

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.19
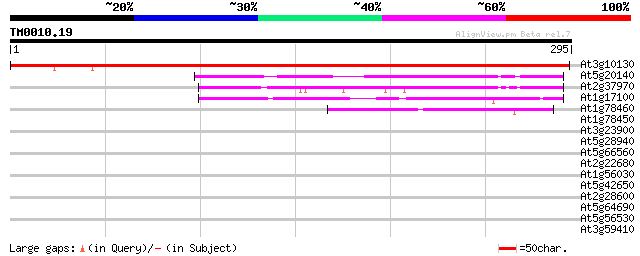
(295 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g10130 unknown protein 355 2e-98
At5g20140 unknown protein 103 1e-22
At2g37970 unknown protein (At2g37970) 100 9e-22
At1g17100 SOUL-like protein 62 3e-10
At1g78460 unknown protein 45 6e-05
At1g78450 unknown protein 39 0.003
At3g23900 unknown protein 33 0.14
At5g28940 32 0.31
At5g66560 unknown protein 30 2.0
At2g22680 copia-like retroelement pol polyprotein 29 3.5
At1g56030 hypothetical protein 29 3.5
At5g42650 allene oxide synthase (emb|CAA73184.1) 28 4.5
At2g28600 putative ATP-dependent RNA helicase 28 4.5
At5g64690 unknown protein 28 5.9
At5g56530 unknown protein (At5g56530) 28 5.9
At3g59410 protein kinase like 28 5.9
>At3g10130 unknown protein
Length = 309
Score = 355 bits (911), Expect = 2e-98
Identities = 182/308 (59%), Positives = 240/308 (77%), Gaps = 14/308 (4%)
Query: 1 MLPLCNASLSSQSSIRTSIPGR-------PNIAITNSASSSERISNRRR------TMSAV 47
M+ + ++S++S S+ +S P + IT S + ++ R +SA
Sbjct: 1 MMMISSSSITSSLSLLSSSPEKLPLINPIQRCPITYSGFRTASVNRAIRRQPQSPAVSAT 60
Query: 48 EARSSLILALASQANSLTQRLVVDVATETARYLFPKRLESRT-LEEALMAVPDLETVKFK 106
E+R SL+LALASQA+S++QRL+ D+A ETA+Y+FPKR +S T LEEA M+VPDLET+ F+
Sbjct: 61 ESRVSLVLALASQASSVSQRLLADLAMETAKYVFPKRFDSSTNLEEAFMSVPDLETMNFR 120
Query: 107 VLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTP 166
VL R D YEIR+VEPYFVAE MPG++GFD GAS+SFNVLAEYLFGKNT+KEKMEMTTP
Sbjct: 121 VLFRTDKYEIRQVEPYFVAETIMPGETGFDSYGASKSFNVLAEYLFGKNTIKEKMEMTTP 180
Query: 167 VYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSVRIKEIPRK 226
V T K QS G KM+MTTPV T+K +DQ++W MSF++PSKYG+NLPLPKD SV+I+++PRK
Sbjct: 181 VVTRKVQSVGEKMEMTTPVITSKAKDQNQWRMSFVMPSKYGSNLPLPKDPSVKIQQVPRK 240
Query: 227 IVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLPFQRRNEI 286
IVAVV+FSG+V DEE+++RE +LR AL++D +F+++ G S EVAQYNPPFTLPF RRNE+
Sbjct: 241 IVAVVAFSGYVTDEEIERRERELRRALQNDKKFRVRDGVSFEVAQYNPPFTLPFMRRNEV 300
Query: 287 ALEVERKD 294
+LEVE K+
Sbjct: 301 SLEVENKE 308
>At5g20140 unknown protein
Length = 378
Score = 103 bits (256), Expect = 1e-22
Identities = 69/196 (35%), Positives = 103/196 (52%), Gaps = 27/196 (13%)
Query: 98 PDLETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTL 157
PDLET K+++L R NYE+R EP+ V E SG S FN +A Y+FGKN+
Sbjct: 203 PDLETPKYQILKRTANYEVRNYEPFIVVETIGDKLSG------SSGFNNVAGYIFGKNST 256
Query: 158 KEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPS-KYGANLPLPKDS 216
EK+ MTTPV+T T + ++ ++PS K ++LP+P +
Sbjct: 257 MEKIPMTTPVFTQ----------------TTDTQLSSDVSVQIVIPSGKDLSSLPMPNEE 300
Query: 217 SVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPP- 275
V +K++ A V FSG ++ V+ +E +LR +L DG + KKG +A+YN P
Sbjct: 301 KVNLKKLEGGFAAAVKFSGKPTEDVVQAKENELRSSLSKDG-LRAKKGCM--LARYNDPG 357
Query: 276 FTLPFQRRNEIALEVE 291
T F RNE+ + +E
Sbjct: 358 RTWNFIMRNEVIIWLE 373
>At2g37970 unknown protein (At2g37970)
Length = 215
Score = 100 bits (249), Expect = 9e-22
Identities = 79/212 (37%), Positives = 108/212 (50%), Gaps = 26/212 (12%)
Query: 100 LETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQS-FNVLAEYL--FGK-- 154
+ET K+ V D YEIRE P AE T +F G F +LA+Y+ FGK
Sbjct: 10 VETPKYTVTKSGDGYEIREYPPAVAAEVTYDAS---EFKGDKDGGFQLLAKYIGVFGKPE 66
Query: 155 NTLKEKMEMTTPVYTSKNQS----------DGVKMDMTTPVYTAKMEDQDKW---TMSFI 201
N EK+ MT PV T + + + K++MT+PV T + + + TM F+
Sbjct: 67 NEKPEKIAMTAPVITKEGEKIAMTAPVITKESEKIEMTSPVVTKEGGGEGRKKLVTMQFL 126
Query: 202 LPSKY--GANLPLPKDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQF 259
LPS Y P P D V IKE + V+ FSG ++ V ++ KL L+ DG F
Sbjct: 127 LPSMYKKAEEAPRPTDERVVIKEEGGRKYGVIKFSGIASESVVSEKVKKLSSHLEKDG-F 185
Query: 260 KIKKGTSVEVAQYNPPFTLPFQRRNEIALEVE 291
KI G V +A+YNPP+TLP R NE+ + VE
Sbjct: 186 KI-TGDFV-LARYNPPWTLPPFRTNEVMIPVE 215
>At1g17100 SOUL-like protein
Length = 232
Score = 62.4 bits (150), Expect = 3e-10
Identities = 60/204 (29%), Positives = 93/204 (45%), Gaps = 31/204 (15%)
Query: 100 LETVKFKVLSRRDNYEIREVE-PYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLK 158
+E ++++ + YEIR +V+ +P S D +F L Y+ GKN
Sbjct: 45 IECPSYELVHSGNGYEIRRYNNTVWVSTEPIPDISLVD--ATRTAFFQLFAYIQGKNEYH 102
Query: 159 EKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDS-S 217
+K+EMT PV + + SDG + +T+SF +P K N P P S +
Sbjct: 103 QKIEMTAPVISQVSPSDGPFC-------------ESSFTVSFYVPKK---NQPDPAPSEN 146
Query: 218 VRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREAL----------KSDGQFKIKKGTSV 267
+ I++ + VAV FSGFV+D+ + ++ L +L KS + ++
Sbjct: 147 LHIQKWNSRYVAVRQFSGFVSDDSIGEQAAALDSSLKGTAWANAIAKSKEDGGVGSDSAY 206
Query: 268 EVAQYNPPFTLPFQRRNEIALEVE 291
VAQYN PF R NEI L E
Sbjct: 207 TVAQYNSPFEFS-GRVNEIWLPFE 229
>At1g78460 unknown protein
Length = 219
Score = 44.7 bits (104), Expect = 6e-05
Identities = 36/124 (29%), Positives = 54/124 (43%), Gaps = 7/124 (5%)
Query: 168 YTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSVRIKEIPRKI 227
Y + VKM+MT PV + +T+S LP K N P D + ++
Sbjct: 92 YIEGDNKSNVKMNMTAPVIAQATPGRSVYTVSLYLPKKNQQNPPQADD--LHVRSTKPTY 149
Query: 228 VAVVSFSGFVNDEEVKQRELKLREALK-SDGQFKIKKG----TSVEVAQYNPPFTLPFQR 282
VAV G+V++ K L E+L+ S+ I+K + +A YNPP +
Sbjct: 150 VAVRQIGGYVSNNVAKDEAAALMESLRDSNWILPIEKSKGKLPAYFLAVYNPPSHTTARV 209
Query: 283 RNEI 286
NEI
Sbjct: 210 INEI 213
>At1g78450 unknown protein
Length = 225
Score = 39.3 bits (90), Expect = 0.003
Identities = 44/204 (21%), Positives = 85/204 (41%), Gaps = 32/204 (15%)
Query: 101 ETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEK 160
E ++V+ + YEI V +T P + + +N L++Y+ G N ++
Sbjct: 35 ECPSYEVVHAGNGYEIHRYNTT-VWISTEPIQDISLNEASGNGWNQLSDYMNGNNDYHQR 93
Query: 161 MEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSVRI 220
+E+ P T +Q + + +SF +P + + P +++ +
Sbjct: 94 IEIALPYITQVSQ------------------NLSTFIVSFFVPKAFQPD--PPPGNNLHV 133
Query: 221 KEIPRKIVAVVSFSGFVNDEEVKQRELKLREAL----------KSDGQFKIKKGTSVEVA 270
+ + VAV SG+V D ++ ++ +L+ +L KS + + VA
Sbjct: 134 QRWDSRYVAVKQISGYVADHKIGKQVAELKASLQGTVWAKAIEKSRETGGVGSAWAYTVA 193
Query: 271 QYNPPFTLPFQRRNEIALEVERKD 294
Q++ PF QR NEI E +D
Sbjct: 194 QFSWPFQWS-QRVNEIWFPFEMED 216
>At3g23900 unknown protein
Length = 989
Score = 33.5 bits (75), Expect = 0.14
Identities = 34/136 (25%), Positives = 55/136 (40%), Gaps = 17/136 (12%)
Query: 169 TSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSV-----RIKEI 223
+S N S + + M V +M+ Q M + ++ AN S+ R EI
Sbjct: 428 SSNNSSSSLPLMMQQAVAMQQMQFQQAILMQQAVATQQAANRAATMKSATELAAARAAEI 487
Query: 224 PRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQ------YNPPFT 277
RK+ G NDE K+ + K R KS + + K + + + Y+PPF
Sbjct: 488 SRKLRP----DGVGNDE--KEADQKSRSPSKSPARSRSKSKSPISYRRRRRSPTYSPPFR 541
Query: 278 LPFQRRNEIALEVERK 293
P R+ L +R+
Sbjct: 542 RPRSHRSRSPLRYQRR 557
>At5g28940
Length = 485
Score = 32.3 bits (72), Expect = 0.31
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 5/60 (8%)
Query: 24 NIAITNSASSSERISNRRRTMSAVEARSSLILALASQANSLTQRLVVDVATETARYLFPK 83
+I + A SER SN T+S +EA + L ++ + +++ LV D AT +R K
Sbjct: 393 DIVMIEDADDSERASNHSHTLSNIEA-----MKLLTRTSEVSEMLVSDTATPKSRVKLEK 447
>At5g66560 unknown protein
Length = 668
Score = 29.6 bits (65), Expect = 2.0
Identities = 15/42 (35%), Positives = 24/42 (56%)
Query: 112 DNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFG 153
D+ E EVE + E P DF G+S+SF ++A++ +G
Sbjct: 85 DDKEEEEVEEQEIEENGYPHIKLEDFPGSSESFEMVAKFCYG 126
>At2g22680 copia-like retroelement pol polyprotein
Length = 683
Score = 28.9 bits (63), Expect = 3.5
Identities = 26/87 (29%), Positives = 37/87 (41%), Gaps = 8/87 (9%)
Query: 18 SIPGRPNIAITNSASSSERISNRRRTMSAVEARSSLILALASQANSLTQRLVVDVATETA 77
S+ GRP T S R M A E R+ L+ + NSLT + T +
Sbjct: 484 SLSGRPAWLGTGSI--------RLGDMYAEEERALLVEIKSPVNNSLTGSRSHKIMTVRS 535
Query: 78 RYLFPKRLESRTLEEALMAVPDLETVK 104
RY+ P E R E+ + +P TV+
Sbjct: 536 RYVDPTTQELRNPEDRALLIPTPLTVR 562
>At1g56030 hypothetical protein
Length = 371
Score = 28.9 bits (63), Expect = 3.5
Identities = 33/172 (19%), Positives = 72/172 (41%), Gaps = 21/172 (12%)
Query: 142 QSFNVLAEYLFGKNTLKEKME-----MTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKW 196
++ VLAE T + + + P+ S+ +D + + ++T A ++ D+
Sbjct: 67 KTVRVLAEVFNNMKTTRTTRKSIIQVLMNPILPSERSTDAMNLFLSTIEKLADLQFSDED 126
Query: 197 TMSFILPSKYGANLPLPKDSSVRIK-------EIPRKIVAVVSFSGFVNDEEVKQRELKL 249
+ S+ L + V +K + RKI VV + + E + + K
Sbjct: 127 FNQLFVSSRLDLQLENKYNDKVEVKLRKEAEDALARKIKEVVDLTERLLQVEALEHKHKA 186
Query: 250 REALKSDGQFKI-------KKGTSVEVAQYNPPFTLPFQRRNEIALEVERKD 294
+ L+++ + + + + +++N L +R +EIAL+ ERK+
Sbjct: 187 KLQLRTETETAVAIERDYMRWKAEIFESEFNNQLVL--RRESEIALDKERKE 236
>At5g42650 allene oxide synthase (emb|CAA73184.1)
Length = 518
Score = 28.5 bits (62), Expect = 4.5
Identities = 14/46 (30%), Positives = 22/46 (47%)
Query: 126 EATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTPVYTSK 171
E ++ GK+ F S +FN LA +G N K++ P +K
Sbjct: 200 ELSLKGKADFGGSSDGTAFNFLARAFYGTNPADTKLKADAPGLITK 245
>At2g28600 putative ATP-dependent RNA helicase
Length = 502
Score = 28.5 bits (62), Expect = 4.5
Identities = 28/126 (22%), Positives = 58/126 (45%), Gaps = 9/126 (7%)
Query: 4 LCNAS-LSSQSSIRTSIPGRPNIAITNSASSSERISNRRRTMSAVEARSSLILALASQAN 62
LC+ L++ SI+ +I + + N++ S+ I + + R ++ ++ASQ +
Sbjct: 282 LCSGGYLNAVKSIKQAISSKHQTIVFNNSFSASIIPAVQSFLGGSVNRVTVNESVASQGS 341
Query: 63 SLTQRLVVDVATETARYLFPKRLESR--------TLEEALMAVPDLETVKFKVLSRRDNY 114
+TQ + V + E F K L+S T EE+ + + +K +S +
Sbjct: 342 CITQTVSVCASEEKKLQKFAKHLDSSSSKLIYIVTKEESFKKIMAILKLKGISVSTSSDS 401
Query: 115 EIREVE 120
++ EV+
Sbjct: 402 KLSEVK 407
>At5g64690 unknown protein
Length = 344
Score = 28.1 bits (61), Expect = 5.9
Identities = 30/126 (23%), Positives = 56/126 (43%), Gaps = 14/126 (11%)
Query: 154 KNTLKEKMEMTTPVYTSKNQSDG----VKMDMTTPVYTAKMEDQDKWTMSFILPSKYGAN 209
K +++++ TP N G V M TT + + +D + F + ++
Sbjct: 87 KTVVEKEVTDLTPTKPDTNNKTGPSSEVSMFDTTALVASNAKDPED---IFDVQTRNDLE 143
Query: 210 LPLPKDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRE----LKLREALKSDGQFKIKKGT 265
+ +PKDS V+ E P+ A +FS EVK E K E +K + K+ + +
Sbjct: 144 VKIPKDSDVKTPETPKANEAEDNFS---ESWEVKFPEELEAKKTSEVVKVAEESKVPEVS 200
Query: 266 SVEVAQ 271
+ E+++
Sbjct: 201 APELSE 206
>At5g56530 unknown protein (At5g56530)
Length = 420
Score = 28.1 bits (61), Expect = 5.9
Identities = 19/68 (27%), Positives = 32/68 (46%)
Query: 149 EYLFGKNTLKEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGA 208
E LFG++ + EK + + T +GV + T PV K ED + + K
Sbjct: 95 ESLFGESKVSEKPKESVNPITQLWHQNGVCSEGTIPVRRTKKEDVLRASSVKRYGKKKHL 154
Query: 209 NLPLPKDS 216
++PLP+ +
Sbjct: 155 SVPLPRSA 162
>At3g59410 protein kinase like
Length = 1271
Score = 28.1 bits (61), Expect = 5.9
Identities = 15/38 (39%), Positives = 20/38 (52%)
Query: 73 ATETARYLFPKRLESRTLEEALMAVPDLETVKFKVLSR 110
ATE ++ FP R+ES L+ AV L + VL R
Sbjct: 751 ATELLKHAFPPRMESELLDSEFFAVLYLCQLHMNVLGR 788
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.129 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,772,695
Number of Sequences: 26719
Number of extensions: 229629
Number of successful extensions: 601
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 576
Number of HSP's gapped (non-prelim): 19
length of query: 295
length of database: 11,318,596
effective HSP length: 99
effective length of query: 196
effective length of database: 8,673,415
effective search space: 1699989340
effective search space used: 1699989340
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0010.19


