

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
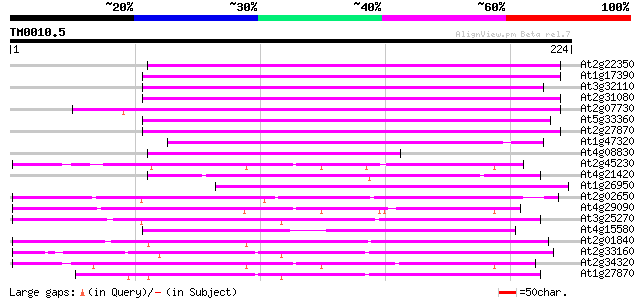
Query= TM0010.5
(224 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g22350 putative non-LTR retroelement reverse transcriptase 122 2e-28
At1g17390 hypothetical protein 116 1e-26
At3g32110 non-LTR reverse transcriptase, putative 115 2e-26
At2g31080 putative non-LTR retroelement reverse transcriptase 112 1e-25
At2g07730 putative non-LTR retroelement reverse transcriptase 109 1e-24
At5g33360 putative protein 108 3e-24
At2g27870 putative non-LTR retroelement reverse transcriptase 104 4e-23
At1g47320 hypothetical protein 87 5e-18
At4g08830 putative protein 86 2e-17
At2g45230 putative non-LTR retroelement reverse transcriptase 85 3e-17
At4g21420 hypothetical protein 80 7e-16
At1g26950 hypothetical protein 75 2e-14
At2g02650 putative reverse transcriptase 70 9e-13
At4g29090 putative protein 69 3e-12
At3g25270 hypothetical protein 68 3e-12
At4g15580 splicing factor like protein 67 6e-12
At2g01840 putative non-LTR retroelement reverse transcriptase 65 2e-11
At2g33160 putative polygalacturonase 64 5e-11
At2g34320 putative non-LTR retroelement reverse transcriptase 64 8e-11
At1g27870 hypothetical protein 62 2e-10
>At2g22350 putative non-LTR retroelement reverse transcriptase
Length = 321
Score = 122 bits (306), Expect = 2e-28
Identities = 62/165 (37%), Positives = 88/165 (52%)
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
WV P D W+K+NTDG+ A GGVLRDHNG ++ GF+ +G CS AELWG+Y+
Sbjct: 153 WVSPEDGWVKLNTDGASRGNPGFATAGGVLRDHNGAWIGGFAVNIGVCSAPLAELWGVYY 212
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFR 175
G +A RG ++V +E DS V + G S P + L++ LS V + HV+R
Sbjct: 213 GLFIAWGRGARRVELEVDSKMVVGFLTTGIADSHPLSFLLRLCYDFLSKGWIVRISHVYR 272
Query: 176 EINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTLYHR 220
E N +AD A L + + ++ P +S L+ D Y R
Sbjct: 273 EANRLADGLANYAFSLSLGLHLLESRPDVVSSILLDDVAGVSYPR 317
>At1g17390 hypothetical protein
Length = 322
Score = 116 bits (290), Expect = 1e-26
Identities = 60/167 (35%), Positives = 87/167 (51%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+ W PP W K+NTDG+ LA GGV+RD +GN+ GFS +G CS AELWG
Sbjct: 102 IAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDGNWCYGFSLNIGICSAPLAELWGA 161
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
Y+G +A +RG ++ +E DS V + G S P + LV+ LS V + HV
Sbjct: 162 YYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHPLSFLVRLCHGLLSKDWSVRISHV 221
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTLYHR 220
+RE N +AD A L + F ++++ P + + D + Y R
Sbjct: 222 YREANRLADGLANYAFFLPLGFHLFNSTPDIVMSIVHDDVAGSAYPR 268
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 115 bits (288), Expect = 2e-26
Identities = 59/160 (36%), Positives = 87/160 (53%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+RW PP + W KINTDG+ LA+ GGVLR+ G + GF+ +GRCS AELWG+
Sbjct: 1741 IRWTPPMEGWYKINTDGASRGNPGLASAGGVLRNSAGAWCGGFAVNIGRCSAPLAELWGV 1800
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
Y+G +A + + +E DS V + G + P + LV+ LS V + HV
Sbjct: 1801 YYGLYMAWAKQLTHLELEVDSEVVVGFLKTGIGETHPLSFLVRLCHNFLSKDWTVRISHV 1860
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDS 213
+RE NS+AD A + L + ++D IP + + L D+
Sbjct: 1861 YREANSLADGLANHAFSLSLGLHVFDEIPISLVMLLSEDN 1900
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 112 bits (281), Expect = 1e-25
Identities = 57/167 (34%), Positives = 90/167 (53%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+RW P+D W+KI TDG+ LAA GG +R+ G +L GF+ +G C+ AELWG
Sbjct: 1061 IRWQVPSDGWVKITTDGASRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGA 1120
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
Y+G +A D+GF++V ++ D V ++ G + P + LV+ + + V + HV
Sbjct: 1121 YYGLLIAWDKGFRRVELDLDCKLVVGFLSTGVSNAHPLSFLVRLCQGFFTRDWLVRVSHV 1180
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTLYHR 220
+RE N +AD A L + +DA P + + L+ D T + R
Sbjct: 1181 YREANRLADGLANYAFTLPLGLHCFDACPEGVRLLLLADVNGTEFPR 1227
>At2g07730 putative non-LTR retroelement reverse transcriptase
Length = 970
Score = 109 bits (273), Expect = 1e-24
Identities = 61/205 (29%), Positives = 102/205 (49%), Gaps = 10/205 (4%)
Query: 26 KKLSQDFLYTLTDATDDAL----------IQRAHGTGDLRWVPPNDDWIKINTDGSYSPV 75
+KL +D L + D ++ ++RA +RW P+D W+K+ TDG+
Sbjct: 762 RKLCRDRLKFIKDIAEEVRKAHVGTLNNHVKRARVERMIRWKAPSDRWVKLTTDGASRGH 821
Query: 76 SNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYHGAKLALDRGFKKVMIESDSA 135
LAA G + + G +L GF+ +G C AELWG Y+G +A D+GF++V + DS
Sbjct: 822 QGLAAASGAILNLQGEWLGGFALNIGSCDAPLAELWGAYYGLLIAWDKGFRRVELNLDSE 881
Query: 136 FAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDF 195
V ++ G + P + LV+ + + V + HV+RE N +AD A L + F
Sbjct: 882 LVVGFLSTGISKAHPLSFLVRLCQGFFTRDWLVRVSHVYREANRLADGLANYAFFLPLGF 941
Query: 196 RIYDAIPSFISVALMHDSCNTLYHR 220
++ P + + L+ D+ T + R
Sbjct: 942 HCFEICPEDVLLFLVEDANGTSFPR 966
>At5g33360 putative protein
Length = 306
Score = 108 bits (269), Expect = 3e-24
Identities = 59/163 (36%), Positives = 84/163 (51%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+RW P+ W K+NTDG+ LA GG LR+ G + GF+ +GRCS AELWG+
Sbjct: 136 VRWSKPSLGWCKLNTDGASHGNPGLAIAGGALRNEYGEWCFGFALNIGRCSAPLAELWGV 195
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
Y+G +A DRG ++ +E DS V + G S P + LV+ LS V + HV
Sbjct: 196 YYGLFMAWDRGITRLELEVDSEMVVGFLRTGIGSSHPLSFLVRMCHGFLSRDWIVRIGHV 255
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNT 216
+RE N +AD A L + + + + PS + L D T
Sbjct: 256 YREANRLADELANYAFDLPLGYHGFASPPSSLDSILRDDELGT 298
>At2g27870 putative non-LTR retroelement reverse transcriptase
Length = 314
Score = 104 bits (259), Expect = 4e-23
Identities = 56/167 (33%), Positives = 83/167 (49%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+ W P + W K+NTDG+ LA+ GGVLRD G + GF+ +G CS AELWG+
Sbjct: 144 IAWSKPEEGWWKLNTDGASRGNPGLASAGGVLRDEEGAWRGGFALNIGVCSAPLAELWGV 203
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
Y+G +A +R ++ IE DS V + G P + LV+ +S +V + HV
Sbjct: 204 YYGLYIAWERRVTRLEIEVDSEIVVGFLKIGINEVHPLSFLVRLCHDFISRDWRVRISHV 263
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTLYHR 220
+RE N +AD A L + F +P + L+ D+ R
Sbjct: 264 YREANRLADGLANYAFSLPLGFHSLSLVPDSLRFILLDDTSGATVSR 310
>At1g47320 hypothetical protein
Length = 259
Score = 87.4 bits (215), Expect = 5e-18
Identities = 52/150 (34%), Positives = 75/150 (49%), Gaps = 3/150 (2%)
Query: 64 IKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYHGAKLALDR 123
+KINTDG+ LA GGVL+D+ G + GFS +GR S AELWG Y+G LA +R
Sbjct: 101 LKINTDGASRGNPGLATAGGVLQDNEGRWCGGFSLNIGRSSAPMAELWGAYYGLYLAWER 160
Query: 124 GFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADA 183
+ +E DS V + G P + LV+ +S +V + HV+RE N AD
Sbjct: 161 KSSHIELEVDSEIVVGFLKTGISDHHPLSFLVRLCHGFISKDWRVRIFHVYREANRFADG 220
Query: 184 FAKNGLGLLMDFRIYDAIPSFISVALMHDS 213
A L + F +P+ + + D+
Sbjct: 221 LANYVFSLPLGFHF---VPNVVDLFWKEDN 247
>At4g08830 putative protein
Length = 947
Score = 85.9 bits (211), Expect = 2e-17
Identities = 41/101 (40%), Positives = 60/101 (58%)
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
W PP+ +W+K+NTDG+ LA GGVLRD G++ GF+ +G CS AELWG+Y+
Sbjct: 841 WKPPDGEWVKLNTDGASRGNLGLATTGGVLRDGIGHWCGGFALDIGVCSAPLAELWGVYY 900
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVK 156
G +A +R F +V +E DS V + G + + LV+
Sbjct: 901 GLYMAWERRFTRVELEVDSELVVGFLTTGISDTHSLSFLVR 941
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 84.7 bits (208), Expect = 3e-17
Identities = 69/213 (32%), Positives = 111/213 (51%), Gaps = 20/213 (9%)
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAHGTGDLR-----W 56
+W RN+LVF+ T V I K ++D DA ++ + T R W
Sbjct: 1164 LWKNRNDLVFKGREFTAPQV---ILKATEDM-----DAWNNRKEPQPQVTSSTRDRCVKW 1215
Query: 57 VPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYL-VGFSALLGRCSINFAELWGIYH 115
PP+ W+K NTDG++S G VLR+H G L +G AL + S+ E+ +
Sbjct: 1216 QPPSHGWVKCNTDGAWSKDLGNCGVGWVLRNHTGRLLWLGLRALPSQQSVLETEVEAL-R 1274
Query: 116 GAKLALDR-GFKKVMIESDSAFAVNLV-NKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
A L+L R +++V+ ESDS + V+L+ N+ +PS A +++I+ L +F++V +
Sbjct: 1275 WAVLSLSRFNYRRVIFESDSQYLVSLIQNEMDIPS--LAPRIQDIRNLLRHFEEVKFQFT 1332
Query: 174 FREINSVADAFAKNGLGLL-MDFRIYDAIPSFI 205
RE N+VAD A+ L L+ D ++Y P +I
Sbjct: 1333 RREGNNVADRTARESLSLMNYDPKMYSITPDWI 1365
>At4g21420 hypothetical protein
Length = 229
Score = 80.5 bits (197), Expect = 7e-16
Identities = 50/158 (31%), Positives = 80/158 (49%), Gaps = 3/158 (1%)
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
W P + IK+N DGS A+ GGV R+H +L+G+S +G + AE +
Sbjct: 63 WKKPENGRIKLNFDGSRGREGQ-ASIGGVFRNHKAEFLLGYSESIGEATSTMAEFAALKR 121
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLVN-KGCLPSKPAAALVKNIKRTLSNFDQVHLKHVF 174
G +LAL+ G + +E D+ +++++ +G L + V IK + + L HV+
Sbjct: 122 GLELALENGLTDLWLEGDAKIIMDIISRRGRLRCEKTNKHVNYIKVVMPELNNCVLSHVY 181
Query: 175 REINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHD 212
RE N VAD AK G D +++ P I + +MHD
Sbjct: 182 REGNRVADKLAKLG-HQFQDPKVWRVQPPEIVLPIMHD 218
>At1g26950 hypothetical protein
Length = 158
Score = 75.5 bits (184), Expect = 2e-14
Identities = 44/141 (31%), Positives = 67/141 (47%)
Query: 83 GVLRDHNGNYLVGFSALLGRCSINFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVN 142
G LRD G++ F+ LGRC+ AELWG+Y+G +A ++G ++ +E DS +
Sbjct: 17 GALRDEYGDWRGSFALNLGRCTAPLAELWGVYYGLVIAWEKGITRLELEVDSKLVAGFLT 76
Query: 143 KGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRIYDAIP 202
G S + LV+ S V + HV+RE N AD A LL+ F +D+
Sbjct: 77 TGIEDSHLLSFLVRLCYGFSSKDWIVRVSHVYREANRFADELANYAFSLLLGFLSFDSGL 136
Query: 203 SFISVALMHDSCNTLYHRGGP 223
F+ + D+ R P
Sbjct: 137 PFVDSIMREDAAGVAGARHVP 157
>At2g02650 putative reverse transcriptase
Length = 365
Score = 70.1 bits (170), Expect = 9e-13
Identities = 60/226 (26%), Positives = 100/226 (43%), Gaps = 19/226 (8%)
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAHGT------GDLR 55
+W +RN +F++ +P K + + ++L + T++ + A +
Sbjct: 147 LWKSRNVFLFQQKCQSPDYEARKGIQDATEWL-NANETTENTNVHVATNPIQTSRRDSSQ 205
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALL--GRCSINFAELWGI 113
W PP + W+K N D Y+ S G +R+ NG+ ++ +A L CS++ AE G
Sbjct: 206 WNPPPEGWVKCNFDSGYTQGSPYTRSGWTIRECNGHIVLCGNAKLQSSTCSLH-AEALGF 264
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
H ++ G + V ESDS V L+N G L+ +I+ + L+ V
Sbjct: 265 LHALQVIWAHGLRYVWFESDSKSLVTLINNG-EDHSLLGTLIYDIRHWMLKLPYCSLEFV 323
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTLYH 219
RE NS ADA A + F+ Y PS++ NTLY+
Sbjct: 324 NRERNSAADALASHVHARDPLFQSYTTPPSWL--------VNTLYY 361
>At4g29090 putative protein
Length = 575
Score = 68.6 bits (166), Expect = 3e-12
Identities = 61/208 (29%), Positives = 102/208 (48%), Gaps = 10/208 (4%)
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAHGTGDLRWVPPND 61
+W RNELVF V + + +++ T+A + + + RW PP
Sbjct: 367 LWKNRNELVFRGREFNAQEVLRRAEDDLEEWRIR-TEAESCGTKPQVNRSSCGRWRPPPH 425
Query: 62 DWIKINTDGSYSPVSNLAACGGVLRDHNGNY-LVGFSALLGRCSINFAELWGIYHGAKLA 120
W+K NTD +++ + G VLR+ G +G AL S+ AEL + A L+
Sbjct: 426 QWVKCNTDATWNRDNERCGIGWVLRNEKGEVKWMGARALPKLKSVLEAELEAM-RWAVLS 484
Query: 121 LDR-GFKKVMIESDSAFAVNLVNKGCL-PS-KPAAALVKNIKRTLSNFDQVHLKHVFREI 177
L R + V+ ESDS + ++N + PS KP +++++R LS F +V + RE
Sbjct: 485 LSRFQYNYVIFESDSQVLIEILNNDEIWPSLKPT---IQDLQRLLSQFTEVKFVFIPREG 541
Query: 178 NSVADAFAKNGLGLL-MDFRIYDAIPSF 204
N++A+ A+ L L D ++Y +PS+
Sbjct: 542 NTLAERVARESLSFLNYDPKLYSIVPSW 569
>At3g25270 hypothetical protein
Length = 343
Score = 68.2 bits (165), Expect = 3e-12
Identities = 56/219 (25%), Positives = 100/219 (45%), Gaps = 11/219 (5%)
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAHGT-------GDL 54
+W +RN+LVF++ + + + Q++ T T +L Q+ H +
Sbjct: 127 LWKSRNQLVFQQKSISWQNTLQRARNDVQEWEDTNTYV--QSLNQQVHSSRHQQPTMART 184
Query: 55 RWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINF-AELWGI 113
+W P WIK N DG+++ + A G ++RD NG Y+ A+ S + +E +
Sbjct: 185 KWQRPPSTWIKYNYDGAFNHQTRNAKAGWLMRDENGVYMGSGQAIGSTTSDSLESEFQAL 244
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
+ A +G++KV+ E DS L+N L + ++ + F++ K V
Sbjct: 245 IIAMQHAWSQGYRKVIFEGDSKQVEELMNNEKL-NFGRFNWIREGRFWQKRFEEAVFKWV 303
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHD 212
R N AD AK+ L F+ + +P+FI+ L +D
Sbjct: 304 PRTNNQPADILAKHHLQPNQSFKFHYYVPAFITSTLYYD 342
>At4g15580 splicing factor like protein
Length = 559
Score = 67.4 bits (163), Expect = 6e-12
Identities = 43/149 (28%), Positives = 65/149 (42%), Gaps = 14/149 (9%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+ W P + W+K++TDG+ AA GGV+RD +G ++ GF+ L + + G
Sbjct: 26 IAWTKPPEGWVKVSTDGASRGNPGPAAAGGVIRDEDGLWVGGFALQLAFVRLRWQSCGG- 84
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
+V +E DS V + G + P A LV+ LS V + HV
Sbjct: 85 -------------RVELEVDSELVVGFLQSGISEAHPLAFLVRLCHGLLSKDWLVRVSHV 131
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIP 202
+RE N +AD A L + A P
Sbjct: 132 YREANRLADGLANYAFSLQFEDATGTAFP 160
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 65.5 bits (158), Expect = 2e-11
Identities = 54/221 (24%), Positives = 96/221 (43%), Gaps = 10/221 (4%)
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAHGTGDL------R 55
+W +RNE +F++L P V K ++ + +++ T+ + D A+ + D +
Sbjct: 1498 LWKSRNEYLFQQLDRFPWKVAQKAEQEATEWVETMVN--DTAISHNTAQSNDRPLSRSKQ 1555
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYL-VGFSALLGRCSINFAELWGIY 114
W P + ++K N D Y + + G +LRD NG L G + L S AE G
Sbjct: 1556 WSSPPEGFLKCNFDSGYVQGRDYTSTGWILRDCNGRVLHSGCAKLQQSYSALQAEALGFL 1615
Query: 115 HGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVF 174
H ++ RG+ V E D+ NL+NK L+ +I+ ++ + +V
Sbjct: 1616 HALQMVWIRGYCYVWFEGDNLELTNLINK-TEDHHLLETLLYDIRFWMTKLPFSSIGYVN 1674
Query: 175 REINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCN 215
RE N AD K + + + P ++ + L + N
Sbjct: 1675 RERNLAADKLTKYANSMSSLYETFHVPPRWLQLYLYYPFTN 1715
>At2g33160 putative polygalacturonase
Length = 664
Score = 64.3 bits (155), Expect = 5e-11
Identities = 59/222 (26%), Positives = 98/222 (43%), Gaps = 13/222 (5%)
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAHGTGDLRWVP--- 58
+W +RN L F++ H P +L+Q L +A + I +HGT +P
Sbjct: 450 LWNSRNILYFQRR-HVP---WETTLQLAQMDLQEWNEAVTPSHIN-SHGTTIRSHIPNWR 504
Query: 59 -PNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINF--AELWGIYH 115
P + W+K N DGS+ + G ++RDHNG Y A +G C N E +
Sbjct: 505 RPTNGWVKCNVDGSFINGNIPGKGGWIVRDHNGRYEFAGQA-IGNCIDNALECEFQALLM 563
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFR 175
+ +G++++ E D+ +L+N G A +++I S F+Q + R
Sbjct: 564 AMQHCWSKGYRRICFEGDNKEMFDLIN-GLKMHFGAFNWIRDILWWASKFEQTQFQWTNR 622
Query: 176 EINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTL 217
+ N AD A+ L F + +P IS AL D +++
Sbjct: 623 QNNKPADILARTPLQHQFLFANHFWVPRVISFALHCDYTSSI 664
>At2g34320 putative non-LTR retroelement reverse transcriptase
Length = 292
Score = 63.5 bits (153), Expect = 8e-11
Identities = 55/214 (25%), Positives = 103/214 (47%), Gaps = 10/214 (4%)
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDF--LYTLTDATDDALIQRAHGTGDLRWVPP 59
+W +RNEL+F+ + V +++ +DF T + A + ++W P
Sbjct: 84 LWKSRNELMFKGKEYDAPEV---LRRAMEDFEEWSTRRELEGKASGPQVERNLSVQWKAP 140
Query: 60 NDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYL-VGFSALLGRCSINFAELWGIYHGAK 118
W+K NTD ++ + G +LR+ +G L +G AL ++ AEL + A
Sbjct: 141 PYQWVKCNTDATWQLENPRCGIGWILRNESGGVLWMGARALPRTKNVLEAELEAL-RWAV 199
Query: 119 LALDR-GFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREI 177
L + R +K+++ ESD+ VNL+N AL ++I++ L +F++V + R
Sbjct: 200 LTMSRFNYKRIIFESDAQALVNLLNSDDFWPTLQPAL-EDIQQLLHHFEEVKFEFTPRGG 258
Query: 178 NSVADAFAKNGLGLL-MDFRIYDAIPSFISVALM 210
N VAD A+ + D +++ +P ++ L+
Sbjct: 259 NKVADRIARESISFSNYDPKLFSIVPQWLRSTLI 292
>At1g27870 hypothetical protein
Length = 213
Score = 62.4 bits (150), Expect = 2e-10
Identities = 56/193 (29%), Positives = 88/193 (45%), Gaps = 9/193 (4%)
Query: 27 KLSQDFLYTLTDATDDALIQ-RAHGTGDL----RWVPPNDDWIKINTDGSYSPVSNLAAC 81
+L+Q + T+A A I R+ G + RW P WIK N DGS+ +
Sbjct: 18 RLAQTDAHEYTEANTVAQINTRSRGVREEDNHDRWRRPERGWIKCNFDGSFVNGDVKSKA 77
Query: 82 GGVLRDHNGNYLVGFSALLGRCSINF--AELWGIYHGAKLALDRGFKKVMIESDSAFAVN 139
G V+RD NG+YL+ A +GR N +E+ + + G+K+V E D+ +
Sbjct: 78 GWVVRDSNGSYLLAGQA-IGRKVDNALESEIQALIISMQHCWSHGYKRVCFEGDNKMLFD 136
Query: 140 LVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRIYD 199
L+N G +++I L F+ +H + R N+ AD AK L F +
Sbjct: 137 LIN-GSKVYFGVHNWIRDIHWWLRKFESIHFNWIRRHHNTPADILAKAPLQNQSLFLNHF 195
Query: 200 AIPSFISVALMHD 212
+P+ I+ AL D
Sbjct: 196 WVPNVITYALHCD 208
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,346,554
Number of Sequences: 26719
Number of extensions: 220497
Number of successful extensions: 538
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 447
Number of HSP's gapped (non-prelim): 78
length of query: 224
length of database: 11,318,596
effective HSP length: 96
effective length of query: 128
effective length of database: 8,753,572
effective search space: 1120457216
effective search space used: 1120457216
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0010.5


