

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.7
(564 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
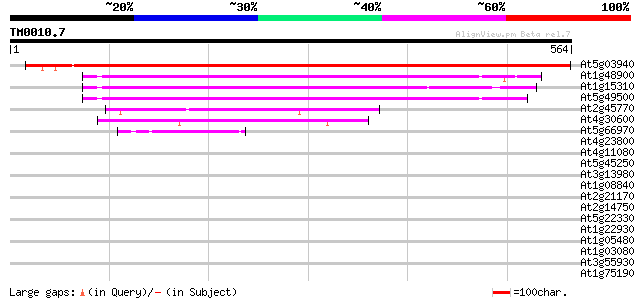
Score E
Sequences producing significant alignments: (bits) Value
At5g03940 signal recognition particle 54CP protein precursor 846 0.0
At1g48900 unknown protein (At1g48900) 227 1e-59
At1g15310 putative signal recognition particle 54 kDa subunit 202 3e-52
At5g49500 SRP54 (signal recognition particle 54 KDa) protein 198 6e-51
At2g45770 putative signal recognition particle receptor (alpha s... 159 5e-39
At4g30600 signal recognition particle receptor-like protein 120 2e-27
At5g66970 putative protein 47 3e-05
At4g23800 98b like protein 39 0.008
At4g11080 98b like protein 37 0.029
At5g45250 disease resistance protein RPS4 37 0.038
At3g13980 hypothetical protein 35 0.15
At1g08840 hypothetical protein,5' partial 35 0.15
At2g21170 putative triosephosphate isomerase 34 0.19
At2g14750 putative adenosine phosphosulfate kinase 34 0.19
At5g22330 Ruv DNA-helicase-like protein 34 0.25
At1g22930 unknown protein 33 0.32
At1g05480 unknown protein 33 0.32
At1g03080 unknown protein 33 0.32
At3g55930 putative protein 33 0.55
At1g75190 unknown protein 33 0.55
>At5g03940 signal recognition particle 54CP protein precursor
Length = 564
Score = 846 bits (2186), Expect = 0.0
Identities = 445/557 (79%), Positives = 496/557 (88%), Gaps = 11/557 (1%)
Query: 17 SSHNRTVCSSSPSPN---SLKLSSPWTNASI------SSRNSFTKEIWGWVQAKSVAVRR 67
SS NR C+ S + N SS +T +I SSRN T+EIW WV++K+V V
Sbjct: 7 SSVNRVPCTLSCTGNRRIKAAFSSAFTGGTINSASLSSSRNLSTREIWSWVKSKTV-VGH 65
Query: 68 RRDMPGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVR 127
R VRAEMFGQLT GLEAAW+KLKGEEVLTK+NI EPMRDIRRALLEADVSLPVVR
Sbjct: 66 GRYRRSQVRAEMFGQLTGGLEAAWSKLKGEEVLTKDNIAEPMRDIRRALLEADVSLPVVR 125
Query: 128 RFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQ 187
RFVQSVSDQAVG+GVIRGVKPDQQLVKIVH+ELVKLMGGEVSEL FAKSG TVILLAGLQ
Sbjct: 126 RFVQSVSDQAVGMGVIRGVKPDQQLVKIVHDELVKLMGGEVSELQFAKSGPTVILLAGLQ 185
Query: 188 GVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSE 247
GVGKTTVCAKL+ YLKKQGKSCML+AGD+YRPAAIDQL ILG+QVGVPVYTAGTDVKP++
Sbjct: 186 GVGKTTVCAKLACYLKKQGKSCMLIAGDVYRPAAIDQLVILGEQVGVPVYTAGTDVKPAD 245
Query: 248 IARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAA 307
IA+QGL+EAKK +DVVI+DTAGRLQIDK MMDELK+VKK LNPTEVLLVVDAMTGQEAA
Sbjct: 246 IAKQGLKEAKKNNVDVVIMDTAGRLQIDKGMMDELKDVKKFLNPTEVLLVVDAMTGQEAA 305
Query: 308 ALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRM 367
ALVTTFN+EIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERM+DLEPFYPDRM
Sbjct: 306 ALVTTFNVEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRM 365
Query: 368 AGRILGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVM 427
AGRILGMGDVLSFVEKA EVM+QEDAE LQ+KIMSAKFDFNDFLKQTRAVA+MGS++RV+
Sbjct: 366 AGRILGMGDVLSFVEKATEVMRQEDAEDLQKKIMSAKFDFNDFLKQTRAVAKMGSMTRVL 425
Query: 428 GMIPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTE 487
GMIPGM KV+PAQIREAEKNL MEAMIE MTPEERE+PELLAESP RRKR+A++SGKTE
Sbjct: 426 GMIPGMGKVSPAQIREAEKNLLVMEAMIEVMTPEERERPELLAESPERRKRIAKDSGKTE 485
Query: 488 QQVSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEKAPPGTARRRKRSEQRS 547
QQVS +VAQ+FQMRV+MKNLMGVM+GGS+P LS LE+ALK E+KAPPGTARR+++++ R
Sbjct: 486 QQVSALVAQIFQMRVKMKNLMGVMEGGSIPALSGLEDALKAEQKAPPGTARRKRKADSRK 545
Query: 548 LFADSA-ARPTPRGFGS 563
F +SA ++P PRGFGS
Sbjct: 546 KFVESASSKPGPRGFGS 562
>At1g48900 unknown protein (At1g48900)
Length = 495
Score = 227 bits (579), Expect = 1e-59
Identities = 138/468 (29%), Positives = 248/468 (52%), Gaps = 14/468 (2%)
Query: 74 VVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSV 133
+V AE+ G++T ++ ++ ++ ++ + E + +I RALL++DVS P+V+ ++
Sbjct: 1 MVLAELGGRITRAIQ----QMSNVTIIDEKALNECLNEITRALLQSDVSFPLVKEMQSNI 56
Query: 134 SDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTT 193
+ G + + + + EL K++ A K+ +V++ GLQG GKTT
Sbjct: 57 KKIVNLEDLAAGHNKRRIIEQAIFSELCKMLDPGKPAFAPKKAKASVVMFVGLQGAGKTT 116
Query: 194 VCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGL 253
C K + Y +K+G LV D +R A DQL + +P Y + T+ P +IA +G+
Sbjct: 117 TCTKYAYYHQKKGYKPALVCADTFRAGAFDQLKQNATKAKIPFYGSYTESDPVKIAVEGV 176
Query: 254 EEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTF 313
+ KK+ D++IVDT+GR + + ++ +E+++V ++ P V+ V+D+ GQ A F
Sbjct: 177 DTFKKENCDLIIVDTSGRHKQEASLFEEMRQVAEATKPDLVIFVMDSSIGQAAFDQAQAF 236
Query: 314 NLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILG 373
+ + I+TK+DG ++GG ALS + P+ +G GE MD+ E F R+LG
Sbjct: 237 KQSVAVGAVIITKMDGHAKGGGALSAVAATKSPVIFIGTGEHMDEFEVFDVKPFVSRLLG 296
Query: 374 MGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGM 433
MGD FV+K QEV+ ++ +L EK+ F Q + + MG + V M+PG+
Sbjct: 297 MGDWSGFVDKLQEVVPKDQQPELLEKLSQGNFTLRIMYDQFQNILNMGPLKEVFSMLPGI 356
Query: 434 S--KVTPAQIREAEKNLKNMEAMIEAMTPEERE--KPELLAESPVRRKRVAQESGKTEQQ 489
S + +E++ +K M+++MT +E + P++ ES R R+A+ SG+ ++
Sbjct: 357 SAEMMPKGHEKESQAKIKRYMTMMDSMTNDELDSSNPKVFNES--RMMRIARGSGRQVRE 414
Query: 490 VSQVVAQ---LFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEKAPP 534
V +++ + L ++ +MK L + + G M LS A + PP
Sbjct: 415 VMEMLEEYKRLAKIWSKMKGLK-IPKNGDMSALSRNMNAQHMSKVLPP 461
>At1g15310 putative signal recognition particle 54 kDa subunit
Length = 479
Score = 202 bits (515), Expect = 3e-52
Identities = 123/458 (26%), Positives = 235/458 (50%), Gaps = 14/458 (3%)
Query: 74 VVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSV 133
+V AE+ G++T ++ ++ ++ ++ + + + +I RALL++DVS +V + ++
Sbjct: 1 MVLAELGGRITRAIQ----QMNNVTIIDEKVLNDFLNEITRALLQSDVSFGLVEKMQTNI 56
Query: 134 SDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTT 193
+ G + + + +EL +++ A K+ +V++ GLQG GKTT
Sbjct: 57 KKIVNLDDLAAGHNKRLIIEQAIFKELCRMLDPGKPAFAPKKAKPSVVMFVGLQGAGKTT 116
Query: 194 VCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGL 253
C K + Y +K+G LV D +R A DQL + +P Y + T+ P +IA +G+
Sbjct: 117 TCTKYAYYHQKKGYKAALVCADTFRAGAFDQLKQNATKAKIPFYGSYTESDPVKIAVEGV 176
Query: 254 EEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTF 313
+ KK+K D++IVDT+GR + ++ +E+++V ++ P V+ V+D+ GQ A F
Sbjct: 177 DRFKKEKCDLIIVDTSGRHKQAASLFEEMRQVAEATEPDLVIFVMDSSIGQAAFEQAEAF 236
Query: 314 NLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILG 373
+ + I+TK+DG ++GG ALS + P+ +G GE MD+ E F R+LG
Sbjct: 237 KETVSVGAVIITKMDGHAKGGGALSAVAATKSPVIFIGTGEHMDEFEVFDVKPFVSRLLG 296
Query: 374 MGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGM 433
GD V+K QEV+ ++ +L E + F Q + ++ ++++ M+PG+
Sbjct: 297 KGDWSGLVDKLQEVVPKDLQNELVENLSQGNFTLRSMYDQFQCSLRI-PLNQLFSMLPGI 355
Query: 434 S--KVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVS 491
S + E+ +K M+++MT +E + P + R R+A+ SG+
Sbjct: 356 SAEMMPKGHGEESRVKMKRYMTMMDSMTNKELDSPNPKIFNESRIMRIARGSGR------ 409
Query: 492 QVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPE 529
+V ++ +M K + M+G +P ++ + + P+
Sbjct: 410 -LVREVMEMLEEYKRIAKTMKGIKIPKNGDMSKVIPPQ 446
>At5g49500 SRP54 (signal recognition particle 54 KDa) protein
Length = 497
Score = 198 bits (504), Expect = 6e-51
Identities = 123/455 (27%), Positives = 236/455 (51%), Gaps = 14/455 (3%)
Query: 74 VVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSV 133
+V AE+ G++ + ++ K+ ++ ++ + + + +I RALL++DVS P+V+ ++
Sbjct: 1 MVLAELGGRIMSAIQ----KMNNVTIIDEKALNDCLNEITRALLQSDVSFPLVKEMQTNI 56
Query: 134 SDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQG--VGK 191
+ G + + + + EL K++ S A K+ +V++ GLQG + K
Sbjct: 57 KKIVNLEDLAAGHNKRRIIEQAIFSELCKMLDPGKSAFAPKKAKPSVVMFVGLQGEVLEK 116
Query: 192 TTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQ 251
+ + ++++G LV D +R A DQL + +P Y + T P +IA +
Sbjct: 117 PQLVPSMLIIIRRKGYKPALVCADTFRAGAFDQLKQNATKSKIPYYGSYTGSDPVKIAVE 176
Query: 252 GLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVT 311
G++ KK+ D++IVDT+GR + ++ +E++++ ++ P V+ V+D+ GQ A
Sbjct: 177 GVDRFKKENCDLIIVDTSGRHKQQASLFEEMRQISEATKPDLVIFVMDSSIGQTAFEQAR 236
Query: 312 TFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRI 371
F + + I+TK+DG ++GG LS + P+ +G GE MD+ E F R+
Sbjct: 237 AFKQTVAVGAVIITKMDGHAKGGGTLSAVAATKSPVIFIGTGEHMDEFEVFDAKPFVSRL 296
Query: 372 LGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIP 431
LG GD+ FV K QEV+ ++ +L E + KF Q + + MG + V M+P
Sbjct: 297 LGNGDMSGFVNKLQEVVPKDQQPELLEMLSHGKFTLRIMYDQFQNMLNMGPLKEVFSMLP 356
Query: 432 GM--SKVTPAQIREAEKNLKNMEAMIEAMTPEERE--KPELLAESPVRRKRVAQESGKTE 487
GM + +E++ +K M+++MT EE + P++ ES R R+A+ SG+
Sbjct: 357 GMRAEMMPEGHEKESQAKIKRYMTMMDSMTNEELDSSNPKVFNES--RIMRIARGSGRIV 414
Query: 488 QQVSQVVAQLFQMRVRMKNLMGV--MQGGSMPTLS 520
++V +++ + ++ + G+ ++ G+M LS
Sbjct: 415 REVMEMLEEYKRLTTMWGKVKGLKNLEKGNMSALS 449
>At2g45770 putative signal recognition particle receptor (alpha
subunit)
Length = 366
Score = 159 bits (401), Expect = 5e-39
Identities = 100/285 (35%), Positives = 153/285 (53%), Gaps = 12/285 (4%)
Query: 97 EEVLTKENIVEPMR---DIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVKPDQQLV 153
+E+L N+ E R ++ ALL +D + R V+ + + + + G + L
Sbjct: 82 DELLLFWNLAETDRVLDELEEALLVSDFGPKITVRIVERLREDIMSGKLKSGSEIKDALK 141
Query: 154 KIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVA 213
+ V E L K +L F K VI++ G+ G GKTT KL++ LK +G ++ A
Sbjct: 142 ESVLEMLAKKNSKTELQLGFRKPA--VIMIVGVNGGGKTTSLGKLAHRLKNEGTKVLMAA 199
Query: 214 GDIYRPAAIDQLTILGKQVGVPVYTA-GTDVKPSEIARQGLEEAKKKKIDVVIVDTAGRL 272
GD +R AA DQL I ++ G + A G K + + + ++ K++ DVV+ DT+GRL
Sbjct: 200 GDTFRAAASDQLEIWAERTGCEIVVAEGDKAKAATVLSKAVKRGKEEGYDVVLCDTSGRL 259
Query: 273 QIDKAMMDELKEVKKSLN------PTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTK 326
+ ++M+EL KK++ P E+LLV+D TG FN +GITG ILTK
Sbjct: 260 HTNYSLMEELIACKKAVGKIVSGAPNEILLVLDGNTGLNMLPQAREFNEVVGITGLILTK 319
Query: 327 LDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRI 371
LDG +RGG +SV E G P+K +G GE ++DL+PF P+ I
Sbjct: 320 LDGSARGGCVVSVVEELGIPVKFIGVGEAVEDLQPFDPEAFVNAI 364
>At4g30600 signal recognition particle receptor-like protein
Length = 634
Score = 120 bits (301), Expect = 2e-27
Identities = 76/290 (26%), Positives = 150/290 (51%), Gaps = 18/290 (6%)
Query: 89 AAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVKP 148
+ + + G+ L + ++ ++ ++ L+ +V+ + + +SV G + +
Sbjct: 332 SVFQSITGKANLERTDLGPALKALKERLMTKNVAEEIAEKLCESVEASLEGKKLSSFTRI 391
Query: 149 DQQLVKIVHEELVKLMGGEVS-------ELAFAKSGTTVILLAGLQGVGKTTVCAKLSNY 201
+ + + LV+++ S A + V++ G+ GVGK+T AK++ +
Sbjct: 392 SSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYW 451
Query: 202 LKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKKKKI 261
L++ S M+ A D +R A++QL +++ +P++ G + P+ +A++ ++EA +
Sbjct: 452 LQQHKVSVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEATRNGS 511
Query: 262 DVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG--- 318
DVV+VDTAGR+Q ++ +M L ++ P VL V +A+ G +A ++ FN ++
Sbjct: 512 DVVLVDTAGRMQDNEPLMRALSKLINLNQPDLVLFVGEALVGNDAVDQLSKFNQKLSDLS 571
Query: 319 -------ITGAILTKLDG-DSRGGAALSVKEVSGKPIKLVGRGERMDDLE 360
I G +LTK D D + GAALS+ +SG P+ VG G+ DL+
Sbjct: 572 TSGNPRLIDGILLTKFDTIDDKVGAALSMVYISGSPVMFVGCGQSYTDLK 621
>At5g66970 putative protein
Length = 171
Score = 47.0 bits (110), Expect = 3e-05
Identities = 37/129 (28%), Positives = 63/129 (48%), Gaps = 7/129 (5%)
Query: 109 MRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEV 168
+ +I ALL+AD+ R V + +++ + I K ++ +++ EL +
Sbjct: 32 LNEITCALLDADIP----RLAVDEIETKSLEI--INLPKAPKKKGALIYMELSSKLDPGK 85
Query: 169 SELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTIL 228
S L K ++++ GL+GV KT CAK + Y +K+G LV D ++ A +L
Sbjct: 86 SALIRGKLEPSIVMFIGLRGVDKTKTCAKYARYHRKKGFRPALVCADTFKFDAFVRLNKA 145
Query: 229 GKQVGVPVY 237
K VPVY
Sbjct: 146 AKD-EVPVY 153
>At4g23800 98b like protein
Length = 456
Score = 38.9 bits (89), Expect = 0.008
Identities = 32/110 (29%), Positives = 49/110 (44%), Gaps = 2/110 (1%)
Query: 431 PGMSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESP--VRRKRVAQESGKTEQ 488
P V + E++L M+ M+E M E+ + ELL E +R+K E+ EQ
Sbjct: 31 PSPVSVKGKSAKSFEQDLMEMQTMLEKMKIEKDKTEELLKEKDEILRKKEEELETRDAEQ 90
Query: 489 QVSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEKAPPGTAR 538
+ +V + Q K M G S T + E+A K ++K P T R
Sbjct: 91 EKLKVELKKLQKMKEFKPNMTFACGQSSLTQAEQEKANKKKKKDCPETKR 140
>At4g11080 98b like protein
Length = 446
Score = 37.0 bits (84), Expect = 0.029
Identities = 29/94 (30%), Positives = 45/94 (47%), Gaps = 12/94 (12%)
Query: 440 QIREAEKNLKNMEAMIEAMTPEEREKPELLAESP--VRRKRVAQESGKTEQQVSQVVAQL 497
+ + EK+L M+AM+E M E+ + +LL E +R+K V QE KTE + Q
Sbjct: 39 ETKSFEKDLMEMQAMLEKMKIEKEKTEDLLKEKDEILRKKEVEQEKLKTELKKLQ----- 93
Query: 498 FQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEK 531
+MK M +L+ EE K ++K
Sbjct: 94 -----KMKEFKPNMTFAFSQSLAQTEEEKKGKKK 122
>At5g45250 disease resistance protein RPS4
Length = 1217
Score = 36.6 bits (83), Expect = 0.038
Identities = 35/120 (29%), Positives = 59/120 (49%), Gaps = 19/120 (15%)
Query: 177 GTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQV---G 233
GT +I + G+ G+GKTT+ +L Y QGK R A IDQ+ + K +
Sbjct: 228 GTRIIGVVGMPGIGKTTLLKEL--YKTWQGK--------FSRHALIDQIRVKSKHLELDR 277
Query: 234 VPVYTAGTDVKPS----EIARQGLEEAKKKKIDVVIVDTAGRLQID--KAMMDELKEVKK 287
+P G K + + + + ++K+ VV+ D + R QID + ++D +KE K+
Sbjct: 278 LPQMLLGELSKLNHPHVDNLKDPYSQLHERKVLVVLDDVSKREQIDALREILDWIKEGKE 337
>At3g13980 hypothetical protein
Length = 357
Score = 34.7 bits (78), Expect = 0.15
Identities = 33/112 (29%), Positives = 50/112 (44%), Gaps = 12/112 (10%)
Query: 7 SRHFSTPTHSSSHNRTVCSSSPSPNSLKLSSPWTNASI--SSRNSFTKEIWGWVQAKSVA 64
S FS+ S H R+ S+SP +S + P +S+ SS KE+ G+++ KS A
Sbjct: 96 SSGFSSSESDSFHGRSKSSASPPSSSRQQPKPIRTSSVDHSSAVQKPKELGGFLRTKSKA 155
Query: 65 VRRRRDM--------PGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEP 108
++ D+ PG A L T AA N K ++ T + EP
Sbjct: 156 LKIYSDLKKVKQPISPGGRLATFLNSLFT--NAATNPKKHKKTTTVAVVEEP 205
>At1g08840 hypothetical protein,5' partial
Length = 874
Score = 34.7 bits (78), Expect = 0.15
Identities = 30/106 (28%), Positives = 45/106 (42%), Gaps = 11/106 (10%)
Query: 182 LLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGT 241
L+ G+ G GKT+ L +G S +L + Y +A+D L I K G+ G
Sbjct: 485 LILGMPGTGKTSTMVHAVKALLIRGSSILLAS---YTNSAVDNLLIKLKAQGIEFLRIGR 541
Query: 242 DVKPSEIARQGLEEAK--------KKKIDVVIVDTAGRLQIDKAMM 279
D E R+ A KKK+D V V + L I+ ++
Sbjct: 542 DEAVHEEVRESCFSAMNMCSVEDIKKKLDQVKVVASTCLGINSPLL 587
>At2g21170 putative triosephosphate isomerase
Length = 315
Score = 34.3 bits (77), Expect = 0.19
Identities = 34/110 (30%), Positives = 51/110 (45%), Gaps = 8/110 (7%)
Query: 22 TVCSSSPSPNSLKLSSPWTNAS-ISSRNSFTKEIWGWVQAKSVAVRRRRDMPGVVRAEMF 80
T ++ PS + L+ SP +A+ +SS SF + + S + R GVV
Sbjct: 4 TSLTAPPSFSGLRRISPKLDAAAVSSHQSFFHRVNSSTRLVSSSSSSHRSPRGVVAMAGS 63
Query: 81 GQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFV 130
G+ G W K G TK++I + + D+ A LEADV + V FV
Sbjct: 64 GKFFVG--GNW-KCNG----TKDSIAKLISDLNSATLEADVDVVVSPPFV 106
>At2g14750 putative adenosine phosphosulfate kinase
Length = 276
Score = 34.3 bits (77), Expect = 0.19
Identities = 16/39 (41%), Positives = 24/39 (61%)
Query: 180 VILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYR 218
VI + GL G GK+T+ L+ L ++GK C ++ GD R
Sbjct: 103 VIWVTGLSGSGKSTLACALNQMLYQKGKLCYILDGDNVR 141
>At5g22330 Ruv DNA-helicase-like protein
Length = 458
Score = 33.9 bits (76), Expect = 0.25
Identities = 52/241 (21%), Positives = 99/241 (40%), Gaps = 36/241 (14%)
Query: 175 KSGTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTIL----GK 230
K +LLAG G GKT + +S L + C +V ++Y + + + +L +
Sbjct: 63 KMAGKALLLAGPPGTGKTALALGISQELGSKVPFCPMVGSEVY-SSEVKKTEVLMENFRR 121
Query: 231 QVGVPVYTAGTDVKPSEIARQGLEEAKK------KKIDVVI-----VDTAGRLQIDKAMM 279
+G+ + +V E+ EE + K I V+ V L++D +
Sbjct: 122 AIGLRIKET-KEVYEGEVTELSPEETESLTGGYGKSISHVVITLKTVKGTKHLKLDPTIY 180
Query: 280 DELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSV 339
D L + K ++ ++ ++A +G +G + A T+ D ++ L
Sbjct: 181 DALIKEKVAVGD---VIYIEANSGAVK---------RVGRSDAFATEFDLEAEEYVPLPK 228
Query: 340 KEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAEKLQEK 399
EV K K + + + DL D R G D+LS + + + + E +KL+++
Sbjct: 229 GEVHKK--KEIVQDVTLQDL-----DAANARPQGGQDILSLMGQMMKPRKTEITDKLRQE 281
Query: 400 I 400
I
Sbjct: 282 I 282
>At1g22930 unknown protein
Length = 1131
Score = 33.5 bits (75), Expect = 0.32
Identities = 37/180 (20%), Positives = 77/180 (42%), Gaps = 4/180 (2%)
Query: 370 RILGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGM 429
R+ + V + V +E+ + + +KL++K+ AK ++FL+Q R Q S+S M
Sbjct: 219 RVQQVRHVANSVSNQREIERSKMRDKLEDKLQRAKRYRSEFLRQRR--RQRDSISLYCDM 276
Query: 430 IPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQ 489
+ + + ++ + + + + + ++ P + + ES T +
Sbjct: 277 MQEDADLLSRKLSRCWRCFVRQKRTTLDLA-KAYDGLKINESLPFEQLAMLLESLNTLKT 335
Query: 490 VSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSN-LEEALKPEEKAPPGTARRRKRSEQRSL 548
V ++ +L KN+ V Q + + + L+ P KA P T R RK + S+
Sbjct: 336 VKSLLDRLEIRLEASKNVTTVSQPSILDNIDHLLKRVATPRRKATPSTLRSRKGKKVSSV 395
>At1g05480 unknown protein
Length = 588
Score = 33.5 bits (75), Expect = 0.32
Identities = 39/166 (23%), Positives = 70/166 (41%), Gaps = 20/166 (12%)
Query: 143 IRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYL 202
+ GVK + + K+V E ++ E+ + G V+ A ++ + LS
Sbjct: 150 LAGVKDEDKKTKMVREVKPDKELDDIREILMGRPGLLVLDEAHTPRNQRSCIWKTLS--- 206
Query: 203 KKQGKSCMLVAGDIY--------------RPAAIDQLTILGKQVGVPVYTAGTDVKPSEI 248
K + + +L++G + RP +++LT K+ G+ V G +EI
Sbjct: 207 KVETQKRILLSGTPFQNNFLELCNVLGLARPKYLERLTSTLKKSGMTVTKRGKKNLGNEI 266
Query: 249 ARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEV 294
+G+EE K + V V LQ + + L+E LNP E+
Sbjct: 267 NNRGIEELKAVMLPFVHVHKGSILQ---SSLPGLRECVVVLNPPEL 309
>At1g03080 unknown protein
Length = 1744
Score = 33.5 bits (75), Expect = 0.32
Identities = 45/217 (20%), Positives = 83/217 (37%), Gaps = 32/217 (14%)
Query: 337 LSVKEVSGKPI--KLVGRGERMDDLEPFYPDRMAGRILGMGDVLSFVEKAQEV------M 388
L+ V GK I K++ ER E I+ + D LS V+ +E
Sbjct: 187 LNFNNVDGKEINAKVLSESERASKAE--------AEIVALKDALSKVQAEKEASLAQFDQ 238
Query: 389 QQEDAEKLQEKIMSAKFDFNDFLKQ-TRAVAQMGSVSRVMGMIPGMSKVTPAQIREAEKN 447
E L+ ++ A+ D +++ TRA A++ ++ + + + + Q ++ +N
Sbjct: 239 NLEKLSNLESEVSRAQEDSRVLIERATRAEAEVETLRESLSKVEVEKESSLLQYQQCLQN 298
Query: 448 LKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLFQMRVRMKNL 507
+ ++E I E E E + + Q +E + Q Q
Sbjct: 299 IADLEDRISLAQKEAGEVDERANRAEAETLALKQSLVSSETDKEAALVQYQQC------- 351
Query: 508 MGVMQGGSMPTLSNLEEALKPEEKAPPGTARRRKRSE 544
+ T+SNLEE L E+ T +R + +E
Sbjct: 352 --------LKTISNLEERLHKAEEDSRLTNQRAENAE 380
>At3g55930 putative protein
Length = 437
Score = 32.7 bits (73), Expect = 0.55
Identities = 30/105 (28%), Positives = 47/105 (44%), Gaps = 19/105 (18%)
Query: 458 MTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLFQMRVRMKNLMGVMQGGSMP 517
MT EER+K L +RVA+E K E +SQ + + +V+M NLM V +
Sbjct: 207 MTKEERKKLRTL-------RRVAKEMEKREM-ISQGRVEPQKSKVKMSNLMKVRASEATQ 258
Query: 518 TLSNLEEALKPE-----------EKAPPGTARRRKRSEQRSLFAD 551
+ LE+ ++ E A T ++ ++R LF D
Sbjct: 259 NPTKLEKEIRTEAAEREQAHMDRNVARKLTPAEKREKKERKLFGD 303
>At1g75190 unknown protein
Length = 131
Score = 32.7 bits (73), Expect = 0.55
Identities = 32/135 (23%), Positives = 57/135 (41%), Gaps = 25/135 (18%)
Query: 412 KQTRAVAQMGSVSRVMGMIPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAE 471
K+T +VA+ S P S+ T + KNL+N E + + E+ + E
Sbjct: 10 KETSSVAEASS--------PTESQATRRRRGRPRKNLENPEDFKKEESEEDEDYEEY--- 58
Query: 472 SPVRRKRVAQESGKTEQQVSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEK 531
E + E + ++VV +++ ++++ G M+ LEE KPE
Sbjct: 59 ----------EDEEEEDEEAEVVINREKLKKKVRSSSGSMEKEQKMKHEELEEEEKPEMT 108
Query: 532 ----APPGTARRRKR 542
PP ++RR +R
Sbjct: 109 VKVVVPPPSSRRSRR 123
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.130 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,456,043
Number of Sequences: 26719
Number of extensions: 472238
Number of successful extensions: 1717
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 1675
Number of HSP's gapped (non-prelim): 80
length of query: 564
length of database: 11,318,596
effective HSP length: 104
effective length of query: 460
effective length of database: 8,539,820
effective search space: 3928317200
effective search space used: 3928317200
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0010.7


