

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.8
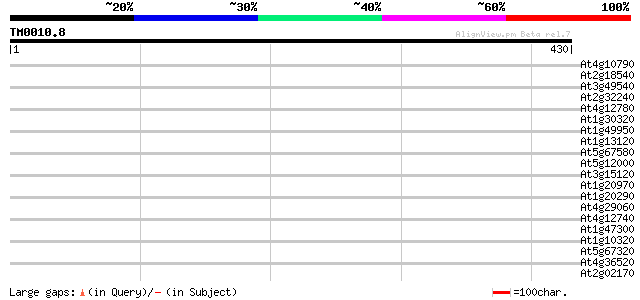
(430 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g10790 predicted protein of unknown function 40 0.002
At2g18540 putative vicilin storage protein (globulin-like) 39 0.007
At3g49540 hypothetical protein 38 0.012
At2g32240 putative myosin heavy chain 38 0.012
At4g12780 auxilin-like protein 37 0.021
At1g30320 unknown protein 37 0.021
At1g49950 telomere repeat binding factor 1 (TRB1)/DNA-binding pr... 37 0.027
At1g13120 unknown protein 36 0.036
At5g67580 telomere repeat binding factor 2 (TRB2) 35 0.061
At5g12000 putative receptor - like kinase 35 0.061
At3g15120 chaperone-like ATPase 35 0.10
At1g20970 hypothetical protein 35 0.10
At1g20290 hypothetical protein 34 0.14
At4g29060 unknown protein 33 0.30
At4g12740 adenine DNA glycosylase like protein 33 0.30
At1g47300 hypothetical protein 33 0.30
At1g10320 unknown protein 33 0.30
At5g67320 unknown protein 33 0.40
At4g36520 trichohyalin like protein 33 0.40
At2g02170 unknown protein 33 0.40
>At4g10790 predicted protein of unknown function
Length = 480
Score = 40.0 bits (92), Expect = 0.002
Identities = 33/82 (40%), Positives = 38/82 (46%), Gaps = 14/82 (17%)
Query: 21 EAARRREEVRQGEEAAAAY----ERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAAV 76
EA RR +R EE AAY E A +Q ER E +AA A + EEE A
Sbjct: 311 EAEERRTNLRLREEQDAAYRAALEADQAREQQRQEEKERLEREAAEAERKLKEEEEA--- 367
Query: 77 EREVEAERERAAAEAEEDAAEK 98
RERAA EAEE A +
Sbjct: 368 -------RERAAREAEERQAAR 382
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 38.5 bits (88), Expect = 0.007
Identities = 32/94 (34%), Positives = 44/94 (46%), Gaps = 7/94 (7%)
Query: 14 ESLKRSHEAARRREE--VRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEE 71
E KR E AR+REE R+ EEA E E+ A +R E + +A EE
Sbjct: 468 ERKKREEEEARKREEERKREEEEAKRREEERKKREEEAEQARKREEEREKEEEMAKKREE 527
Query: 72 NAAAVEREVEAERERAAAE----AEEDAAEKYEQ 101
ERE E ER+R + EE+A ++ E+
Sbjct: 528 ERQRKERE-EVERKRREEQERKRREEEARKREEE 560
Score = 38.1 bits (87), Expect = 0.009
Identities = 31/92 (33%), Positives = 41/92 (43%), Gaps = 5/92 (5%)
Query: 11 RLDESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEE 70
R ++ KR E AR+REE R+ EE A E E+ E E K EE
Sbjct: 542 REEQERKRREEEARKREEERKREEEMAKRR----EQERQRKEREEVERKIREEQERKREE 597
Query: 71 ENAAAVERE-VEAERERAAAEAEEDAAEKYEQ 101
E A E+E + ERE + E+ A K E+
Sbjct: 598 EMAKRREQERQKKEREEMERKKREEEARKREE 629
Score = 37.0 bits (84), Expect = 0.021
Identities = 29/93 (31%), Positives = 42/93 (44%), Gaps = 4/93 (4%)
Query: 13 DESLKRSHEAARRREEVR--QGEEAAAAYERHVAEYEQYMAAYERYE--AKAAAAAVAMA 68
+E KR E A+RREE R + EEA A +R ++ A +R E + V
Sbjct: 481 EEERKREEEEAKRREEERKKREEEAEQARKREEEREKEEEMAKKREEERQRKEREEVERK 540
Query: 69 EEENAAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E RE EA + + EE+ A++ EQ
Sbjct: 541 RREEQERKRREEEARKREEERKREEEMAKRREQ 573
Score = 36.2 bits (82), Expect = 0.036
Identities = 30/92 (32%), Positives = 39/92 (41%), Gaps = 4/92 (4%)
Query: 11 RLDESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEE 70
R +E R E A+RREE ER E E+ A +R E + A E
Sbjct: 439 RKEEEEARKREEAKRREEEEAKRREEEETERKKREEEE---ARKREEERKREEEEAKRRE 495
Query: 71 ENAAAVEREVEAERER-AAAEAEEDAAEKYEQ 101
E E E E R+R E EE+ A+K E+
Sbjct: 496 EERKKREEEAEQARKREEEREKEEEMAKKREE 527
Score = 35.4 bits (80), Expect = 0.061
Identities = 27/90 (30%), Positives = 37/90 (41%), Gaps = 3/90 (3%)
Query: 14 ESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENA 73
E ++ E AR+REE ++ EE A + E E E EA+ EEE
Sbjct: 436 ERRRKEEEEARKREEAKRREEEEA---KRREEEETERKKREEEEARKREEERKREEEEAK 492
Query: 74 AAVEREVEAERERAAAEAEEDAAEKYEQSA 103
E + E E A E+ EK E+ A
Sbjct: 493 RREEERKKREEEAEQARKREEEREKEEEMA 522
Score = 35.0 bits (79), Expect = 0.080
Identities = 31/98 (31%), Positives = 38/98 (38%), Gaps = 14/98 (14%)
Query: 11 RLDESLKRSHEAARRREEVRQG-----------EEAAAAYERHVA---EYEQYMAAYERY 56
R ++ KR E A+RRE+ RQ EE A E +A E E+ E
Sbjct: 588 REEQERKREEEMAKRREQERQKKEREEMERKKREEEARKREEEMAKIREEERQRKEREDV 647
Query: 57 EAKAAAAAVAMAEEENAAAVEREVEAERERAAAEAEED 94
E K EEE E AE ER E EE+
Sbjct: 648 ERKRREEEAMRREEERKREEEAAKRAEEERRKKEEEEE 685
Score = 35.0 bits (79), Expect = 0.080
Identities = 32/97 (32%), Positives = 43/97 (43%), Gaps = 8/97 (8%)
Query: 7 KALDRLDESLKRSHEA--ARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAA 64
+A R +E KR EA AR+REE R+ EE A E E+ E E K
Sbjct: 490 EAKRREEERKKREEEAEQARKREEEREKEEEMAKKR----EEERQRKEREEVERKRREEQ 545
Query: 65 VAMAEEENAAAVEREVEAERERAAAEAEEDAAEKYEQ 101
EE A +RE E +RE A+ E ++ E+
Sbjct: 546 ERKRREEEAR--KREEERKREEEMAKRREQERQRKER 580
Score = 33.1 bits (74), Expect = 0.30
Identities = 31/101 (30%), Positives = 43/101 (41%), Gaps = 18/101 (17%)
Query: 7 KALDRLDESLKRSHEAARRR----EEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAA 62
K + ++E +R E RR EE R+ EEA E E+ E E K
Sbjct: 419 KLMREIEERKRREEEEIERRRKEEEEARKREEAKRREEEEAKRREE-----EETERKKR- 472
Query: 63 AAVAMAEEENAAAVEREVEAERERAAAEAEEDAAEKYEQSA 103
EEE A +RE E +RE A+ E+ +K E+ A
Sbjct: 473 ------EEEEAR--KREEERKREEEEAKRREEERKKREEEA 505
Score = 29.6 bits (65), Expect = 3.4
Identities = 35/114 (30%), Positives = 48/114 (41%), Gaps = 24/114 (21%)
Query: 11 RLDESLKRSHEAARR--REEV---------RQGEEAAA---AYERHVAEYEQYMAAYERY 56
R +E KR + +R REEV R+ EE A ER E E+
Sbjct: 563 REEEMAKRREQERQRKEREEVERKIREEQERKREEEMAKRREQERQKKEREEMERKKREE 622
Query: 57 EAKAAAAAVAMAEEENAAAVEREVEAERERAAAEA---------EEDAAEKYEQ 101
EA+ +A EE ERE + ER+R EA EE+AA++ E+
Sbjct: 623 EARKREEEMAKIREEERQRKERE-DVERKRREEEAMRREEERKREEEAAKRAEE 675
>At3g49540 hypothetical protein
Length = 166
Score = 37.7 bits (86), Expect = 0.012
Identities = 33/88 (37%), Positives = 43/88 (48%), Gaps = 10/88 (11%)
Query: 29 VRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAAVEREVEAERERAA 88
V + A A VA + AA E EAK A AVA A E+ AVE E +AE E
Sbjct: 80 VEESSSADAGEAAVVAPEKVENAATENAEAKVEAVAVA-APEKVEVAVEAEKKAEAEPVK 138
Query: 89 AEAE---------EDAAEKYEQSATVTV 107
AEAE ++ +++ E+ A VTV
Sbjct: 139 AEAEPVKAEAEPVKEESKQEEKEAVVTV 166
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 37.7 bits (86), Expect = 0.012
Identities = 31/116 (26%), Positives = 54/116 (45%), Gaps = 31/116 (26%)
Query: 7 KALDRLDESLKRSHEAARRREEV-----------------------------RQGEEAAA 37
K L L+E LKRS E A++ EE+ ++ EE A
Sbjct: 198 KKLIELEEGLKRSAEEAQKFEELHKQSASHADSESQKALEFSELLKSTKESAKEMEEKMA 257
Query: 38 AYERHVAEYEQYMAAYERYEA--KAAAAAVAMAEEENAAAVEREVEAERERAAAEA 91
+ ++ + E + M+ E+ EA K++A +A +EE A + R +E E++ ++ EA
Sbjct: 258 SLQQEIKELNEKMSENEKVEAALKSSAGELAAVQEELALSKSRLLETEQKVSSTEA 313
>At4g12780 auxilin-like protein
Length = 904
Score = 37.0 bits (84), Expect = 0.021
Identities = 38/114 (33%), Positives = 47/114 (40%), Gaps = 19/114 (16%)
Query: 17 KRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAAV 76
K A + + R+ E AA H E+ A AKAAA A AE+ A A
Sbjct: 571 KVQRAAVGKATDARERAERAAVQRAHAEARERAAAGARDKAAKAAAEAREKAEKAAAEAK 630
Query: 77 ER----------EVEAER---ERAAAEAEEDAA------EKYEQSATVTVFFGS 111
ER V AER ERAAAEA AA ++ E + + FF S
Sbjct: 631 ERANAEAREKETRVRAERAAVERAAAEARGRAAAQAKAKQQQENTNDLDSFFSS 684
Score = 36.2 bits (82), Expect = 0.036
Identities = 31/94 (32%), Positives = 40/94 (41%), Gaps = 7/94 (7%)
Query: 7 KALDRLDESLKRSHEAARRREE---VRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAA 63
KA +R E +R + R E RQ E A R A E A+ + + A
Sbjct: 524 KAQEREKEEREREQKRIERERERLVARQAVERATREARERAATE----AHAKVQRAAVGK 579
Query: 64 AVAMAEEENAAAVEREVEAERERAAAEAEEDAAE 97
A E AAV+R RERAAA A + AA+
Sbjct: 580 ATDARERAERAAVQRAHAEARERAAAGARDKAAK 613
Score = 34.3 bits (77), Expect = 0.14
Identities = 36/108 (33%), Positives = 50/108 (45%), Gaps = 10/108 (9%)
Query: 7 KALDRLDESL--KRSHEAARRREEVRQGEEAAAAYERHV------AEYEQYMAAYERYEA 58
K ++R E L +++ E A R R EA A +R A AA +R A
Sbjct: 538 KRIERERERLVARQAVERATREARERAATEAHAKVQRAAVGKATDARERAERAAVQRAHA 597
Query: 59 KAAAAAVAMAEEENAAAVEREVEAERERAAAEAEEDA-AEKYEQSATV 105
+A A A A ++ AA E + E+AAAEA+E A AE E+ V
Sbjct: 598 EARERAAAGARDK-AAKAAAEAREKAEKAAAEAKERANAEAREKETRV 644
Score = 30.8 bits (68), Expect = 1.5
Identities = 30/106 (28%), Positives = 47/106 (44%), Gaps = 12/106 (11%)
Query: 10 DRLDESLKRSHEAARRREEV-------RQGE--EAAAAYERHVAEYEQYMAAYERYEAKA 60
D +D++ + A RRE+ R+G+ E + ER + E +Q ER E +A
Sbjct: 462 DAMDKAEAKFRHAKERREKENLKASRSREGDHTENYDSRERELRE-KQVRLDRERAEREA 520
Query: 61 AAAAVAMAEEENAAAVEREVEAERERAAAE--AEEDAAEKYEQSAT 104
E+E ++ +E ERER A E E E++AT
Sbjct: 521 EMEKAQEREKEEREREQKRIERERERLVARQAVERATREARERAAT 566
Score = 28.5 bits (62), Expect = 7.5
Identities = 33/111 (29%), Positives = 55/111 (48%), Gaps = 18/111 (16%)
Query: 9 LDRLDE-SLKRSHEAARRREEVRQGEEA---------AAAYERHVAEYE-QYMAAYERYE 57
+D LD+ S+ R+ AA + GE++ AAA + + + E ++ A ER E
Sbjct: 420 MDELDDFSIGRNQTAANGYPDPSSGEDSDVFSTAAASAAAMKDAMDKAEAKFRHAKERRE 479
Query: 58 AKAAAAAVAMAEE--ENAAAVERE-----VEAERERAAAEAEEDAAEKYEQ 101
+ A+ + + EN + ERE V +RERA EAE + A++ E+
Sbjct: 480 KENLKASRSREGDHTENYDSRERELREKQVRLDRERAEREAEMEKAQEREK 530
>At1g30320 unknown protein
Length = 509
Score = 37.0 bits (84), Expect = 0.021
Identities = 31/97 (31%), Positives = 47/97 (47%), Gaps = 8/97 (8%)
Query: 3 VDWWKALDRLDESLKRSHEAARRREEVR----QGEEAA---AAYERHVAEYEQYMAAYER 55
+++ K +E+ K H A +REE+R + +E A A R A+ EQ A E
Sbjct: 393 IEFEKRATAWEEAEKSKHNARYKREEIRIQAWESQEKAKLEAEMRRIEAKVEQMKAEAEA 452
Query: 56 YEAKAAAAAVAMAEEENAAAVEREVEAERERAAAEAE 92
K A A +EE+ A A R+ + E+A AEA+
Sbjct: 453 KIMKKIALAKQRSEEKRALAEARKTR-DAEKAVAEAQ 488
Score = 30.4 bits (67), Expect = 2.0
Identities = 27/98 (27%), Positives = 50/98 (50%), Gaps = 7/98 (7%)
Query: 1 MDVDWWKALDRLDESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKA 60
M++ W + + +E K+++ A +++ + E+ A A+E AE ++ A Y+R E +
Sbjct: 367 MNIAAWASKE--EEENKKNNGDAEEAQKI-EFEKRATAWEE--AEKSKHNARYKREEIRI 421
Query: 61 AAAAVAMAEEENAAAVEREVEAERERAAAEAEEDAAEK 98
A E+ A R +EA+ E+ AEAE +K
Sbjct: 422 QAWE--SQEKAKLEAEMRRIEAKVEQMKAEAEAKIMKK 457
>At1g49950 telomere repeat binding factor 1 (TRB1)/DNA-binding
protein PcMYB1, putative
Length = 300
Score = 36.6 bits (83), Expect = 0.027
Identities = 21/64 (32%), Positives = 34/64 (52%)
Query: 44 AEYEQYMAAYERYEAKAAAAAVAMAEEENAAAVEREVEAERERAAAEAEEDAAEKYEQSA 103
++ + +A + AAA A A E AA+ EA +E AAEAE +AA+ + + A
Sbjct: 225 SQIDTEIARMKSMNVHEAAAVAAQAVAEAEAAMAEAEEAAKEAEAAEAEAEAAQAFAEEA 284
Query: 104 TVTV 107
+ T+
Sbjct: 285 SKTL 288
>At1g13120 unknown protein
Length = 611
Score = 36.2 bits (82), Expect = 0.036
Identities = 35/112 (31%), Positives = 54/112 (47%), Gaps = 11/112 (9%)
Query: 7 KALDRLDESLKRSHEAARRREEVR-QGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAV 65
+ALD +++R H+ + EE + + EEA R +++ E+ A+A A
Sbjct: 197 EALDTHLTAVQREHKIKSQIEERKIRSEEAQEEARRKERAHQEEKIRQEKARAEAQMLAK 256
Query: 66 AMAEEENAAAVEREVEAERERAAAE---AEEDAAEK--YEQSATVTVFFGSS 112
AEEE ++EVE + R AE A+ AAE+ EQ A + GSS
Sbjct: 257 IRAEEE-----KKEVERKAAREVAEKEVADRKAAEQKLAEQKAVIESVTGSS 303
>At5g67580 telomere repeat binding factor 2 (TRB2)
Length = 299
Score = 35.4 bits (80), Expect = 0.061
Identities = 27/89 (30%), Positives = 42/89 (46%), Gaps = 2/89 (2%)
Query: 21 EAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAAVEREV 80
E +++ + E A + + + E YM + A AAA A+AE E A +
Sbjct: 206 EGNNKKDPTKPEENGANSLTKFRVDGELYMIKGMTAQEAAEAAARAVAEAEFAITEAEQA 265
Query: 81 EAERERAAAEAEEDAAEKYEQSATVTVFF 109
E ERA AEAE AA+ + ++A + F
Sbjct: 266 AKEAERAEAEAE--AAQIFAKAAMKALKF 292
>At5g12000 putative receptor - like kinase
Length = 703
Score = 35.4 bits (80), Expect = 0.061
Identities = 29/65 (44%), Positives = 34/65 (51%), Gaps = 7/65 (10%)
Query: 21 EAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAAVEREV 80
E ARR EE R EEAA A VAE E+ A R +AA A MAE E + E+
Sbjct: 332 EEARRFEEARNAEEAALA----VAEMEK---AKCRAALEAAEKAQRMAELEGQRRKQAEM 384
Query: 81 EAERE 85
+A RE
Sbjct: 385 KARRE 389
>At3g15120 chaperone-like ATPase
Length = 1954
Score = 34.7 bits (78), Expect = 0.10
Identities = 24/83 (28%), Positives = 43/83 (50%), Gaps = 5/83 (6%)
Query: 27 EEVRQGEEAAAAYERHVAEYEQYM----AAYERYEAKAAAAAVAMAEEENAAAVER-EVE 81
E+V++ + + + + +E E+ M AA E+Y+ K A E EN A V+R E E
Sbjct: 192 EDVQKESDTSNSEDESASESEESMQADSAAREKYQEKKATKRSVFLESENEAEVDRTETE 251
Query: 82 AERERAAAEAEEDAAEKYEQSAT 104
+E + + E D +++ +S T
Sbjct: 252 SEDGTDSTDNEIDDSDEEGESET 274
>At1g20970 hypothetical protein
Length = 1498
Score = 34.7 bits (78), Expect = 0.10
Identities = 24/67 (35%), Positives = 37/67 (54%), Gaps = 7/67 (10%)
Query: 17 KRSHEAARRREEVRQGEEAAAAYERH----VAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
K E ++ EE R+ +EAA E+H +A+ ++ M ++ E KA A AV A++E
Sbjct: 1213 KEEEELIKKEEEKRKQKEAAKMKEQHRLEEIAKAKEAMERKKKREEKAKARAVLKAQKE- 1271
Query: 73 AAAVERE 79
A ERE
Sbjct: 1272 --AEERE 1276
>At1g20290 hypothetical protein
Length = 497
Score = 34.3 bits (77), Expect = 0.14
Identities = 24/87 (27%), Positives = 31/87 (35%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 398 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 457
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKY 99
E E E E E E EE+ +KY
Sbjct: 458 EEEEEEEEEEEEEEEEEEEEEEEEKKY 484
Score = 33.9 bits (76), Expect = 0.18
Identities = 25/95 (26%), Positives = 34/95 (35%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 393 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 452
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQSATVTV 107
E E E E E E EE+ E+ E+ + V
Sbjct: 453 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKYMIV 487
Score = 33.9 bits (76), Expect = 0.18
Identities = 25/95 (26%), Positives = 35/95 (36%)
Query: 7 KALDRLDESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVA 66
+A + +E + E EE + EE E E E+ E E +
Sbjct: 362 QAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 421
Query: 67 MAEEENAAAVEREVEAERERAAAEAEEDAAEKYEQ 101
EEE E E E E E E EE+ E+ E+
Sbjct: 422 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 456
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 391 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 450
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 451 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 479
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 380 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 439
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 440 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 468
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 384 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 443
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 444 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 472
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 388 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 447
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 448 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 476
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 369 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 428
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 429 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 457
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 373 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 432
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 433 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 461
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 387 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 446
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 447 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 475
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 382 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 441
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 442 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 470
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 381 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 440
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 441 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 469
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 392 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 451
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 452 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 480
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 386 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 445
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 446 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 474
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 383 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 442
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 443 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 471
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 371 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 430
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 431 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 459
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 374 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 433
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 434 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 462
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 375 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 434
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 435 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 463
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 379 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 438
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 439 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 467
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 389 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 448
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 449 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 477
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 385 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 444
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 445 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 473
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 390 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 449
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 450 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 478
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 378 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 437
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 438 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 466
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 376 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 435
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 436 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 464
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 377 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 436
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 437 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 465
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 370 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 429
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 430 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 458
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/89 (26%), Positives = 32/89 (34%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEEN 72
+E + E EE + EE E E E+ E E + EEE
Sbjct: 372 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 431
Query: 73 AAAVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 432 EEEEEEEEEEEEEEEEEEEEEEEEEEEEE 460
Score = 32.7 bits (73), Expect = 0.40
Identities = 23/87 (26%), Positives = 33/87 (37%)
Query: 15 SLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAA 74
++ ++ E EE + EE E E E+ E E + EEE
Sbjct: 359 TIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 418
Query: 75 AVEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 419 EEEEEEEEEEEEEEEEEEEEEEEEEEE 445
Score = 32.0 bits (71), Expect = 0.68
Identities = 23/86 (26%), Positives = 31/86 (35%)
Query: 16 LKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAA 75
++ E EE + EE E E E+ E E + EEE
Sbjct: 361 VQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 420
Query: 76 VEREVEAERERAAAEAEEDAAEKYEQ 101
E E E E E E EE+ E+ E+
Sbjct: 421 EEEEEEEEEEEEEEEEEEEEEEEEEE 446
Score = 28.5 bits (62), Expect = 7.5
Identities = 21/73 (28%), Positives = 26/73 (34%)
Query: 29 VRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAAVEREVEAERERAA 88
V GE E E E+ E E + EEE E E E E E
Sbjct: 353 VHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 412
Query: 89 AEAEEDAAEKYEQ 101
E EE+ E+ E+
Sbjct: 413 EEEEEEEEEEEEE 425
Score = 28.1 bits (61), Expect = 9.8
Identities = 21/73 (28%), Positives = 27/73 (36%)
Query: 29 VRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAAVEREVEAERERAA 88
V +G A E E E+ E E + EEE E E E E E
Sbjct: 355 VGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 414
Query: 89 AEAEEDAAEKYEQ 101
E EE+ E+ E+
Sbjct: 415 EEEEEEEEEEEEE 427
>At4g29060 unknown protein
Length = 953
Score = 33.1 bits (74), Expect = 0.30
Identities = 28/88 (31%), Positives = 40/88 (44%), Gaps = 14/88 (15%)
Query: 30 RQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAAVEREVEAER----- 84
R+ EE AA ++ E E+ E A A+V AE E + V EV +E
Sbjct: 346 RKNEEIAAFLDKREEEAEK--PPVETPVEPEAEASVTSAEVEESVCVPAEVTSEEVPSSE 403
Query: 85 -------ERAAAEAEEDAAEKYEQSATV 105
E A +AE+D+ EK EQ+ T+
Sbjct: 404 TPKVVEEEVIATKAEDDSPEKEEQTETL 431
>At4g12740 adenine DNA glycosylase like protein
Length = 608
Score = 33.1 bits (74), Expect = 0.30
Identities = 20/50 (40%), Positives = 29/50 (58%), Gaps = 1/50 (2%)
Query: 54 ERYEAKAAAAAVAMAE-EENAAAVEREVEAERERAAAEAEEDAAEKYEQS 102
E+ EA+ A A E EE A E E EA++E A E+EE+ E+ E++
Sbjct: 59 EKKEAEREAEREAEREAEEEEKAEEAEAEADKEEAEEESEEEEEEEEEEA 108
Score = 30.0 bits (66), Expect = 2.6
Identities = 26/77 (33%), Positives = 33/77 (42%), Gaps = 6/77 (7%)
Query: 19 SHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAAVER 78
S A R + +R+ E A E E+ E+ E A A AEEE+
Sbjct: 44 SQSFAPREKLMRKCREKKEAEREAEREAEREAEEEEKAEEAEAEADKEEAEEES------ 97
Query: 79 EVEAERERAAAEAEEDA 95
E E E E AEAEE+A
Sbjct: 98 EEEEEEEEEEAEAEEEA 114
Score = 28.1 bits (61), Expect = 9.8
Identities = 17/44 (38%), Positives = 21/44 (47%)
Query: 57 EAKAAAAAVAMAEEENAAAVEREVEAERERAAAEAEEDAAEKYE 100
EA+ A AEE A A + E E E E E EE+A + E
Sbjct: 70 EAEREAEEEEKAEEAEAEADKEEAEEESEEEEEEEEEEAEAEEE 113
>At1g47300 hypothetical protein
Length = 306
Score = 33.1 bits (74), Expect = 0.30
Identities = 20/55 (36%), Positives = 26/55 (46%)
Query: 69 EEENAAAVEREVEAERERAAAEAEEDAAEKYEQSATVTVFFGSSKAFSFGFLNKS 123
EEE E E E E E E EE++ E+ ++ TV G K + FL KS
Sbjct: 245 EEEEEEEEEEEEEEEEEEEEEEEEEESKEREKEKKIETVIGGWIKQYWACFLRKS 299
>At1g10320 unknown protein
Length = 757
Score = 33.1 bits (74), Expect = 0.30
Identities = 28/90 (31%), Positives = 39/90 (43%), Gaps = 12/90 (13%)
Query: 14 ESLKRSHEAARRREEVRQGEEAAAAYERHVA----------EYEQYMAAYERYEAKAAAA 63
E +R EAA +E + +E AA R + + +AA ER EAKA
Sbjct: 8 EEEERHEEAAGEKESFEESKEKAAEMSRKEKRKAMKKLKRKQVRKEIAAKEREEAKAKLN 67
Query: 64 AVAMAEEENAAAVEREVEAERERAAAEAEE 93
AE+E A+E E RE+ + EE
Sbjct: 68 --DPAEQERLKAIEEEDARRREKELKDFEE 95
Score = 30.4 bits (67), Expect = 2.0
Identities = 26/88 (29%), Positives = 36/88 (40%), Gaps = 7/88 (7%)
Query: 21 EAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEENAAAVEREV 80
E A +EE + EEAA E E+ + + KA + AA ERE
Sbjct: 2 EQANEKEEEERHEEAAGEKESFEESKEKAAEMSRKEKRKAMKKLKRKQVRKEIAAKEREE 61
Query: 81 -------EAERERAAAEAEEDAAEKYEQ 101
AE+ER A EEDA + ++
Sbjct: 62 AKAKLNDPAEQERLKAIEEEDARRREKE 89
>At5g67320 unknown protein
Length = 613
Score = 32.7 bits (73), Expect = 0.40
Identities = 26/84 (30%), Positives = 33/84 (38%), Gaps = 5/84 (5%)
Query: 10 DRLDESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAE 69
DR K HE + RE R+ E ER E E+ ER K +
Sbjct: 129 DRNRAKEKDRHEKQKEREREREKLEREKEREREKIEREK-----EREREKMEREIFEREK 183
Query: 70 EENAAAVEREVEAERERAAAEAEE 93
+ ERE+E ERER E E+
Sbjct: 184 DRLKLEKEREIEREREREKIEREK 207
Score = 29.3 bits (64), Expect = 4.4
Identities = 21/92 (22%), Positives = 37/92 (39%), Gaps = 6/92 (6%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERH------VAEYEQYMAAYERYEAKAAAAAVA 66
D+ ++R HE R R + + E ER E E+ E+ +
Sbjct: 119 DKGVEREHEGDRNRAKEKDRHEKQKEREREREKLEREKEREREKIEREKEREREKMEREI 178
Query: 67 MAEEENAAAVEREVEAERERAAAEAEEDAAEK 98
E++ +E+E E ERER + E + + +
Sbjct: 179 FEREKDRLKLEKEREIEREREREKIEREKSHE 210
>At4g36520 trichohyalin like protein
Length = 1432
Score = 32.7 bits (73), Expect = 0.40
Identities = 27/85 (31%), Positives = 36/85 (41%), Gaps = 10/85 (11%)
Query: 11 RLDESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEE 70
R+ E+ E RR +E R+ EE ER + E A E+ E + A EE
Sbjct: 725 RMREAFALEQEKERRIKEAREKEEN----ERRIKE------AREKAELEQRLKATLEQEE 774
Query: 71 ENAAAVEREVEAERERAAAEAEEDA 95
+ ER+ E ER A E E A
Sbjct: 775 KERQIKERQEREENERRAKEVLEQA 799
Score = 32.7 bits (73), Expect = 0.40
Identities = 26/92 (28%), Positives = 40/92 (43%), Gaps = 4/92 (4%)
Query: 7 KALDRLDESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVA 66
K ++ +S + +E +R EE + +EA E E A E+ E + A
Sbjct: 613 KKMEMRSQSETKLNEPLKRMEEETRIKEARLREENDRRE----RVAVEKAENEKRLKAAL 668
Query: 67 MAEEENAAAVEREVEAERERAAAEAEEDAAEK 98
EE+ E +AE ER A EA E A ++
Sbjct: 669 EQEEKERKIKEAREKAENERRAVEAREKAEQE 700
Score = 32.0 bits (71), Expect = 0.68
Identities = 30/107 (28%), Positives = 43/107 (40%), Gaps = 10/107 (9%)
Query: 7 KALDRLDESLKRSHEAARRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVA 66
K +RL E R+ EE R+ E +R + A ER E A A
Sbjct: 1133 KEAERLKRERDLEMEQLRKVEEEREREREREK-DRMAFDQRALADARERLEKACAEAREK 1191
Query: 67 MAEEE---------NAAAVEREVEAERERAAAEAEEDAAEKYEQSAT 104
++ AAVER R+RAA +A +A E+ E+S +
Sbjct: 1192 SLPDKLSMEARLRAERAAVERATSEARDRAAEKAAFEARERMERSVS 1238
Score = 28.5 bits (62), Expect = 7.5
Identities = 19/80 (23%), Positives = 33/80 (40%), Gaps = 2/80 (2%)
Query: 24 RRREEVRQGEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMAEEE--NAAAVEREVE 81
R EE + + + + H ++ YE +A A E + +V+RE E
Sbjct: 1075 RNPEESKSAPKTSYGFRNHEYKFTHQQERGNIYETQAGLNQDAKVERPLPSRVSVQREKE 1134
Query: 82 AERERAAAEAEEDAAEKYEQ 101
AER + + E + K E+
Sbjct: 1135 AERLKRERDLEMEQLRKVEE 1154
Score = 28.1 bits (61), Expect = 9.8
Identities = 25/97 (25%), Positives = 40/97 (40%), Gaps = 4/97 (4%)
Query: 13 DESLKRSHEAA---RRREEVRQ-GEEAAAAYERHVAEYEQYMAAYERYEAKAAAAAVAMA 68
+ +K + E A RR E R+ E+ E+ E + A + E + A A+
Sbjct: 674 ERKIKEAREKAENERRAVEAREKAEQERKMKEQQELELQLKEAFEKEEENRRMREAFALE 733
Query: 69 EEENAAAVEREVEAERERAAAEAEEDAAEKYEQSATV 105
+E+ E + E ER EA E A + AT+
Sbjct: 734 QEKERRIKEAREKEENERRIKEAREKAELEQRLKATL 770
>At2g02170 unknown protein
Length = 486
Score = 32.7 bits (73), Expect = 0.40
Identities = 32/97 (32%), Positives = 49/97 (49%), Gaps = 18/97 (18%)
Query: 13 DESLKRSHEAARRREEVRQGEEAAAAYERHV-----AEYEQYMAAYERYEAKAAAAAVAM 67
+E+ K H A RREE++ A+E H AE ++ ER + +A +
Sbjct: 381 EEAEKAKHMARFRREEMK-----IQAWENHQKAKSEAEMKKTEVKVERIKGRAQDRLM-- 433
Query: 68 AEEENAAAVEREVEAERERAAAEAEED-AAEKYEQSA 103
+ A +ER+ AE +RAAAEA++D A K E+ A
Sbjct: 434 ---KKLATIERK--AEEKRAAAEAKKDHQAAKTEKQA 465
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,863,138
Number of Sequences: 26719
Number of extensions: 533977
Number of successful extensions: 2129
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 1835
Number of HSP's gapped (non-prelim): 229
length of query: 430
length of database: 11,318,596
effective HSP length: 102
effective length of query: 328
effective length of database: 8,593,258
effective search space: 2818588624
effective search space used: 2818588624
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0010.8


