

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.21
(161 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
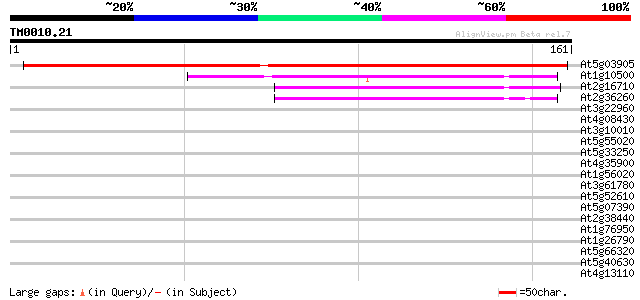
At5g03905 unknown protein 200 3e-52
At1g10500 unknown protein 65 2e-11
At2g16710 putative HesB-like protein 59 9e-10
At2g36260 hypothetical protein 56 7e-09
At3g22960 pyruvate kinase like protein 30 0.57
At4g08430 putative protein 30 0.74
At3g10010 hypothetical protein 30 0.74
At5g55020 putative transcription factor MYB120 (MYB120) 29 1.3
At5g33250 putative protein 28 1.6
At4g35900 bZIP transcription factor AtbZIP14 28 1.6
At1g56020 unknown protein 28 1.6
At3g61780 putative protein 28 2.2
At5g52610 putative protein 28 2.8
At5g07390 respiratory burst oxidase protein A 28 2.8
At2g38440 unknown protein 28 2.8
At1g76950 zinc finger protein (PRAF1) 28 2.8
At1g26790 H-protein promoter binding factor-2b, putative 28 2.8
At5g66320 GATA-binding transcription factor-like protein 27 3.7
At5g40630 unknown protein 27 3.7
At4g13110 putative protein 27 3.7
>At5g03905 unknown protein
Length = 158
Score = 200 bits (508), Expect = 3e-52
Identities = 99/156 (63%), Positives = 123/156 (78%), Gaps = 2/156 (1%)
Query: 5 SLFRRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKE 64
SL +R+AP LA R R+N +L +FSS+S+ +++S SS + VH+++NC+RRMKE
Sbjct: 4 SLVKRVAPYLAGRIRENHRLLNFSSASAIKEASSSSSSQPESSSNDVVHLSDNCIRRMKE 63
Query: 65 LEASESATAPKTLRLSVETGGCSGFQYAFDLDDRINSDDRVFEREGIKLVVDNISYDFVK 124
L++SE K LRL VETGGCSGFQY F+LD+R N DDRVFE+ G+KLVVDN+SYD VK
Sbjct: 64 LQSSEPEK--KMLRLGVETGGCSGFQYKFELDNRTNPDDRVFEKNGVKLVVDNVSYDLVK 121
Query: 125 GATVDYVEELIRSAFVVTENPSAVGGCSCKSSFMVK 160
GAT+DY EELIR+AFVV NPSAVGGCSCKSSFMVK
Sbjct: 122 GATIDYEEELIRAAFVVAVNPSAVGGCSCKSSFMVK 157
>At1g10500 unknown protein
Length = 180
Score = 64.7 bits (156), Expect = 2e-11
Identities = 36/108 (33%), Positives = 56/108 (51%), Gaps = 5/108 (4%)
Query: 52 VHMTENCLRRMKELEASESATAPKTLRLSVETGGCSGFQYAFDLDDRINS--DDRVFERE 109
+ ++EN L+ + ++ + LR+ V+ GGCSG Y D ++R N+ DD E +
Sbjct: 72 ISLSENALKHLSKMRSERGEDL--CLRIGVKQGGCSGMSYTMDFENRANARPDDSTIEYQ 129
Query: 110 GIKLVVDNISYDFVKGATVDYVEELIRSAFVVTENPSAVGGCSCKSSF 157
G +V D S F+ G +DY + LI F + NP+A C C SF
Sbjct: 130 GFTIVCDPKSMLFLFGMQLDYSDALIGGGFSFS-NPNATQTCGCGKSF 176
>At2g16710 putative HesB-like protein
Length = 137
Score = 59.3 bits (142), Expect = 9e-10
Identities = 29/82 (35%), Positives = 47/82 (56%), Gaps = 1/82 (1%)
Query: 77 LRLSVETGGCSGFQYAFDLDDRINSDDRVFEREGIKLVVDNISYDFVKGATVDYVEELIR 136
LRL V+ GC+G Y + D D + E +G++++V+ + V G +D+V++ +R
Sbjct: 45 LRLGVKARGCNGLSYTLNYADEKGKFDELVEEKGVRILVEPKALMHVIGTKMDFVDDKLR 104
Query: 137 SAFVVTENPSAVGGCSCKSSFM 158
S FV NP++ G C C SFM
Sbjct: 105 SEFVFI-NPNSQGQCGCGESFM 125
>At2g36260 hypothetical protein
Length = 109
Score = 56.2 bits (134), Expect = 7e-09
Identities = 31/81 (38%), Positives = 45/81 (55%), Gaps = 2/81 (2%)
Query: 77 LRLSVETGGCSGFQYAFDLDDRINSDDRVFEREGIKLVVDNISYDFVKGATVDYVEELIR 136
LRL+VE GC+G Y + D V E +G+K++VD + V G +D+V++ +R
Sbjct: 28 LRLAVEAKGCNGLSYVLNYAQEKGKFDEVVEEKGVKILVDPKAVMHVIGTEMDFVDDKLR 87
Query: 137 SAFVVTENPSAVGGCSCKSSF 157
S FV NP+A C C SF
Sbjct: 88 SEFVFV-NPNAT-KCGCGESF 106
>At3g22960 pyruvate kinase like protein
Length = 596
Score = 30.0 bits (66), Expect = 0.57
Identities = 25/60 (41%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Query: 23 KLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAPKTLRLSVE 82
K TS +SSSSS D +SSSSSS V ++ N +K E S ATA T +E
Sbjct: 42 KYTSIRASSSSSPSPDLDSSSSSSS--SQVLLSPNGTGAVKSDERSVVATAVTTDTSGIE 99
>At4g08430 putative protein
Length = 808
Score = 29.6 bits (65), Expect = 0.74
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 4/45 (8%)
Query: 8 RRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPV 52
RR P LA +QK S +S+ ++ D +SSS + H++PV
Sbjct: 485 RRRVPQLAG----SQKYPSVGNSTVKRIITDVPSSSSVTEHLQPV 525
>At3g10010 hypothetical protein
Length = 1309
Score = 29.6 bits (65), Expect = 0.74
Identities = 16/60 (26%), Positives = 30/60 (49%)
Query: 1 MQTSSLFRRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLR 60
++T +L R+ R + +K +S + SS+ AT +S S++V P +T +R
Sbjct: 198 LETPTLKRKKIRPKVVREGKTKKASSKAGIKKSSIAATATKTSEESNYVRPKRLTRRSIR 257
>At5g55020 putative transcription factor MYB120 (MYB120)
Length = 523
Score = 28.9 bits (63), Expect = 1.3
Identities = 17/42 (40%), Positives = 24/42 (56%), Gaps = 1/42 (2%)
Query: 13 SLAARFRQNQKLTSFSSSSSS-SVLHDATTSSSSSSHVEPVH 53
S + F N +S+ +S+SS S LH T SS+S H PV+
Sbjct: 344 SSSPSFSLNPSSSSYPTSTSSPSFLHSHYTPSSTSFHTNPVY 385
>At5g33250 putative protein
Length = 279
Score = 28.5 bits (62), Expect = 1.6
Identities = 16/45 (35%), Positives = 24/45 (52%), Gaps = 4/45 (8%)
Query: 8 RRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPV 52
RR P LA +QK +S+ S++ D +SSS H++PV
Sbjct: 42 RRRVPQLAG----SQKYPYVGNSTVKSIITDVPSSSSVHEHLQPV 82
>At4g35900 bZIP transcription factor AtbZIP14
Length = 285
Score = 28.5 bits (62), Expect = 1.6
Identities = 18/47 (38%), Positives = 27/47 (57%)
Query: 20 QNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELE 66
+NQ LT SS SSSS +++SS+SSS P ++ R + +E
Sbjct: 18 KNQTLTKVSSISSSSPSSSSSSSSTSSSSPLPSQDSQAQKRSLVTME 64
>At1g56020 unknown protein
Length = 398
Score = 28.5 bits (62), Expect = 1.6
Identities = 27/100 (27%), Positives = 41/100 (41%), Gaps = 11/100 (11%)
Query: 1 MQTSSLFRRLAPSLAARFRQ----------NQKLTSFSSSSSSSVLHDATTSSSSSSHVE 50
M+ S LF AP R+R+ ++ S +SSSSS + T+S H
Sbjct: 120 MEISDLFSPKAPRCTTRWRELLGLKRLVNAKEQEESIKASSSSSSTNPKTSSFKQFLHRG 179
Query: 51 PVHMTENCLRRMKELEASESATAPKTLRLSVETGGCSGFQ 90
T KE + SES + + RLS+ + S +
Sbjct: 180 SKSSTAASSPLQKESDISESISVASS-RLSLSSSSSSSHE 218
>At3g61780 putative protein
Length = 1121
Score = 28.1 bits (61), Expect = 2.2
Identities = 18/69 (26%), Positives = 31/69 (44%), Gaps = 1/69 (1%)
Query: 17 RFRQNQKLTSFSS-SSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAPK 75
+F +N+ +TS S S L + V+ +H + RR +E+EA +
Sbjct: 310 KFDRNELMTSISKVKGSEKKLELVNSPHVELDFVDKIHEIKAMARRAREIEAGIELNEKQ 369
Query: 76 TLRLSVETG 84
L ++ ETG
Sbjct: 370 KLDVNKETG 378
>At5g52610 putative protein
Length = 351
Score = 27.7 bits (60), Expect = 2.8
Identities = 17/46 (36%), Positives = 24/46 (51%)
Query: 27 FSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESAT 72
F SSS S++ AT S+++SSH C+ M +L S AT
Sbjct: 68 FFSSSQPSLVTKATCSANNSSHTPDCVNGLICVEYMSQLWISNPAT 113
>At5g07390 respiratory burst oxidase protein A
Length = 902
Score = 27.7 bits (60), Expect = 2.8
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 5/71 (7%)
Query: 19 RQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAPKTLR 78
R N L +SS S+ + A++ SSSS+ P LRR K S + A K L+
Sbjct: 87 RSNSVLKRLASSVSTGITRVASSVSSSSARKPPRPQLAK-LRRSK----SRAELALKGLK 141
Query: 79 LSVETGGCSGF 89
+T G +G+
Sbjct: 142 FITKTDGVTGW 152
>At2g38440 unknown protein
Length = 1421
Score = 27.7 bits (60), Expect = 2.8
Identities = 24/92 (26%), Positives = 44/92 (47%), Gaps = 6/92 (6%)
Query: 3 TSSLFRRLAPSLAARFRQNQKLT---SFSSSSSSSVLHDATTSSSSSSHVEPVHMTEN-- 57
TSS L S++ + K++ + S SSS + T+ SS+ + P +++N
Sbjct: 482 TSSFKSELVDSMSHVTPEANKVSHDLNVQESVSSSNVDGQTSLSSNGTCSSPRPVSQNDQ 541
Query: 58 -CLRRMKELEASESATAPKTLRLSVETGGCSG 88
C ++ L + T+P+ +RL + GG G
Sbjct: 542 SCSLTVQSLASEVVETSPELVRLDLMKGGNDG 573
>At1g76950 zinc finger protein (PRAF1)
Length = 1103
Score = 27.7 bits (60), Expect = 2.8
Identities = 15/70 (21%), Positives = 34/70 (48%)
Query: 21 NQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAPKTLRLS 80
+++LTS S SSSS+ +S + +++P+ ++ + ++ +S A +
Sbjct: 146 SRELTSSSPSSSSASASRGHSSPGTPFNIDPITSPKSAEPEVPPTDSEKSHVALDNKNMQ 205
Query: 81 VETGGCSGFQ 90
+ G GF+
Sbjct: 206 TKVSGSDGFR 215
>At1g26790 H-protein promoter binding factor-2b, putative
Length = 396
Score = 27.7 bits (60), Expect = 2.8
Identities = 16/28 (57%), Positives = 19/28 (67%)
Query: 30 SSSSSVLHDATTSSSSSSHVEPVHMTEN 57
SSSS VL D+++SSSSSS HM N
Sbjct: 55 SSSSHVLPDSSSSSSSSSLSLRPHMMNN 82
>At5g66320 GATA-binding transcription factor-like protein
Length = 339
Score = 27.3 bits (59), Expect = 3.7
Identities = 17/41 (41%), Positives = 21/41 (50%)
Query: 11 APSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEP 51
A + + R R K+ S SSSSS +TSSSSS P
Sbjct: 166 AKARSKRNRNGLKVWSLGSSSSSGPSSSGSTSSSSSGPSSP 206
>At5g40630 unknown protein
Length = 165
Score = 27.3 bits (59), Expect = 3.7
Identities = 16/27 (59%), Positives = 20/27 (73%)
Query: 21 NQKLTSFSSSSSSSVLHDATTSSSSSS 47
N K +S SSSSSSS +++SSSSSS
Sbjct: 22 NNKRSSSSSSSSSSFSSLSSSSSSSSS 48
>At4g13110 putative protein
Length = 365
Score = 27.3 bits (59), Expect = 3.7
Identities = 13/21 (61%), Positives = 16/21 (75%)
Query: 27 FSSSSSSSVLHDATTSSSSSS 47
F SS S +H++TTSSSSSS
Sbjct: 51 FLIGSSDSPMHESTTSSSSSS 71
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.126 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,274,684
Number of Sequences: 26719
Number of extensions: 127774
Number of successful extensions: 1050
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 958
Number of HSP's gapped (non-prelim): 85
length of query: 161
length of database: 11,318,596
effective HSP length: 91
effective length of query: 70
effective length of database: 8,887,167
effective search space: 622101690
effective search space used: 622101690
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0010.21


