

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
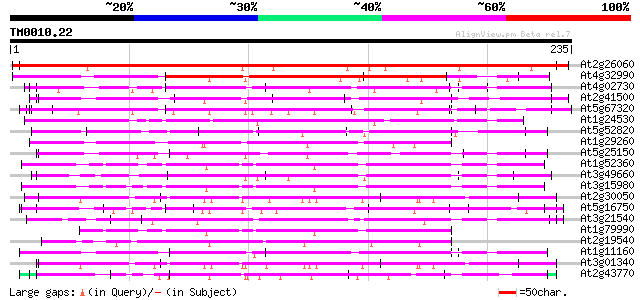
Query= TM0010.22
(235 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g26060 unknown protein 365 e-101
At4g32990 putative protein 141 3e-34
At4g02730 putative WD-repeat protein 84 8e-17
At2g41500 putative U4/U6 small nuclear ribonucleoprotein 84 8e-17
At5g67320 unknown protein 83 1e-16
At1g24530 unknown protein 80 1e-15
At5g52820 Notchless protein homolog 78 5e-15
At1g29260 peroxisomal targeting signal type 2 receptor 77 6e-15
At5g25150 transcription initiation factor IID-associated factor-... 76 1e-14
At1g52360 coatomer complex subunit, putative 76 2e-14
At3g49660 putative WD-40 repeat - protein 75 3e-14
At3g15980 putative coatomer complex subunit 75 3e-14
At2g30050 putative protein transport protein SEC13 74 5e-14
At5g16750 WD40-repeat protein 73 1e-13
At3g21540 WD repeat like protein 72 3e-13
At1g79990 coatomer protein like complex, subunit beta 2 (beta pr... 70 1e-12
At2g19540 putative WD-40 repeat protein 69 2e-12
At1g11160 hypothetical protein 68 5e-12
At3g01340 transport protein SEC13, putative 67 8e-12
At2g43770 putative splicing factor 66 1e-11
>At2g26060 unknown protein
Length = 352
Score = 365 bits (937), Expect = e-101
Identities = 164/234 (70%), Positives = 194/234 (82%), Gaps = 1/234 (0%)
Query: 2 ERLELKEVQRLEGHNDRVWSLDWNPATGHA-GTPLVFASCSGDKTVRIWEQDLSSGLWAC 60
+ LEL E+Q+LEGH DRVWS+ WNP + HA G + ASCSGD TVRIWEQ S W C
Sbjct: 6 KNLELVEIQKLEGHTDRVWSVAWNPVSSHADGVSPILASCSGDNTVRIWEQSSLSRSWTC 65
Query: 61 KAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVS 120
K VL+ETHTRTVRSCAWSPSG+LLATASFD TT IW+N G +FEC++TLEGHENEVKSVS
Sbjct: 66 KTVLEETHTRTVRSCAWSPSGQLLATASFDGTTGIWKNYGSEFECISTLEGHENEVKSVS 125
Query: 121 WNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDN 180
WNASG+ LATCSRDKSVWIWE+ NE++C +VL GHTQDVKMV+WHPT D+L SCSYDN
Sbjct: 126 WNASGSCLATCSRDKSVWIWEVLEGNEYDCAAVLTGHTQDVKMVQWHPTMDVLFSCSYDN 185
Query: 181 SIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSNLLMAV 234
+IKVW E D ++QCVQTLG+ NNGH+STVW++SFNA+GDKMVTCS +L + +
Sbjct: 186 TIKVWWSEDDDGEYQCVQTLGESNNGHSSTVWSISFNAAGDKMVTCSDDLTLKI 239
Score = 103 bits (258), Expect = 6e-23
Identities = 78/242 (32%), Positives = 129/242 (53%), Gaps = 26/242 (10%)
Query: 5 ELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVL 64
E + + LEGH + V S+ WN A+G A+CS DK+V IWE L + C AVL
Sbjct: 107 EFECISTLEGHENEVKSVSWN-ASGSC-----LATCSRDKSVWIWEV-LEGNEYDCAAVL 159
Query: 65 DETHTRTVRSCAWSPSGKLLATASFDATTAIW--ENVGGDFECVATL----EGHENEVKS 118
HT+ V+ W P+ +L + S+D T +W E+ G+++CV TL GH + V S
Sbjct: 160 TG-HTQDVKMVQWHPTMDVLFSCSYDNTIKVWWSEDDDGEYQCVQTLGESNNGHSSTVWS 218
Query: 119 VSWNASGTLLATCSRDKSVWIW-----EMQPVNEFE---CVSVLQG-HTQDVKMVKWHPT 169
+S+NA+G + TCS D ++ IW +MQ E+ + L G H + + W
Sbjct: 219 ISFNAAGDKMVTCSDDLTLKIWGTDIAKMQSGEEYAPWIHLCTLSGYHDRTIYSAHW-SR 277
Query: 170 EDILISCSYDNSIKVWAD-EGDSDDWQCVQTLGQPNNGHTSTVWALSFN-ASGDKMVTCS 227
+DI+ S + DN+I+++ D + DS D L + N H + V ++ ++ G++++ +
Sbjct: 278 DDIIASGAGDNAIRLFVDSKHDSVDGPSYNLLLKKNKAHENDVNSVQWSPGEGNRLLASA 337
Query: 228 SN 229
S+
Sbjct: 338 SD 339
Score = 34.7 bits (78), Expect = 0.045
Identities = 25/93 (26%), Positives = 42/93 (44%), Gaps = 14/93 (15%)
Query: 12 LEGHNDR-VWSLDWNPATGHAGTPLVFASCSGDKTVRIW----EQDLSSGLWACKAVLDE 66
L G++DR ++S W+ + AS +GD +R++ + + ++
Sbjct: 262 LSGYHDRTIYSAHWSRDD-------IIASGAGDNAIRLFVDSKHDSVDGPSYNLLLKKNK 314
Query: 67 THTRTVRSCAWSPS--GKLLATASFDATTAIWE 97
H V S WSP +LLA+AS D IW+
Sbjct: 315 AHENDVNSVQWSPGEGNRLLASASDDGMVKIWQ 347
>At4g32990 putative protein
Length = 243
Score = 141 bits (355), Expect = 3e-34
Identities = 70/121 (57%), Positives = 88/121 (71%), Gaps = 5/121 (4%)
Query: 66 ETHTRTVRSCAWSPSGK-LLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNAS 124
E HT V + AW+P+ ++A+ S D T IWE T +GHE+EVKSVSWNAS
Sbjct: 17 EGHTDRVWNVAWNPAADGVIASCSADKTVRIWEQ--SSLTRSWTCKGHESEVKSVSWNAS 74
Query: 125 GTLLATCSRDKSVWIWEMQPV--NEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSI 182
G+LLATC RDKSVWIWE+QP +EF+ ++VL GH++DVKMV WHPT D+L SCSYDN+I
Sbjct: 75 GSLLATCGRDKSVWIWEIQPEEDDEFDTIAVLTGHSEDVKMVLWHPTMDVLFSCSYDNTI 134
Query: 183 K 183
K
Sbjct: 135 K 135
Score = 117 bits (294), Expect = 4e-27
Identities = 81/232 (34%), Positives = 117/232 (49%), Gaps = 30/232 (12%)
Query: 2 ERLELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACK 61
+ L L+EVQ+LEGH DRVW++ WNPA V ASCS DKTVRIWEQ + W CK
Sbjct: 6 KNLGLEEVQKLEGHTDRVWNVAWNPAADG-----VIASCSADKTVRIWEQSSLTRSWTCK 60
Query: 62 AVLDETHTRTVRSCAWSPSGKLLATASFDATTAIWE---NVGGDFECVATLEGHENEVKS 118
H V+S +W+ SG LLAT D + IWE +F+ +A L GH +VK
Sbjct: 61 G-----HESEVKSVSWNASGSLLATCGRDKSVWIWEIQPEEDDEFDTIAVLTGHSEDVKM 115
Query: 119 VSWNASGTLLATCSRDKSV-WIWEMQPVNEFECVSVLQGHTQDVKMVK-----------W 166
V W+ + +L +CS D ++ I + P +S L VK+ K +
Sbjct: 116 VLWHPTMDVLFSCSYDNTIKEISSLLPGLLTFGLSELCSDDLAVKIWKTDISRMQSGEGY 175
Query: 167 HPTEDILISCSYDNSIKVWAD-EGDSDDWQCVQTLGQPNNGH----TSTVWA 213
P + ++ S + D++I+++ D + DS D + L + H S WA
Sbjct: 176 VPWDGVIASGAGDDTIQLFVDSDSDSVDGPSYKLLVKKEKAHEMDVNSVQWA 227
Score = 57.4 bits (137), Expect = 6e-09
Identities = 29/79 (36%), Positives = 42/79 (52%), Gaps = 9/79 (11%)
Query: 149 ECVSVLQGHTQDVKMVKWHPTED-ILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGH 207
E V L+GHT V V W+P D ++ SCS D ++++W + W C GH
Sbjct: 11 EEVQKLEGHTDRVWNVAWNPAADGVIASCSADKTVRIWEQSSLTRSWTC--------KGH 62
Query: 208 TSTVWALSFNASGDKMVTC 226
S V ++S+NASG + TC
Sbjct: 63 ESEVKSVSWNASGSLLATC 81
>At4g02730 putative WD-repeat protein
Length = 333
Score = 83.6 bits (205), Expect = 8e-17
Identities = 44/162 (27%), Positives = 86/162 (52%), Gaps = 12/162 (7%)
Query: 66 ETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASG 125
E HT + +S G LLA+AS D T +W ++ + EGH + + ++W++
Sbjct: 40 EGHTAAISCVKFSNDGNLLASASVDKTMILWSAT--NYSLIHRYEGHSSGISDLAWSSDS 97
Query: 126 TLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
+ S D ++ IW+ + + +EC+ VL+GHT V V ++P ++++S S+D +I++W
Sbjct: 98 HYTCSASDDCTLRIWDAR--SPYECLKVLRGHTNFVFCVNFNPPSNLIVSGSFDETIRIW 155
Query: 186 ADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCS 227
+ +CV+ + H+ + ++ FN G +V+ S
Sbjct: 156 ----EVKTGKCVRMI----KAHSMPISSVHFNRDGSLIVSAS 189
Score = 82.8 bits (203), Expect = 1e-16
Identities = 56/198 (28%), Positives = 95/198 (47%), Gaps = 27/198 (13%)
Query: 7 KEVQRLEGHNDRV----WSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKA 62
+ ++ LEGH + +S D N + AS S DKT+ +W S +
Sbjct: 34 RHLKTLEGHTAAISCVKFSNDGN----------LLASASVDKTMILWSATNYSLIHRY-- 81
Query: 63 VLDETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWN 122
E H+ + AWS +AS D T IW+ +EC+ L GH N V V++N
Sbjct: 82 ---EGHSSGISDLAWSSDSHYTCSASDDCTLRIWD-ARSPYECLKVLRGHTNFVFCVNFN 137
Query: 123 ASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSI 182
L+ + S D+++ IWE V +CV +++ H+ + V ++ +++S S+D S
Sbjct: 138 PPSNLIVSGSFDETIRIWE---VKTGKCVRMIKAHSMPISSVHFNRDGSLIVSASHDGSC 194
Query: 183 KVWADEGDSDDWQCVQTL 200
K+W D+ + C++TL
Sbjct: 195 KIW----DAKEGTCLKTL 208
Score = 73.9 bits (180), Expect = 7e-14
Identities = 49/177 (27%), Positives = 85/177 (47%), Gaps = 16/177 (9%)
Query: 9 VQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETH 68
+ R EGH+ + L W+ + + S S D T+RIW+ + + C VL H
Sbjct: 78 IHRYEGHSSGISDLAWSSDSHYT------CSASDDCTLRIWD---ARSPYECLKVL-RGH 127
Query: 69 TRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLL 128
T V ++P L+ + SFD T IWE G +CV ++ H + SV +N G+L+
Sbjct: 128 TNFVFCVNFNPPSNLIVSGSFDETIRIWEVKTG--KCVRMIKAHSMPISSVHFNRDGSLI 185
Query: 129 ATCSRDKSVWIWEMQPVNEFECV-SVLQGHTQDVKMVKWHPTEDILISCSYDNSIKV 184
+ S D S IW+ E C+ +++ + V K+ P ++ + D+++K+
Sbjct: 186 VSASHDGSCKIWD---AKEGTCLKTLIDDKSPAVSFAKFSPNGKFILVATLDSTLKL 239
Score = 61.6 bits (148), Expect = 3e-10
Identities = 55/220 (25%), Positives = 91/220 (41%), Gaps = 54/220 (24%)
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWE--------------------- 50
L GH + V+ +++NP + + S S D+T+RIWE
Sbjct: 124 LRGHTNFVFCVNFNPPSN------LIVSGSFDETIRIWEVKTGKCVRMIKAHSMPISSVH 177
Query: 51 ------------QDLSSGLW-----ACKAVLDETHTRTVRSCAWSPSGKLLATASFDATT 93
D S +W C L + + V +SP+GK + A+ D+T
Sbjct: 178 FNRDGSLIVSASHDGSCKIWDAKEGTCLKTLIDDKSPAVSFAKFSPNGKFILVATLDSTL 237
Query: 94 AIWENVGGDFECVATLEGHENEVKSVSWNAS---GTLLATCSRDKSVWIWEMQPVNEFEC 150
+ G F V T GH N+V ++ S G + + S D V++W++Q N
Sbjct: 238 KLSNYATGKFLKVYT--GHTNKVFCITSAFSVTNGKYIVSGSEDNCVYLWDLQARN---I 292
Query: 151 VSVLQGHTQDVKMVKWHPTEDILISCS--YDNSIKVWADE 188
+ L+GHT V V HP ++ + S D +I++W +
Sbjct: 293 LQRLEGHTDAVISVSCHPVQNEISSSGNHLDKTIRIWKQD 332
Score = 55.5 bits (132), Expect = 2e-08
Identities = 31/120 (25%), Positives = 59/120 (48%), Gaps = 10/120 (8%)
Query: 108 TLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWH 167
TLEGH + V ++ G LLA+ S DK++ +W + + +GH+ + + W
Sbjct: 38 TLEGHTAAISCVKFSNDGNLLASASVDKTMILWS---ATNYSLIHRYEGHSSGISDLAWS 94
Query: 168 PTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCS 227
S S D ++++W ++C++ L GHT+ V+ ++FN + +V+ S
Sbjct: 95 SDSHYTCSASDDCTLRIW---DARSPYECLKVL----RGHTNFVFCVNFNPPSNLIVSGS 147
>At2g41500 putative U4/U6 small nuclear ribonucleoprotein
Length = 554
Score = 83.6 bits (205), Expect = 8e-17
Identities = 62/220 (28%), Positives = 106/220 (48%), Gaps = 25/220 (11%)
Query: 9 VQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETH 68
+Q EGH DR+ + ++P+ + GT S DKT R+W D+++G + +L E H
Sbjct: 332 LQTFEGHLDRLARVAFHPSGKYLGT------TSYDKTWRLW--DINTG---AELLLQEGH 380
Query: 69 TRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLL 128
+R+V A+ G L A+ D+ +W+ G + +GH V SV+++ +G L
Sbjct: 381 SRSVYGIAFQQDGALAASCGLDSLARVWDLRTG--RSILVFQGHIKPVFSVNFSPNGYHL 438
Query: 129 ATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTED-ILISCSYDNSIKVWAD 187
A+ D IW+++ + + ++ H V VK+ P E L + SYD + +W+
Sbjct: 439 ASGGEDNQCRIWDLR---MRKSLYIIPAHANLVSQVKYEPQEGYFLATASYDMKVNIWSG 495
Query: 188 EGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCS 227
D+ V++L GH S V +L A + T S
Sbjct: 496 R----DFSLVKSLA----GHESKVASLDITADSSCIATVS 527
Score = 72.4 bits (176), Expect = 2e-13
Identities = 47/175 (26%), Positives = 77/175 (43%), Gaps = 19/175 (10%)
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRT 71
L+ H +R + ++P A+ S D+T ++W+ D E H
Sbjct: 294 LKDHKERATDVVFSPVDD------CLATASADRTAKLWKTD------GTLLQTFEGHLDR 341
Query: 72 VRSCAWSPSGKLLATASFDATTAIWE-NVGGDFECVATLEGHENEVKSVSWNASGTLLAT 130
+ A+ PSGK L T S+D T +W+ N G + + EGH V +++ G L A+
Sbjct: 342 LARVAFHPSGKYLGTTSYDKTWRLWDINTGAE---LLLQEGHSRSVYGIAFQQDGALAAS 398
Query: 131 CSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
C D +W+++ + V QGH + V V + P L S DN ++W
Sbjct: 399 CGLDSLARVWDLRTGRS---ILVFQGHIKPVFSVNFSPNGYHLASGGEDNQCRIW 450
Score = 64.7 bits (156), Expect = 4e-11
Identities = 42/129 (32%), Positives = 63/129 (48%), Gaps = 14/129 (10%)
Query: 13 EGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTV 72
+GH V+S++++P H AS D RIW+ + L+ A H V
Sbjct: 420 QGHIKPVFSVNFSPNGYH------LASGGEDNQCRIWDLRMRKSLYIIPA-----HANLV 468
Query: 73 RSCAWSPS-GKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATC 131
+ P G LATAS+D IW G DF V +L GHE++V S+ A + +AT
Sbjct: 469 SQVKYEPQEGYFLATASYDMKVNIWS--GRDFSLVKSLAGHESKVASLDITADSSCIATV 526
Query: 132 SRDKSVWIW 140
S D+++ +W
Sbjct: 527 SHDRTIKLW 535
Score = 64.3 bits (155), Expect = 5e-11
Identities = 43/165 (26%), Positives = 76/165 (46%), Gaps = 13/165 (7%)
Query: 70 RTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLA 129
R + C++S GK+LAT S T +WE + +A L+ H+ V ++ LA
Sbjct: 256 RPLTGCSFSRDGKILATCSLSGVTKLWE-MPQVTNTIAVLKDHKERATDVVFSPVDDCLA 314
Query: 130 TCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEG 189
T S D++ +W+ + + +GH + V +HP+ L + SYD + ++W
Sbjct: 315 TASADRTAKLWK----TDGTLLQTFEGHLDRLARVAFHPSGKYLGTTSYDKTWRLW---- 366
Query: 190 DSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSNLLMAV 234
D + L Q GH+ +V+ ++F G +C + L V
Sbjct: 367 --DINTGAELLLQ--EGHSRSVYGIAFQQDGALAASCGLDSLARV 407
Score = 53.9 bits (128), Expect = 7e-08
Identities = 34/117 (29%), Positives = 55/117 (46%), Gaps = 11/117 (9%)
Query: 111 GHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTE 170
G + + S++ G +LATCS +WEM V ++VL+ H + V + P +
Sbjct: 253 GDDRPLTGCSFSRDGKILATCSLSGVTKLWEMPQVT--NTIAVLKDHKERATDVVFSPVD 310
Query: 171 DILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCS 227
D L + S D + K+W +G TL Q GH + ++F+ SG + T S
Sbjct: 311 DCLATASADRTAKLWKTDG---------TLLQTFEGHLDRLARVAFHPSGKYLGTTS 358
>At5g67320 unknown protein
Length = 613
Score = 82.8 bits (203), Expect = 1e-16
Identities = 58/228 (25%), Positives = 91/228 (39%), Gaps = 59/228 (25%)
Query: 8 EVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWE------------QDLSS 55
+V+ LEGH V + W+P+ + AS SGD T RIW +++++
Sbjct: 257 DVRILEGHTSEVCACAWSPSAS------LLASGSGDATARIWSIPEGSFKAVHTGRNINA 310
Query: 56 GLWACKAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENE 115
+ ++ V + W+ G LLAT S D IW G E ++TL H+
Sbjct: 311 LILKHAKGKSNEKSKDVTTLDWNGEGTLLATGSCDGQARIWTLNG---ELISTLSKHKGP 367
Query: 116 VKSVSWNASGTLLATCSRDKSVWIWEMQ-------------------------------- 143
+ S+ WN G L T S D++ +W+++
Sbjct: 368 IFSLKWNKKGDYLLTGSVDRTAVVWDVKAEEWKQQFEFHSGPTLDVDWRNNVSFATSSTD 427
Query: 144 ------PVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
+ E GH +V VKW PT +L SCS D++ K+W
Sbjct: 428 SMIYLCKIGETRPAKTFTGHQGEVNCVKWDPTGSLLASCSDDSTAKIW 475
Score = 82.0 bits (201), Expect = 2e-16
Identities = 52/145 (35%), Positives = 71/145 (48%), Gaps = 21/145 (14%)
Query: 66 ETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVAT-----------LEGHEN 114
E HT V +CAWSPS LLA+ S DAT IW G F+ V T +G N
Sbjct: 262 EGHTSEVCACAWSPSASLLASGSGDATARIWSIPEGSFKAVHTGRNINALILKHAKGKSN 321
Query: 115 E----VKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTE 170
E V ++ WN GTLLAT S D IW + E +S L H + +KW+
Sbjct: 322 EKSKDVTTLDWNGEGTLLATGSCDGQARIWTLNG----ELISTLSKHKGPIFSLKWNKKG 377
Query: 171 DILISCSYDNSIKVWADEGDSDDWQ 195
D L++ S D + VW + +++W+
Sbjct: 378 DYLLTGSVDRTAVVW--DVKAEEWK 400
Score = 80.1 bits (196), Expect = 9e-16
Identities = 65/257 (25%), Positives = 103/257 (39%), Gaps = 71/257 (27%)
Query: 19 VWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWS 78
V +LDWN GT L SC G + +L S L H + S W+
Sbjct: 327 VTTLDWNGE----GTLLATGSCDGQARIWTLNGELISTL--------SKHKGPIFSLKWN 374
Query: 79 PSGKLLATASFDATTAIWE------------------------NVG-------------- 100
G L T S D T +W+ NV
Sbjct: 375 KKGDYLLTGSVDRTAVVWDVKAEEWKQQFEFHSGPTLDVDWRNNVSFATSSTDSMIYLCK 434
Query: 101 -GDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQ 159
G+ T GH+ EV V W+ +G+LLA+CS D + IW + + V L+ HT+
Sbjct: 435 IGETRPAKTFTGHQGEVNCVKWDPTGSLLASCSDDSTAKIWN---IKQSTFVHDLREHTK 491
Query: 160 DVKMVKWHPT---------EDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTST 210
++ ++W PT + L S S+D+++K+W + + C NGH
Sbjct: 492 EIYTIRWSPTGPGTNNPNKQLTLASASFDSTVKLW--DAELGKMLC------SFNGHREP 543
Query: 211 VWALSFNASGDKMVTCS 227
V++L+F+ +G+ + + S
Sbjct: 544 VYSLAFSPNGEYIASGS 560
Score = 76.6 bits (187), Expect = 1e-14
Identities = 54/189 (28%), Positives = 89/189 (46%), Gaps = 26/189 (13%)
Query: 5 ELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVL 64
E + + GH V + W+P T + ASCS D T +IW S+ + +
Sbjct: 437 ETRPAKTFTGHQGEVNCVKWDP------TGSLLASCSDDSTAKIWNIKQSTFVHDLRE-- 488
Query: 65 DETHTRTVRSCAWSPSGK---------LLATASFDATTAIWENVGGDFECVATLEGHENE 115
HT+ + + WSP+G LA+ASFD+T +W+ G C + GH
Sbjct: 489 ---HTKEIYTIRWSPTGPGTNNPNKQLTLASASFDSTVKLWDAELGKMLC--SFNGHREP 543
Query: 116 VKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILIS 175
V S++++ +G +A+ S DKS+ IW + E + V G+ + V W+ + + +
Sbjct: 544 VYSLAFSPNGEYIASGSLDKSIHIWS---IKEGKIVKTYTGN-GGIFEVCWNKEGNKIAA 599
Query: 176 CSYDNSIKV 184
C DNS+ V
Sbjct: 600 CFADNSVCV 608
Score = 64.7 bits (156), Expect = 4e-11
Identities = 41/138 (29%), Positives = 65/138 (46%), Gaps = 11/138 (7%)
Query: 9 VQRLEGHNDRVWSLDWNPA---TGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLD 65
V L H ++++ W+P T + L AS S D TV++W+ +L L +
Sbjct: 483 VHDLREHTKEIYTIRWSPTGPGTNNPNKQLTLASASFDSTVKLWDAELGKMLCSFNG--- 539
Query: 66 ETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASG 125
H V S A+SP+G+ +A+ S D + IW G + V T G+ + V WN G
Sbjct: 540 --HREPVYSLAFSPNGEYIASGSLDKSIHIWSIKEG--KIVKTYTGNGG-IFEVCWNKEG 594
Query: 126 TLLATCSRDKSVWIWEMQ 143
+A C D SV + + +
Sbjct: 595 NKIAACFADNSVCVLDFR 612
Score = 62.0 bits (149), Expect = 3e-10
Identities = 61/243 (25%), Positives = 101/243 (41%), Gaps = 51/243 (20%)
Query: 10 QRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDET-- 67
Q+ E H+ +DW + FA+ S D S ++ CK + ET
Sbjct: 401 QQFEFHSGPTLDVDWR-------NNVSFATSSTD-----------SMIYLCK--IGETRP 440
Query: 68 ------HTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSW 121
H V W P+G LLA+ S D+T IW F V L H E+ ++ W
Sbjct: 441 AKTFTGHQGEVNCVKWDPTGSLLASCSDDSTAKIWNIKQSTF--VHDLREHTKEIYTIRW 498
Query: 122 NASGT---------LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDI 172
+ +G LA+ S D +V +W+ + + + C GH + V + + P +
Sbjct: 499 SPTGPGTNNPNKQLTLASASFDSTVKLWDAE-LGKMLCS--FNGHREPVYSLAFSPNGEY 555
Query: 173 LISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSNLLM 232
+ S S D SI +W+ + + + V+T N G W N G+K+ C ++ +
Sbjct: 556 IASGSLDKSIHIWSIK----EGKIVKTY-TGNGGIFEVCW----NKEGNKIAACFADNSV 606
Query: 233 AVM 235
V+
Sbjct: 607 CVL 609
>At1g24530 unknown protein
Length = 418
Score = 79.7 bits (195), Expect = 1e-15
Identities = 61/212 (28%), Positives = 105/212 (48%), Gaps = 16/212 (7%)
Query: 7 KEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDE 66
K ++ H R+W + T A + S S DKT++IW +S L CK + +
Sbjct: 178 KNYVQVRRHKKRLWIEHADAVTALAVSDGFIYSVSWDKTLKIWR---ASDL-RCKESI-K 232
Query: 67 THTRTVRSCAWSPSGKLLATASFDATTAIWENVGGD--FECVATLEGHENEVKSVSWNAS 124
H V + A S +G + T S D +W G+ VATLE H++ V +++ N
Sbjct: 233 AHDDAVNAIAVSTNGTVY-TGSADRRIRVWAKPTGEKRHTLVATLEKHKSAVNALALNDD 291
Query: 125 GTLLATCSRDKSVWIWEMQPVNEFECV-SVLQGHTQDVKMVKWHPTEDILISCSYDNSIK 183
G++L + S D+S+ +WE + + + V L+GH D ++ D+L+S S D +++
Sbjct: 292 GSVLFSGSCDRSILVWEREDTSNYMAVRGALRGH--DKAILSLFNVSDLLLSGSADRTVR 349
Query: 184 VWADEGDSDDWQCVQTLGQPNNGHTSTVWALS 215
+W G + C++ L +GHT V +L+
Sbjct: 350 IWR-RGPDSSYSCLEVL----SGHTKPVKSLA 376
>At5g52820 Notchless protein homolog
Length = 473
Score = 77.8 bits (190), Expect = 5e-15
Identities = 50/153 (32%), Positives = 74/153 (47%), Gaps = 9/153 (5%)
Query: 33 TPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWSPSGKLLATASFDAT 92
+P S S D T+ +WE +S K L H + V +SP GK +A+ASFD +
Sbjct: 328 SPERLVSGSDDFTMFLWEPSVSK---QPKKRLTG-HQQLVNHVYFSPDGKWIASASFDKS 383
Query: 93 TAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVS 152
+W + G F V GH V VSW+A LL + S+D ++ IWE + +
Sbjct: 384 VRLWNGITGQF--VTVFRGHVGPVYQVSWSADSRLLLSGSKDSTLKIWE---IRTKKLKQ 438
Query: 153 VLQGHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
L GH +V V W P + ++S D +K+W
Sbjct: 439 DLPGHADEVFAVDWSPDGEKVVSGGKDRVLKLW 471
Score = 76.3 bits (186), Expect = 1e-14
Identities = 57/181 (31%), Positives = 84/181 (45%), Gaps = 21/181 (11%)
Query: 10 QRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHT 69
Q + GH + V + ++P G L AS SGD TVR+W+ + L+ CK H
Sbjct: 103 QTIAGHAEAVLCVSFSPD----GKQL--ASGSGDTTVRLWDLYTETPLFTCKG-----HK 151
Query: 70 RTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSW-----NAS 124
V + AWSP GK L + S W G+ E + L GH+ + +SW ++
Sbjct: 152 NWVLTVAWSPDGKHLVSGSKSGEICCWNPKKGELE-GSPLTGHKKWITGISWEPVHLSSP 210
Query: 125 GTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKV 184
T S+D IW++ C+S GHT V VKW + I+ + S D +IK+
Sbjct: 211 CRRFVTSSKDGDARIWDITLKKSIICLS---GHTLAVTCVKW-GGDGIIYTGSQDCTIKM 266
Query: 185 W 185
W
Sbjct: 267 W 267
Score = 58.5 bits (140), Expect = 3e-09
Identities = 43/146 (29%), Positives = 70/146 (47%), Gaps = 12/146 (8%)
Query: 80 SGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWI 139
S + L + S D T +WE + L GH+ V V ++ G +A+ S DKSV +
Sbjct: 328 SPERLVSGSDDFTMFLWEPSVSK-QPKKRLTGHQQLVNHVYFSPDGKWIASASFDKSVRL 386
Query: 140 WEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSDDWQCVQT 199
W +F V+V +GH V V W +L+S S D+++K+W +
Sbjct: 387 WN-GITGQF--VTVFRGHVGPVYQVSWSADSRLLLSGSKDSTLKIWEIR--------TKK 435
Query: 200 LGQPNNGHTSTVWALSFNASGDKMVT 225
L Q GH V+A+ ++ G+K+V+
Sbjct: 436 LKQDLPGHADEVFAVDWSPDGEKVVS 461
Score = 52.4 bits (124), Expect = 2e-07
Identities = 37/132 (28%), Positives = 61/132 (46%), Gaps = 13/132 (9%)
Query: 10 QRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHT 69
+RL GH V + ++P AS S DK+VR+W +G+ + H
Sbjct: 354 KRLTGHQQLVNHVYFSPDGKW------IASASFDKSVRLW-----NGITGQFVTVFRGHV 402
Query: 70 RTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLA 129
V +WS +LL + S D+T IWE + L GH +EV +V W+ G +
Sbjct: 403 GPVYQVSWSADSRLLLSGSKDSTLKIWEIRTKKLK--QDLPGHADEVFAVDWSPDGEKVV 460
Query: 130 TCSRDKSVWIWE 141
+ +D+ + +W+
Sbjct: 461 SGGKDRVLKLWK 472
Score = 43.5 bits (101), Expect = 1e-04
Identities = 33/114 (28%), Positives = 49/114 (42%), Gaps = 14/114 (12%)
Query: 105 CVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNE-FECVSVLQGHTQDVKM 163
C T+ GH V VS++ G LA+ S D +V +W++ F C +GH V
Sbjct: 101 CSQTIAGHAEAVLCVSFSPDGKQLASGSGDTTVRLWDLYTETPLFTC----KGHKNWVLT 156
Query: 164 VKWHPTEDILISCSYDNSIKVW-ADEGDSDDWQCVQTLGQPNNGHTSTVWALSF 216
V W P L+S S I W +G+ + G P GH + +S+
Sbjct: 157 VAWSPDGKHLVSGSKSGEICCWNPKKGELE--------GSPLTGHKKWITGISW 202
>At1g29260 peroxisomal targeting signal type 2 receptor
Length = 317
Score = 77.4 bits (189), Expect = 6e-15
Identities = 54/180 (30%), Positives = 87/180 (48%), Gaps = 17/180 (9%)
Query: 9 VQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETH 68
++ + H V S+D+NP + F + S D TV++W D + + K H
Sbjct: 99 IRSFQEHAREVQSVDYNPTRRDS-----FLTSSWDDTVKLWAMDRPASVRTFK-----EH 148
Query: 69 TRTVRSCAWSPS-GKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNA-SGT 126
V W+P G + A+AS D T IW+ + + H+ E+ S WN
Sbjct: 149 AYCVYQAVWNPKHGDVFASASGDCTLRIWD--VREPGSTMIIPAHDFEILSCDWNKYDDC 206
Query: 127 LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHP-TEDILISCSYDNSIKVW 185
+LAT S DK+V +W+++ ++VL GH V+ VK+ P ++ SCSYD S+ +W
Sbjct: 207 ILATSSVDKTVKVWDVRSYR--VPLAVLNGHGYAVRKVKFSPHRRSLIASCSYDMSVCLW 264
Score = 63.2 bits (152), Expect = 1e-10
Identities = 54/184 (29%), Positives = 85/184 (45%), Gaps = 24/184 (13%)
Query: 9 VQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETH 68
V+ + H V+ WNP G VFAS SGD T+RIW+ ++ H
Sbjct: 142 VRTFKEHAYCVYQAVWNPKHGD-----VFASASGDCTLRIWDVREPG-----STMIIPAH 191
Query: 69 TRTVRSCAWSP-SGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNA-SGT 126
+ SC W+ +LAT+S D T +W+ V +A L GH V+ V ++ +
Sbjct: 192 DFEILSCDWNKYDDCILATSSVDKTVKVWD-VRSYRVPLAVLNGHGYAVRKVKFSPHRRS 250
Query: 127 LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQ-----DVKMVKWHPTEDILISCSYDNS 181
L+A+CS D SV +W+ + E V HT+ D+ ++ E ++ S +D
Sbjct: 251 LIASCSYDMSVCLWDY--MVEDALVGRYDHHTEFAVGIDMSVL----VEGLMASTGWDEL 304
Query: 182 IKVW 185
+ VW
Sbjct: 305 VYVW 308
Score = 34.7 bits (78), Expect = 0.045
Identities = 21/73 (28%), Positives = 39/73 (52%), Gaps = 2/73 (2%)
Query: 116 VKSVSWNAS-GTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTE-DIL 173
V V W+ S ++L D SV I++ + Q H ++V+ V ++PT D
Sbjct: 63 VYDVCWSESHDSVLIAAIGDGSVKIYDTALPPPSNPIRSFQEHAREVQSVDYNPTRRDSF 122
Query: 174 ISCSYDNSIKVWA 186
++ S+D+++K+WA
Sbjct: 123 LTSSWDDTVKLWA 135
>At5g25150 transcription initiation factor IID-associated
factor-like protein
Length = 700
Score = 76.3 bits (186), Expect = 1e-14
Identities = 59/205 (28%), Positives = 89/205 (42%), Gaps = 27/205 (13%)
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRT 71
L GH+ V+S ++P S S D T+R+W L++ L K H
Sbjct: 414 LLGHSGPVYSATFSPPGDFV------LSSSADTTIRLWSTKLNANLVCYKG-----HNYP 462
Query: 72 VRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATC 131
V +SP G A+ S D T IW + + + GH + V W+ + +AT
Sbjct: 463 VWDAQFSPFGHYFASCSHDRTARIWSM--DRIQPLRIMAGH---LSDVDWHPNCNYIATG 517
Query: 132 SRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDS 191
S DK+V +W++Q ECV + GH V + P + S D +I +W D
Sbjct: 518 SSDKTVRLWDVQTG---ECVRIFIGHRSMVLSLAMSPDGRYMASGDEDGTIMMW----DL 570
Query: 192 DDWQCVQTLGQPNNGHTSTVWALSF 216
+C+ P GH S VW+LS+
Sbjct: 571 STARCI----TPLMGHNSCVWSLSY 591
Score = 64.3 bits (155), Expect = 5e-11
Identities = 42/159 (26%), Positives = 74/159 (46%), Gaps = 18/159 (11%)
Query: 68 HTRTVRSCAWSPSGKLLATASFDATTAIWE-NVGGDFECVATLEGHENEVKSVSWNASGT 126
H+ V S +SP G + ++S D T +W + + C +GH V ++ G
Sbjct: 417 HSGPVYSATFSPPGDFVLSSSADTTIRLWSTKLNANLVC---YKGHNYPVWDAQFSPFGH 473
Query: 127 LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWA 186
A+CS D++ IW M + + + ++ GH D V WHP + + + S D ++++W
Sbjct: 474 YFASCSHDRTARIWSM---DRIQPLRIMAGHLSD---VDWHPNCNYIATGSSDKTVRLW- 526
Query: 187 DEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVT 225
D +CV+ GH S V +L+ + G M +
Sbjct: 527 ---DVQTGECVRIF----IGHRSMVLSLAMSPDGRYMAS 558
Score = 54.3 bits (129), Expect = 5e-08
Identities = 55/248 (22%), Positives = 90/248 (36%), Gaps = 78/248 (31%)
Query: 13 EGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQD-----------LSSGLWA-- 59
+GHN VW ++P GH FASCS D+T RIW D LS W
Sbjct: 457 KGHNYPVWDAQFSPF-GH-----YFASCSHDRTARIWSMDRIQPLRIMAGHLSDVDWHPN 510
Query: 60 CKAVLDETHTRTVR---------------------SCAWSPSGKLLATASFDATTAIWEN 98
C + + +TVR S A SP G+ +A+ D T +W+
Sbjct: 511 CNYIATGSSDKTVRLWDVQTGECVRIFIGHRSMVLSLAMSPDGRYMASGDEDGTIMMWDL 570
Query: 99 VGGDFECVATLEGHENEVKSVSWNA----------------------------------- 123
C+ L GH + V S+S+
Sbjct: 571 --STARCITPLMGHNSCVWSLSYRVVQGYDIEWYGFLSWVVECCIGLMCWVVNGMTGFGC 628
Query: 124 -SGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSI 182
G+LLA+ S D +V +W++ + G++ ++ ++ PT+ + + S+
Sbjct: 629 GEGSLLASGSADCTVKLWDVTSSTKLTKAEEKNGNSNRLRSLRTFPTKSTPVHALRERSL 688
Query: 183 KVWADEGD 190
D+ +
Sbjct: 689 NPQTDQNN 696
Score = 38.5 bits (88), Expect = 0.003
Identities = 31/141 (21%), Positives = 56/141 (38%), Gaps = 29/141 (20%)
Query: 108 TLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPV---------------------N 146
T N + S + G+L+A D S+ +W+M + N
Sbjct: 347 TFVNTHNGLNCSSISHDGSLVAGGFSDSSIKVWDMAKIGQAGSGALQAENDSSDQSIGPN 406
Query: 147 EFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNG 206
++L GH+ V + P D ++S S D +I++W+ + ++ + C + G
Sbjct: 407 GRRSYTLLLGHSGPVYSATFSPPGDFVLSSSADTTIRLWSTKLNA-NLVCYK-------G 458
Query: 207 HTSTVWALSFNASGDKMVTCS 227
H VW F+ G +CS
Sbjct: 459 HNYPVWDAQFSPFGHYFASCS 479
Score = 26.9 bits (58), Expect = 9.4
Identities = 26/86 (30%), Positives = 33/86 (38%), Gaps = 17/86 (19%)
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRT 71
L GHN VWSL + G+ F S W + GL C V + T
Sbjct: 579 LMGHNSCVWSLSYRVVQGYDIEWYGFLS---------WVVECCIGL-MCWVV----NGMT 624
Query: 72 VRSCAWSPSGKLLATASFDATTAIWE 97
C G LLA+ S D T +W+
Sbjct: 625 GFGCG---EGSLLASGSADCTVKLWD 647
>At1g52360 coatomer complex subunit, putative
Length = 926
Score = 75.9 bits (185), Expect = 2e-14
Identities = 62/222 (27%), Positives = 105/222 (46%), Gaps = 26/222 (11%)
Query: 6 LKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLD 65
+ +V+ E H+D + + +P P V +S S D +++W+ + WAC +
Sbjct: 89 MDKVKVFEAHSDYIRCVAVHPTL-----PYVLSS-SDDMLIKLWDWEKG---WACTQIF- 138
Query: 66 ETHTRTVRSCAWSPSG-KLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNAS 124
E H+ V ++P A+AS D T IW N+G + TL+ H+ V V +
Sbjct: 139 EGHSHYVMQVTFNPKDTNTFASASLDRTIKIW-NLGSP-DPNFTLDAHQKGVNCVDYFTG 196
Query: 125 GT--LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSI 182
G L T S D + +W+ Q + CV L+GHT +V V +HP I+I+ S D ++
Sbjct: 197 GDKPYLITGSDDHTAKVWDYQTKS---CVQTLEGHTHNVSAVCFHPELPIIITGSEDGTV 253
Query: 183 KVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMV 224
++W + ++ TL N VWA+ + S ++V
Sbjct: 254 RIW----HATTYRLENTL----NYGLERVWAIGYIKSSRRVV 287
Score = 39.7 bits (91), Expect = 0.001
Identities = 36/147 (24%), Positives = 62/147 (41%), Gaps = 14/147 (9%)
Query: 72 VRSCAWSPSGKLLATASFDATTAIWENVGGDFECVA-TLEGHENEVKSVSWNASGTLLAT 130
V+S P+ + + + T IW + +A + E E V+S + A +
Sbjct: 18 VKSVDLHPTEPWILASLYSGTLCIWNY---QTQVMAKSFEVTELPVRSAKFVARKQWVVA 74
Query: 131 CSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGD 190
+ D + ++ N + V V + H+ ++ V HPT ++S S D IK+W E
Sbjct: 75 GADDMYIRVYNY---NTMDKVKVFEAHSDYIRCVAVHPTLPYVLSSSDDMLIKLWDWE-- 129
Query: 191 SDDWQCVQTLGQPNNGHTSTVWALSFN 217
W C Q GH+ V ++FN
Sbjct: 130 -KGWACTQIF----EGHSHYVMQVTFN 151
>At3g49660 putative WD-40 repeat - protein
Length = 317
Score = 75.1 bits (183), Expect = 3e-14
Identities = 53/180 (29%), Positives = 85/180 (46%), Gaps = 18/180 (10%)
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRT 71
L H+D V ++D+N G+ +V S S D RIW+ SG C L +
Sbjct: 151 LPAHSDPVTAVDFN----RDGSLIV--SSSYDGLCRIWD----SGTGHCVKTLIDDENPP 200
Query: 72 VRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENE---VKSVSWNASGTLL 128
V +SP+GK + + D T +W F + T GH N + S +G +
Sbjct: 201 VSFVRFSPNGKFILVGTLDNTLRLWNISSAKF--LKTYTGHVNAQYCISSAFSVTNGKRI 258
Query: 129 ATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADE 188
+ S D V +WE+ N + + L+GHT+ V V HPTE+++ S S D ++++W +
Sbjct: 259 VSGSEDNCVHMWEL---NSKKLLQKLEGHTETVMNVACHPTENLIASGSLDKTVRIWTQK 315
Score = 74.7 bits (182), Expect = 4e-14
Identities = 47/164 (28%), Positives = 85/164 (51%), Gaps = 14/164 (8%)
Query: 67 THTRTVRSCAWSPSGKLLATASFDATTAIWE-NVGGD--FECVATLEGHENEVKSVSWNA 123
+H R V S +S G+LLA+AS D T + N D E V GHEN + V++++
Sbjct: 22 SHNRAVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVAFSS 81
Query: 124 SGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIK 183
+ + S DK++ +W+++ + + L GHT V ++P ++++S S+D +++
Sbjct: 82 DARFIVSASDDKTLKLWDVETGS---LIKTLIGHTNYAFCVNFNPQSNMIVSGSFDETVR 138
Query: 184 VWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCS 227
+W D +C++ L H+ V A+ FN G +V+ S
Sbjct: 139 IW----DVTTGKCLKVL----PAHSDPVTAVDFNRDGSLIVSSS 174
Score = 74.7 bits (182), Expect = 4e-14
Identities = 56/202 (27%), Positives = 91/202 (44%), Gaps = 15/202 (7%)
Query: 10 QRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHT 69
Q L HN V S+ ++ + AS S DKT+R + + + A H
Sbjct: 18 QTLTSHNRAVSSVKFS------SDGRLLASASADKTIRTYTINTINDPIAEPVQEFTGHE 71
Query: 70 RTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLA 129
+ A+S + + +AS D T +W+ G + TL GH N V++N ++
Sbjct: 72 NGISDVAFSSDARFIVSASDDKTLKLWDVETGSL--IKTLIGHTNYAFCVNFNPQSNMIV 129
Query: 130 TCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEG 189
+ S D++V IW+ V +C+ VL H+ V V ++ +++S SYD ++W
Sbjct: 130 SGSFDETVRIWD---VTTGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIW---- 182
Query: 190 DSDDWQCVQTLGQPNNGHTSTV 211
DS CV+TL N S V
Sbjct: 183 DSGTGHCVKTLIDDENPPVSFV 204
Score = 54.3 bits (129), Expect = 5e-08
Identities = 34/122 (27%), Positives = 61/122 (49%), Gaps = 10/122 (8%)
Query: 108 TLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNE--FECVSVLQGHTQDVKMVK 165
TL H V SV +++ G LLA+ S DK++ + + +N+ E V GH + V
Sbjct: 19 TLTSHNRAVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVA 78
Query: 166 WHPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVT 225
+ ++S S D ++K+W D + ++TL GHT+ + ++FN + +V+
Sbjct: 79 FSSDARFIVSASDDKTLKLW----DVETGSLIKTL----IGHTNYAFCVNFNPQSNMIVS 130
Query: 226 CS 227
S
Sbjct: 131 GS 132
Score = 34.3 bits (77), Expect = 0.059
Identities = 20/45 (44%), Positives = 27/45 (59%), Gaps = 6/45 (13%)
Query: 7 KEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQ 51
K +Q+LEGH + V ++ +P T + AS S DKTVRIW Q
Sbjct: 276 KLLQKLEGHTETVMNVACHP------TENLIASGSLDKTVRIWTQ 314
>At3g15980 putative coatomer complex subunit
Length = 921
Score = 75.1 bits (183), Expect = 3e-14
Identities = 64/222 (28%), Positives = 105/222 (46%), Gaps = 26/222 (11%)
Query: 6 LKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLD 65
+ +V+ E H+D + + +P P V +S S D +++W D +G WAC +
Sbjct: 92 MDKVKVFEAHSDYIRCVAVHPTL-----PYVLSS-SDDMLIKLW--DWENG-WACTQIF- 141
Query: 66 ETHTRTVRSCAWSPSG-KLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNAS 124
E H+ V ++P A+AS D T IW N+G + TL+ H+ V V +
Sbjct: 142 EGHSHYVMQVVFNPKDTNTFASASLDRTIKIW-NLGSP-DPNFTLDAHQKGVNCVDYFTG 199
Query: 125 GT--LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSI 182
G L T S D + +W+ Q + CV L GHT +V V +HP I+I+ S D ++
Sbjct: 200 GDKPYLITGSDDHTAKVWDYQTKS---CVQTLDGHTHNVSAVCFHPELPIIITGSEDGTV 256
Query: 183 KVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMV 224
++W + ++ TL N VWA+ + S ++V
Sbjct: 257 RIW----HATTYRLENTL----NYGLERVWAIGYIKSSRRVV 290
Score = 38.5 bits (88), Expect = 0.003
Identities = 34/150 (22%), Positives = 58/150 (38%), Gaps = 20/150 (13%)
Query: 72 VRSCAWSPSGKLLATASFDATTAIW----ENVGGDFECVATLEGHENEVKSVSWNASGTL 127
V+S P+ + + + T IW + + FE + W +G
Sbjct: 21 VKSVDLHPTEPWILASLYSGTVCIWNYQTQTITKSFEVTELPVRSAKFIPRKQWVVAG-- 78
Query: 128 LATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWAD 187
+ D + ++ N + V V + H+ ++ V HPT ++S S D IK+W
Sbjct: 79 ----ADDMYIRVYNY---NTMDKVKVFEAHSDYIRCVAVHPTLPYVLSSSDDMLIKLWDW 131
Query: 188 EGDSDDWQCVQTLGQPNNGHTSTVWALSFN 217
E + W C Q GH+ V + FN
Sbjct: 132 E---NGWACTQIF----EGHSHYVMQVVFN 154
>At2g30050 putative protein transport protein SEC13
Length = 302
Score = 74.3 bits (181), Expect = 5e-14
Identities = 62/245 (25%), Positives = 104/245 (42%), Gaps = 33/245 (13%)
Query: 7 KEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDE 66
+++ L GH VW + W H + ASCS D V +W++ + W V +
Sbjct: 47 QQLATLTGHRGPVWEVAW----AHPKYGSILASCSYDGQVILWKEG-NQNQWTQDHVFTD 101
Query: 67 THTRTVRSCAWSPS--GKLLATASFDATTAIW-ENVGGDFECVATLEGHENEVKSVSW-- 121
H +V S AW+P G LA S D +++ G ++ + H V SVSW
Sbjct: 102 -HKSSVNSIAWAPHDIGLSLACGSSDGNISVFTARADGGWDTSRIDQAHPVGVTSVSWAP 160
Query: 122 -NASGTL-----------LATCSRDKSVWIWEMQPVN-EFECVSVLQGHTQDVKMVKWHP 168
A G L LA+ D +V +W++ + + +C LQ HT V+ V W P
Sbjct: 161 ATAPGALVSSGLLDPVYKLASGGCDNTVKVWKLANGSWKMDCFPALQKHTDWVRDVAWAP 220
Query: 169 T----EDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMV 224
+ + S S D + +W + + W+ G+ + VW +S++ +G+ +
Sbjct: 221 NLGLPKSTIASGSQDGKVIIWTVGKEGEQWE-----GKVLKDFMTPVWRVSWSLTGNLLA 275
Query: 225 TCSSN 229
N
Sbjct: 276 VSDGN 280
Score = 57.8 bits (138), Expect = 5e-09
Identities = 48/156 (30%), Positives = 69/156 (43%), Gaps = 8/156 (5%)
Query: 64 LDETHTRTVRSCAWSPSGKLLATASFDATTAIW--ENVGGDFECVATLEGHENEVKSVSW 121
++ H V GK +ATAS D T I N GG + +ATL GH V V+W
Sbjct: 6 IETGHEDIVHDVQMDYYGKRIATASSDCTIKITGVSNNGGSQQ-LATLTGHRGPVWEVAW 64
Query: 122 NAS--GTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTE-DILISC-S 177
G++LA+CS D V +W+ N++ V H V + W P + + ++C S
Sbjct: 65 AHPKYGSILASCSYDGQVILWKEGNQNQWTQDHVFTDHKSSVNSIAWAPHDIGLSLACGS 124
Query: 178 YDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWA 213
D +I V+ D W + G TS WA
Sbjct: 125 SDGNISVFTARADG-GWDTSRIDQAHPVGVTSVSWA 159
Score = 54.7 bits (130), Expect = 4e-08
Identities = 49/159 (30%), Positives = 73/159 (45%), Gaps = 20/159 (12%)
Query: 13 EGHNDRVWSLDWNPAT--------GHAGTPLVFASCSGDKTVRIWEQDLSSGLWA--CKA 62
+ H V S+ W PAT G AS D TV++W+ L++G W C
Sbjct: 147 QAHPVGVTSVSWAPATAPGALVSSGLLDPVYKLASGGCDNTVKVWK--LANGSWKMDCFP 204
Query: 63 VLDETHTRTVRSCAWSPSGKL----LATASFDATTAIWENVGGDFECVA--TLEGHENEV 116
L + HT VR AW+P+ L +A+ S D IW VG + E L+ V
Sbjct: 205 ALQK-HTDWVRDVAWAPNLGLPKSTIASGSQDGKVIIW-TVGKEGEQWEGKVLKDFMTPV 262
Query: 117 KSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQ 155
VSW+ +G LLA + +V +W+ E+E V+ ++
Sbjct: 263 WRVSWSLTGNLLAVSDGNNNVTVWKEAVDGEWEQVTAVE 301
>At5g16750 WD40-repeat protein
Length = 876
Score = 72.8 bits (177), Expect = 1e-13
Identities = 62/230 (26%), Positives = 106/230 (45%), Gaps = 28/230 (12%)
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRT 71
L GH + V SLD +G L+ S DKTVR+W S C V H
Sbjct: 397 LAGHKEVVLSLD--TCVSSSGNVLIVTG-SKDKTVRLWNATSKS----CIGV-GTGHNGD 448
Query: 72 VRSCAWSP-SGKLLATASFDATTAIW--ENVGGDFECVATLE------GHENEVKSVSWN 122
+ + A++ S + S D T +W + + D E L+ H+ ++ SV+
Sbjct: 449 ILAVAFAKKSFSFFVSGSGDRTLKVWSLDGISEDSEEPINLKTRSVVAAHDKDINSVAVA 508
Query: 123 ASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSI 182
+ +L+ T S D++ IW + + V L+GH + + V++ + +++ S D ++
Sbjct: 509 RNDSLVCTGSEDRTASIWRLPDLVH---VVTLKGHKRRIFSVEFSTVDQCVMTASGDKTV 565
Query: 183 KVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSNLLM 232
K+WA D C++T GHTS+V SF G + V+C ++ L+
Sbjct: 566 KIWA----ISDGSCLKTF----EGHTSSVLRASFITDGTQFVSCGADGLL 607
Score = 70.9 bits (172), Expect = 6e-13
Identities = 51/167 (30%), Positives = 85/167 (50%), Gaps = 15/167 (8%)
Query: 66 ETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASG 125
E + T+ + A SP KLL +A +W+ +C+ + +GHE V ++ +ASG
Sbjct: 57 EGESDTLTALALSPDDKLLFSAGHSRQIRVWDLE--TLKCIRSWKGHEGPVMGMACHASG 114
Query: 126 TLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHP--TEDILISCSYDNSIK 183
LLAT D+ V +W+ V+ C +GH V + +HP ++ILIS S D +++
Sbjct: 115 GLLATAGADRKVLVWD---VDGGFCTHYFRGHKGVVSSILFHPDSNKNILISGSDDATVR 171
Query: 184 VWADEGDSDDWQCVQTLGQPNNGHTSTVWALS------FNASGDKMV 224
VW + + +C+ + + + TS ALS F+A DK+V
Sbjct: 172 VWDLNAKNTEKKCLAIMEKHFSAVTSI--ALSEDGLTLFSAGRDKVV 216
Score = 58.5 bits (140), Expect = 3e-09
Identities = 45/156 (28%), Positives = 66/156 (41%), Gaps = 14/156 (8%)
Query: 40 CSG--DKTVRIWE-QDLSSGLWACKAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIW 96
C+G D+T IW DL V + H R + S +S + + TAS D T IW
Sbjct: 515 CTGSEDRTASIWRLPDL------VHVVTLKGHKRRIFSVEFSTVDQCVMTASGDKTVKIW 568
Query: 97 ENVGGDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQG 156
G C+ T EGH + V S+ GT +C D + +W VN EC++
Sbjct: 569 AISDGS--CLKTFEGHTSSVLRASFITDGTQFVSCGADGLLKLWN---VNTSECIATYDQ 623
Query: 157 HTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSD 192
H V + +++ + D I +W D SD
Sbjct: 624 HEDKVWALAVGKKTEMIATGGGDAVINLWHDSTASD 659
Score = 52.8 bits (125), Expect = 2e-07
Identities = 43/180 (23%), Positives = 83/180 (45%), Gaps = 17/180 (9%)
Query: 5 ELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVL 64
+L V L+GH R++S++++ T SGDKTV+IW +S G +C
Sbjct: 530 DLVHVVTLKGHKRRIFSVEFSTVDQCVMT------ASGDKTVKIWA--ISDG--SCLKTF 579
Query: 65 DETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNAS 124
E HT +V ++ G + D +W NV EC+AT + HE++V +++
Sbjct: 580 -EGHTSSVLRASFITDGTQFVSCGADGLLKLW-NVNTS-ECIATYDQHEDKVWALAVGKK 636
Query: 125 GTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKV 184
++AT D + +W ++ E ++ +++ E+ ++ Y +I++
Sbjct: 637 TEMIATGGGDAVINLWHDSTASDKED----DFRKEEEAILRGQELENAVLDAEYTKAIRL 692
Score = 46.6 bits (109), Expect = 1e-05
Identities = 38/149 (25%), Positives = 68/149 (45%), Gaps = 18/149 (12%)
Query: 6 LKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLD 65
LK ++ +GH V + + + G + A+ D+ V +W D+ G C
Sbjct: 92 LKCIRSWKGHEGPVMGMACHASGG------LLATAGADRKVLVW--DVDGGF--CTHYF- 140
Query: 66 ETHTRTVRSCAWSPSGK--LLATASFDATTAIWENVGGDFE--CVATLEGHENEVKSVSW 121
H V S + P +L + S DAT +W+ + E C+A +E H + V S++
Sbjct: 141 RGHKGVVSSILFHPDSNKNILISGSDDATVRVWDLNAKNTEKKCLAIMEKHFSAVTSIAL 200
Query: 122 NASGTLLATCSRDKSVWIWEMQPVNEFEC 150
+ G L + RDK V +W++ +++ C
Sbjct: 201 SEDGLTLFSAGRDKVVNLWDL---HDYSC 226
Score = 42.0 bits (97), Expect = 3e-04
Identities = 30/152 (19%), Positives = 62/152 (40%), Gaps = 14/152 (9%)
Query: 78 SPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSV 137
S G +A A D + D +T+EG + + +++ + LL + + +
Sbjct: 28 SSDGSFIACACGDVINIVDST---DSSVKSTIEGESDTLTALALSPDDKLLFSAGHSRQI 84
Query: 138 WIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSDDWQCV 197
+W+++ + +C+ +GH V + H + +L + D + VW D D C
Sbjct: 85 RVWDLETL---KCIRSWKGHEGPVMGMACHASGGLLATAGADRKVLVW----DVDGGFCT 137
Query: 198 QTLGQPNNGHTSTVWALSFNASGDKMVTCSSN 229
GH V ++ F+ +K + S +
Sbjct: 138 HYF----RGHKGVVSSILFHPDSNKNILISGS 165
>At3g21540 WD repeat like protein
Length = 949
Score = 72.0 bits (175), Expect = 3e-13
Identities = 60/225 (26%), Positives = 100/225 (43%), Gaps = 20/225 (8%)
Query: 8 EVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDET 67
+V+ ++ H +WS+ P +G F + S D V+ WE + L V
Sbjct: 479 KVEEVKAHGGTIWSI--TPIPNDSG----FVTVSADHEVKFWEYQATKKLTVSN-VKSMK 531
Query: 68 HTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTL 127
V + A SP K +A A D+T ++ F +L GH+ V + ++ G L
Sbjct: 532 MNDDVLAVAISPDAKHIAVALLDSTVKVFYMDSLKF--YLSLYGHKLPVMCIDISSDGEL 589
Query: 128 LATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWAD 187
+ T S+DK++ IW + + +C + H V VK+ L S D +K W
Sbjct: 590 IVTGSQDKNLKIWGL---DFGDCHKSIFAHGDSVMGVKFVRNTHYLFSIGKDRLVKYW-- 644
Query: 188 EGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSNLLM 232
D+D ++ + TL GH + +W L+ + GD +VT S + M
Sbjct: 645 --DADKFEHLLTL----EGHHAEIWCLAISNRGDFLVTGSHDRSM 683
Score = 60.1 bits (144), Expect = 1e-09
Identities = 47/207 (22%), Positives = 89/207 (42%), Gaps = 34/207 (16%)
Query: 43 DKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGD 102
D ++RIW+ + + C+ + +H V + ++ G +LA+ S D +W+ VG
Sbjct: 85 DGSIRIWDTEKGT----CEVNFN-SHKGAVTALRYNKVGSMLASGSKDNDIILWDVVGES 139
Query: 103 FECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVK 162
+ L GH ++V + + G L + S+DK + +W+++ + C+ ++ GH +V
Sbjct: 140 G--LFRLRGHRDQVTDLVFLDGGKKLVSSSKDKFLRVWDLETQH---CMQIVSGHHSEVW 194
Query: 163 MVKWHPTEDI-----------------------LISCSYDNSIKVWADEGDSDDWQCVQT 199
V P E L+S S N IK + + W+ ++
Sbjct: 195 SVDTDPEERYVVTGSADQELRFYAVKEYSSNGSLVSDSNANEIKASEEHSTENKWEILKL 254
Query: 200 LGQPNNGHTSTVWALSFNASGDKMVTC 226
G+ V + FN SG K++ C
Sbjct: 255 FGEIQRQTKDRVARVRFNVSG-KLLAC 280
Score = 55.1 bits (131), Expect = 3e-08
Identities = 47/183 (25%), Positives = 83/183 (44%), Gaps = 22/183 (12%)
Query: 56 GLW-----ACKAVLDETHTRTVRSCAW----SPSGKLLATASFDATTAIWENVGGDFECV 106
G+W C L + +R S A S + L+A D + IW+ G C
Sbjct: 42 GIWHVRQGVCSKTLTPSSSRGGPSLAVTSIASSASSLVAVGYADGSIRIWDTEKGT--CE 99
Query: 107 ATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKW 166
H+ V ++ +N G++LA+ S+D + +W++ V E + L+GH V + +
Sbjct: 100 VNFNSHKGAVTALRYNKVGSMLASGSKDNDIILWDV--VGE-SGLFRLRGHRDQVTDLVF 156
Query: 167 HPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTC 226
L+S S D ++VW D + C+Q + +GH S VW++ + +VT
Sbjct: 157 LDGGKKLVSSSKDKFLRVW----DLETQHCMQIV----SGHHSEVWSVDTDPEERYVVTG 208
Query: 227 SSN 229
S++
Sbjct: 209 SAD 211
Score = 35.8 bits (81), Expect = 0.020
Identities = 27/101 (26%), Positives = 44/101 (42%), Gaps = 7/101 (6%)
Query: 41 SGDKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIWENVG 100
S DK ++IW D K++ H +V + + L + D W+
Sbjct: 594 SQDKNLKIWGLDFGD---CHKSIF--AHGDSVMGVKFVRNTHYLFSIGKDRLVKYWD--A 646
Query: 101 GDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWE 141
FE + TLEGH E+ ++ + G L T S D+S+ W+
Sbjct: 647 DKFEHLLTLEGHHAEIWCLAISNRGDFLVTGSHDRSMRRWD 687
Score = 29.6 bits (65), Expect = 1.4
Identities = 28/108 (25%), Positives = 42/108 (37%), Gaps = 20/108 (18%)
Query: 9 VQRLEGHNDRVWSLDWNP-----ATGHAGTPLVF------------ASCSGDKTVRIWEQ 51
+Q + GH+ VWS+D +P TG A L F S S ++ E+
Sbjct: 183 MQIVSGHHSEVWSVDTDPEERYVVTGSADQELRFYAVKEYSSNGSLVSDSNANEIKASEE 242
Query: 52 DLSSGLWACKAVLDETHTRT---VRSCAWSPSGKLLATASFDATTAIW 96
+ W + E +T V ++ SGKLLA T I+
Sbjct: 243 HSTENKWEILKLFGEIQRQTKDRVARVRFNVSGKLLACQMAGKTIEIF 290
>At1g79990 coatomer protein like complex, subunit beta 2 (beta
prime)
Length = 920
Score = 69.7 bits (169), Expect = 1e-12
Identities = 49/159 (30%), Positives = 77/159 (47%), Gaps = 13/159 (8%)
Query: 30 HAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWSPSG-KLLATAS 88
H P V +S S D +++W+ + W C + E H+ V ++P A+AS
Sbjct: 108 HPTLPYVLSS-SDDMLIKLWDWEKG---WLCTQIF-EGHSHYVMQVTFNPKDTNTFASAS 162
Query: 89 FDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGT--LLATCSRDKSVWIWEMQPVN 146
D T IW N+G + TL+ H V V + G L T S D + +W+ Q +
Sbjct: 163 LDRTIKIW-NLGSP-DPNFTLDAHLKGVNCVDYFTGGDKPYLITGSDDHTAKVWDYQTKS 220
Query: 147 EFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
CV L+GHT +V V +HP I+I+ S D ++++W
Sbjct: 221 ---CVQTLEGHTHNVSAVSFHPELPIIITGSEDGTVRIW 256
Score = 39.3 bits (90), Expect = 0.002
Identities = 34/146 (23%), Positives = 59/146 (40%), Gaps = 12/146 (8%)
Query: 72 VRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATC 131
V+S P+ + + + T IW V + + E V+S + A +
Sbjct: 18 VKSVDLHPTEPWILASLYSGTLCIWNYQTQTM--VKSFDVTELPVRSAKFIARKQWVVAG 75
Query: 132 SRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDS 191
+ D + ++ N + + V + H ++ V HPT ++S S D IK+W E
Sbjct: 76 ADDMFIRVYNY---NTMDKIKVFEAHADYIRCVAVHPTLPYVLSSSDDMLIKLWDWE--- 129
Query: 192 DDWQCVQTLGQPNNGHTSTVWALSFN 217
W C Q GH+ V ++FN
Sbjct: 130 KGWLCTQIF----EGHSHYVMQVTFN 151
>At2g19540 putative WD-40 repeat protein
Length = 469
Score = 68.9 bits (167), Expect = 2e-12
Identities = 51/178 (28%), Positives = 88/178 (48%), Gaps = 16/178 (8%)
Query: 14 GHNDRVWSLDWNPATGHAGTPLVFASCSGD--KTVRIWEQDLSSGLWACKAVLDETHTRT 71
GH D +++DW+PAT AG L SGD + +WE +SG WA + HT +
Sbjct: 221 GHKDEGYAIDWSPAT--AGRLL-----SGDCKSMIHLWEP--ASGSWAVDPIPFAGHTAS 271
Query: 72 VRSCAWSPSGK-LLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWN-ASGTLLA 129
V WSP+ + + A+ S D + A+W+ G + + + H +V +SWN + +LA
Sbjct: 272 VEDLQWSPAEENVFASCSVDGSVAVWDIRLGKSPAL-SFKAHNADVNVISWNRLASCMLA 330
Query: 130 TCSRDKSVWIWEMQPVNEFEC-VSVLQGHTQDVKMVKWHPTE-DILISCSYDNSIKVW 185
+ S D + I +++ + + V+ + H + ++W E L S DN + +W
Sbjct: 331 SGSDDGTFSIRDLRLIKGGDAVVAHFEYHKHPITSIEWSAHEASTLAVTSGDNQLTIW 388
>At1g11160 hypothetical protein
Length = 974
Score = 67.8 bits (164), Expect = 5e-12
Identities = 46/188 (24%), Positives = 85/188 (44%), Gaps = 25/188 (13%)
Query: 5 ELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVL 64
E K V+ GH +++++P AS S D +R+W+ + K
Sbjct: 38 ESKMVRAFTGHRSNCSAVEFHPFGEF------LASGSSDTNLRVWDTRKKGCIQTYKG-- 89
Query: 65 DETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGG----DFECVATLEGHENEVKSVS 120
HTR + + +SP G+ + + D +W+ G +F+C HE ++S+
Sbjct: 90 ---HTRGISTIEFSPDGRWVVSGGLDNVVKVWDLTAGKLLHEFKC------HEGPIRSLD 140
Query: 121 WNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDN 180
++ LLAT S D++V W+++ FE + + V+ + +HP L C D+
Sbjct: 141 FHPLEFLLATGSADRTVKFWDLE---TFELIGTTRPEATGVRAIAFHPDGQTLF-CGLDD 196
Query: 181 SIKVWADE 188
+KV++ E
Sbjct: 197 GLKVYSWE 204
Score = 52.8 bits (125), Expect = 2e-07
Identities = 28/118 (23%), Positives = 56/118 (46%), Gaps = 5/118 (4%)
Query: 68 HTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTL 127
HT V S A++ L+ + +W+ + + V GH + +V ++ G
Sbjct: 6 HTSPVDSVAFNSEEVLVLAGASSGVIKLWDLE--ESKMVRAFTGHRSNCSAVEFHPFGEF 63
Query: 128 LATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
LA+ S D ++ +W+ + C+ +GHT+ + +++ P ++S DN +KVW
Sbjct: 64 LASGSSDTNLRVWDTRKKG---CIQTYKGHTRGISTIEFSPDGRWVVSGGLDNVVKVW 118
Score = 52.0 bits (123), Expect = 3e-07
Identities = 32/118 (27%), Positives = 59/118 (49%), Gaps = 11/118 (9%)
Query: 108 TLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWH 167
+L GH + V SV++N+ L+ + + +W+++ E + V GH + V++H
Sbjct: 2 SLCGHTSPVDSVAFNSEEVLVLAGASSGVIKLWDLE---ESKMVRAFTGHRSNCSAVEFH 58
Query: 168 PTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVT 225
P + L S S D +++VW D+ C+QT GHT + + F+ G +V+
Sbjct: 59 PFGEFLASGSSDTNLRVW----DTRKKGCIQTY----KGHTRGISTIEFSPDGRWVVS 108
>At3g01340 transport protein SEC13, putative
Length = 302
Score = 67.0 bits (162), Expect = 8e-12
Identities = 58/240 (24%), Positives = 99/240 (41%), Gaps = 33/240 (13%)
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRT 71
L GH VW + W H + ASCS D + +W++ + W V + H +
Sbjct: 52 LTGHRGPVWQVAW----AHPKFGSLLASCSYDGQIILWKEG-NQNQWTQAHVFTD-HKVS 105
Query: 72 VRSCAWSPS--GKLLATASFDATTAIWE-NVGGDFECVATLEGHENEVKSVSWN------ 122
V S AW+P G LA + D +++ G ++ + H V SVSW
Sbjct: 106 VNSIAWAPHELGLSLACGASDGNISVFSARADGGWDTTKIDQAHPVGVTSVSWAPATEPG 165
Query: 123 ---ASGTL-----LATCSRDKSVWIWEMQPVN-EFECVSVLQGHTQDVKMVKWHPT---- 169
+SG + LA+ D +V +W+ + + +C L HT V+ V W P
Sbjct: 166 ALVSSGMIDPVYKLASGGCDSTVKVWKFSNGSWKMDCFPALNKHTDWVRDVAWAPNLGLP 225
Query: 170 EDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSN 229
+ + S S D + +W + + W+ G + VW +S++ +G+ + N
Sbjct: 226 KSTIASGSEDGKVIIWTIGKEGEQWE-----GTVLKDFKTPVWRVSWSLTGNLLAVSDGN 280
Score = 62.8 bits (151), Expect = 2e-10
Identities = 48/156 (30%), Positives = 72/156 (45%), Gaps = 8/156 (5%)
Query: 64 LDETHTRTVRSCAWSPSGKLLATASFDATTAIW--ENVGGDFECVATLEGHENEVKSVSW 121
++ H+ T+ GK +ATAS D T I N GG + +ATL GH V V+W
Sbjct: 6 IETGHSDTIHDVVMDYYGKRVATASSDCTIKITGVSNSGGS-QHLATLTGHRGPVWQVAW 64
Query: 122 NAS--GTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTE-DILISC-S 177
G+LLA+CS D + +W+ N++ V H V + W P E + ++C +
Sbjct: 65 AHPKFGSLLASCSYDGQIILWKEGNQNQWTQAHVFTDHKVSVNSIAWAPHELGLSLACGA 124
Query: 178 YDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWA 213
D +I V++ D W + G TS WA
Sbjct: 125 SDGNISVFSARADG-GWDTTKIDQAHPVGVTSVSWA 159
Score = 58.2 bits (139), Expect = 4e-09
Identities = 49/159 (30%), Positives = 75/159 (46%), Gaps = 20/159 (12%)
Query: 13 EGHNDRVWSLDWNPAT--------GHAGTPLVFASCSGDKTVRIWEQDLSSGLWA--CKA 62
+ H V S+ W PAT G AS D TV++W+ S+G W C
Sbjct: 147 QAHPVGVTSVSWAPATEPGALVSSGMIDPVYKLASGGCDSTVKVWK--FSNGSWKMDCFP 204
Query: 63 VLDETHTRTVRSCAWSPSGKL----LATASFDATTAIWENVGGDFECV--ATLEGHENEV 116
L++ HT VR AW+P+ L +A+ S D IW +G + E L+ + V
Sbjct: 205 ALNK-HTDWVRDVAWAPNLGLPKSTIASGSEDGKVIIW-TIGKEGEQWEGTVLKDFKTPV 262
Query: 117 KSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQ 155
VSW+ +G LLA + +V +W+ E+E V+V++
Sbjct: 263 WRVSWSLTGNLLAVSDGNNNVTVWKESVDGEWEQVTVVE 301
>At2g43770 putative splicing factor
Length = 343
Score = 66.2 bits (160), Expect = 1e-11
Identities = 47/147 (31%), Positives = 64/147 (42%), Gaps = 16/147 (10%)
Query: 43 DKTVRIWEQDLSSGLWACKAVLD-ETHTRTVRSCAWSPSGKLLATASFDATTAIWEN--V 99
D V++W DL G +A + E H T+ + SP G L T D +W+
Sbjct: 201 DNDVKVW--DLRKG----EATMTLEGHQDTITGMSLSPDGSYLLTNGMDNKLCVWDMRPY 254
Query: 100 GGDFECVATLEGH----ENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQ 155
CV EGH E + SW+ GT + S D+ V IW+ + L
Sbjct: 255 APQNRCVKIFEGHQHNFEKNLLKCSWSPDGTKVTAGSSDRMVHIWD---TTSRRTIYKLP 311
Query: 156 GHTQDVKMVKWHPTEDILISCSYDNSI 182
GHT V +HPTE I+ SCS D +I
Sbjct: 312 GHTGSVNECVFHPTEPIIGSCSSDKNI 338
Score = 63.5 bits (153), Expect = 9e-11
Identities = 64/270 (23%), Positives = 107/270 (38%), Gaps = 64/270 (23%)
Query: 5 ELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVL 64
+ K L+GH + + L W G+ +V S S DKTVR W D+ +G K
Sbjct: 85 DCKNFMVLKGHKNAILDLHWTSD----GSQIV--SASPDKTVRAW--DVETGKQIKKMA- 135
Query: 65 DETHTRTVRSCAWSPSGK-LLATASFDATTAIWE-------------------------- 97
H+ V SC + G L+ + S D T +W+
Sbjct: 136 --EHSSFVNSCCPTRRGPPLIISGSDDGTAKLWDMRQRGAIQTFPDKYQITAVSFSDAAD 193
Query: 98 -----NVGGDF--------ECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQP 144
V D E TLEGH++ + +S + G+ L T D + +W+M+P
Sbjct: 194 KIFTGGVDNDVKVWDLRKGEATMTLEGHQDTITGMSLSPDGSYLLTNGMDNKLCVWDMRP 253
Query: 145 -VNEFECVSVLQGH----TQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSDDWQCVQT 199
+ CV + +GH +++ W P + + S D + +W D+ + +
Sbjct: 254 YAPQNRCVKIFEGHQHNFEKNLLKCSWSPDGTKVTAGSSDRMVHIW----DTTSRRTIYK 309
Query: 200 LGQPNNGHTSTVWALSFNASGDKMVTCSSN 229
L GHT +V F+ + + +CSS+
Sbjct: 310 L----PGHTGSVNECVFHPTEPIIGSCSSD 335
Score = 57.8 bits (138), Expect = 5e-09
Identities = 39/158 (24%), Positives = 74/158 (46%), Gaps = 12/158 (7%)
Query: 68 HTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTL 127
H V + ++P+G L+A+ S D +W V GD + L+GH+N + + W + G+
Sbjct: 52 HPSAVYTMKFNPAGTLIASGSHDREIFLWR-VHGDCKNFMVLKGHKNAILDLHWTSDGSQ 110
Query: 128 LATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWAD 187
+ + S DK+V W+++ + + ++ + P ++IS S D + K+W
Sbjct: 111 IVSASPDKTVRAWDVETGKQIKKMAEHSSFVNSCCPTRRGP--PLIISGSDDGTAKLW-- 166
Query: 188 EGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVT 225
D +QT + A+SF+ + DK+ T
Sbjct: 167 --DMRQRGAIQTFPDKYQ-----ITAVSFSDAADKIFT 197
Score = 50.4 bits (119), Expect = 8e-07
Identities = 50/219 (22%), Positives = 89/219 (39%), Gaps = 27/219 (12%)
Query: 9 VQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACK-AVLDET 67
+ L GH V+++ +NP AGT + AS S D+ + +W CK ++ +
Sbjct: 46 IMLLSGHPSAVYTMKFNP----AGT--LIASGSHDREIFLWRVH-----GDCKNFMVLKG 94
Query: 68 HTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASG-T 126
H + W+ G + +AS D T W+ G + + + H + V S G
Sbjct: 95 HKNAILDLHWTSDGSQIVSASPDKTVRAWDVETG--KQIKKMAEHSSFVNSCCPTRRGPP 152
Query: 127 LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWA 186
L+ + S D + +W+M+ + + V + D + + DN +KVW
Sbjct: 153 LIISGSDDGTAKLWDMRQRGAIQTFP----DKYQITAVSFSDAADKIFTGGVDNDVKVW- 207
Query: 187 DEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVT 225
D + TL GH T+ +S + G ++T
Sbjct: 208 ---DLRKGEATMTL----EGHQDTITGMSLSPDGSYLLT 239
Score = 47.0 bits (110), Expect = 9e-06
Identities = 28/134 (20%), Positives = 56/134 (40%), Gaps = 11/134 (8%)
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLD---ETH 68
LEGH D + + +P + T + D + +W+ + C + +
Sbjct: 218 LEGHQDTITGMSLSPDGSYLLTNGM------DNKLCVWDMRPYAPQNRCVKIFEGHQHNF 271
Query: 69 TRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLL 128
+ + C+WSP G + S D IW+ + L GH V ++ + ++
Sbjct: 272 EKNLLKCSWSPDGTKVTAGSSDRMVHIWDTTSR--RTIYKLPGHTGSVNECVFHPTEPII 329
Query: 129 ATCSRDKSVWIWEM 142
+CS DK++++ E+
Sbjct: 330 GSCSSDKNIYLGEI 343
Score = 37.7 bits (86), Expect = 0.005
Identities = 19/77 (24%), Positives = 39/77 (49%), Gaps = 7/77 (9%)
Query: 151 VSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTST 210
+ +L GH V +K++P ++ S S+D I +W GD ++ ++ GH +
Sbjct: 46 IMLLSGHPSAVYTMKFNPAGTLIASGSHDREIFLWRVHGDCKNFMVLK-------GHKNA 98
Query: 211 VWALSFNASGDKMVTCS 227
+ L + + G ++V+ S
Sbjct: 99 ILDLHWTSDGSQIVSAS 115
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.129 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,029,622
Number of Sequences: 26719
Number of extensions: 253462
Number of successful extensions: 1914
Number of sequences better than 10.0: 192
Number of HSP's better than 10.0 without gapping: 119
Number of HSP's successfully gapped in prelim test: 73
Number of HSP's that attempted gapping in prelim test: 700
Number of HSP's gapped (non-prelim): 694
length of query: 235
length of database: 11,318,596
effective HSP length: 96
effective length of query: 139
effective length of database: 8,753,572
effective search space: 1216746508
effective search space used: 1216746508
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0010.22


