NVVDSSPSNNSESIRKF 616
+ P QP T+ P + P ++T A + P+ V +S+ S S +
Sbjct: 534 KPMPVSQPPTTSKPLPVSQPPPTFQSTCPSQPPAASSSLSPLPPVFNSTQSFQSPPV--- 590
Query: 617 MEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADP 665
SA+ E T PSP P P + P P E P
Sbjct: 591 ----STTPSAVPE-ASTIPSP-----PAPAPVAQPTHVFNQTPPPEQTP 629
Score = 40.8 bits (94), Expect = 0.004
Identities = 54/203 (26%), Positives = 84/203 (40%), Gaps = 21/203 (10%)
Query: 445 TEPPARVTRSSAHSSKPALTSDDDL----NLFDALPISALLQHSSNPLTPIPESQPAEQT 500
++PPA S S+P TS + N +P+S +S PL P+ + P Q+
Sbjct: 504 SQPPA--ASKSFPISQPPTTSKPFVSQPPNTSKPMPVSQP-PTTSKPL-PVSQPPPTFQS 559
Query: 501 TSPPHSPR-SSFFQPSPTEAPLWNLLQN--QPSRSDEPTSL---LTIPYDPLSSEPIIHE 554
T P P SS P P P++N Q+ P S P+++ TIP P + P+
Sbjct: 560 TCPSQPPAASSSLSPLP---PVFNSTQSFQSPPVSTTPSAVPEASTIPSPPAPA-PVAQP 615
Query: 555 QPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIR 614
NQT P +T A R + P + T + +PI +S ++S R
Sbjct: 616 THVFNQTPPPEQTPKSGAARTDSEPEVSSFWTTCPYCYVLYEYPIIYEESVLKCQTKSCR 675
Query: 615 K---FMEVRKEKVSALEEYYLTC 634
+ ++V A E+ Y C
Sbjct: 676 RAYQAVKVPSPPPVAEEDSYYCC 698
>At5g53870 predicted GPI-anchored protein
Length = 370
Score = 45.1 bits (105), Expect = 2e-04
Identities = 49/216 (22%), Positives = 79/216 (35%), Gaps = 19/216 (8%)
Query: 499 QTTSPPHSPRSSFF--QPSPTEAPLWNLLQ-NQPSRSDEPTSLLTIPYDPLSSEPIIHEQ 555
Q ++P HSP S QP + +P+ + + PS+S P S ++ P SS PI H
Sbjct: 133 QPSAPAHSPVPSVSPTQPPKSHSPVSPVAPASAPSKSQPPRSSVSPAQPPKSSSPISHTP 192
Query: 556 P---------EPNQTEPQPRTSDHSAPRASARPAARTTDTDSST-AFTPVSFPINVVDSS 605
P P P++ + S PA + + + T + +P P + +
Sbjct: 193 ALSPSHATSHSPATPSPSPKSPSPVSHSPSHSPAHTPSHSPAHTPSHSPAHAPSHSPAHA 252
Query: 606 PSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADP 665
PS++ S PSP P P P P +P+ P
Sbjct: 253 PSHSPAHAPSHSPAHSPSHSPATP---KSPSPSSSPAQSPAT-PSPMTPQSPSPVSSPSP 308
Query: 666 LVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHP 701
Q+ DQ P+ P P ++ + +P P
Sbjct: 309 --DQSAAPSDQSTPLAPSPSETTPTADNITAPAPSP 342
Score = 40.8 bits (94), Expect = 0.004
Identities = 55/236 (23%), Positives = 85/236 (35%), Gaps = 49/236 (20%)
Query: 421 KKKKKVRIVVKPTRVEPAAEV------VRRTEPPARVTRSS--AHSSKPALTSDDDLNLF 472
+K +K+ +VV R +P+A V T+PP + S A +S P+ + ++
Sbjct: 118 QKGQKLIVVVLAVRNQPSAPAHSPVPSVSPTQPPKSHSPVSPVAPASAPSKSQPPRSSVS 177
Query: 473 DALPISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRS 532
A P SS+P++ P P+ T+ P +P S PSP + + PS S
Sbjct: 178 PAQP-----PKSSSPISHTPALSPSHATSHSPATPSPSPKSPSPVSHSPSHSPAHTPSHS 232
Query: 533 DEPTSLLTIPYDPL-----------------------SSEPIIHEQPEPNQT------EP 563
T + + P S P + P P+ + P
Sbjct: 233 PAHTPSHSPAHAPSHSPAHAPSHSPAHAPSHSPAHSPSHSPATPKSPSPSSSPAQSPATP 292
Query: 564 QPRTSDHSAPRASARPAARTTDTDSSTAF-------TPVSFPINVVDSSPSNNSES 612
P T +P +S P +D ST TP + I SP NS S
Sbjct: 293 SPMTPQSPSPVSSPSPDQSAAPSDQSTPLAPSPSETTPTADNITAPAPSPRTNSAS 348
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,198,208
Number of Sequences: 26719
Number of extensions: 1070736
Number of successful extensions: 6568
Number of sequences better than 10.0: 324
Number of HSP's better than 10.0 without gapping: 73
Number of HSP's successfully gapped in prelim test: 258
Number of HSP's that attempted gapping in prelim test: 5199
Number of HSP's gapped (non-prelim): 973
length of query: 952
length of database: 11,318,596
effective HSP length: 109
effective length of query: 843
effective length of database: 8,406,225
effective search space: 7086447675
effective search space used: 7086447675
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0012a.3


