

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.15
(268 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
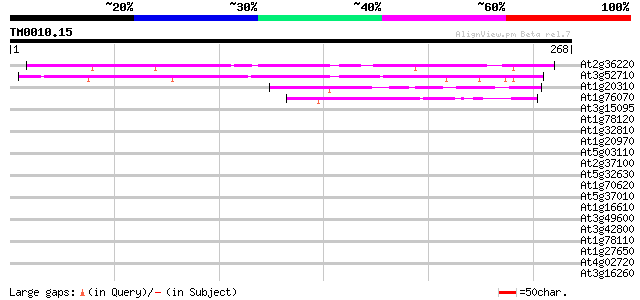
Score E
Sequences producing significant alignments: (bits) Value
At2g36220 unknown protein 166 9e-42
At3g52710 unknown protein 164 6e-41
At1g20310 unknown protein 43 2e-04
At1g76070 unknown protein 42 3e-04
At3g15095 unknown protein, 3' partial 38 0.005
At1g78120 hypothetical protein 36 0.019
At1g32810 hypothetical protein 35 0.032
At1g20970 hypothetical protein 33 0.16
At5g03110 putative protein 33 0.21
At2g37100 unknown protein 33 0.21
At5g32630 putative protein 32 0.27
At1g70620 unknown protein 32 0.27
At5g37010 serine-rich protein - like 32 0.35
At1g16610 arginine/serine-rich protein 32 0.46
At3g49600 putative protein 30 1.0
At3g42800 putative protein 30 1.0
At1g78110 unknown protein 30 1.0
At1g27650 U2 snRNP auxiliary factor, putative 30 1.0
At4g02720 unknown protein 30 1.3
At3g16260 unknown protein 30 1.8
>At2g36220 unknown protein
Length = 263
Score = 166 bits (421), Expect = 9e-42
Identities = 116/269 (43%), Positives = 150/269 (55%), Gaps = 36/269 (13%)
Query: 9 SACAGGSIDRKITCETLANKTDPPERHPDS-------PPESFWLSKDEEHDWFDRNALYE 61
S C GGS RKI CETLA+ + P + +S PPES++LS D + +W NA ++
Sbjct: 12 SVCIGGSDHRKIVCETLADDSTIPPYYNNSAVSPSDFPPESYFLSNDAQLEWLSDNAFFD 71
Query: 62 RKESTKG------STSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIK 115
RK+S KG S +S +SQRF L K+ IIGLPKPQK F +AK RR H N +
Sbjct: 72 RKDSQKGNSGILNSNPNSNPSSQRFLLKSKASIIGLPKPQKTCFNEAKQRR-HAGKNRVI 130
Query: 116 LFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSSIDAGEENPG 175
L KR S K+ S EPSSPKVSC+GRVRS+R+RSR R+ + S ++ P
Sbjct: 131 L--KRVGSRIKTDISLLEPSSPKVSCIGRVRSRRERSR----RMHRQKSSRVE-----PV 179
Query: 176 RIGRKHGFFERFRAIFR--SGRRKKAHRETDSAAEDSTVTNSVVKARDSTASVNDASFAE 233
+K GF FRAIFR G + + RET ++ ++ S + A SV D
Sbjct: 180 NRVKKPGFMASFRAIFRIKGGCKDVSARETHTSTRNTHDIRSRLPAEADEKSVFD----- 234
Query: 234 SISSEP--PGLGGMMRFASGRRSESWGVG 260
EP PGLGGM RFASGRR++ G G
Sbjct: 235 --GGEPVVPGLGGMTRFASGRRADLLGGG 261
>At3g52710 unknown protein
Length = 289
Score = 164 bits (414), Expect = 6e-41
Identities = 117/289 (40%), Positives = 155/289 (53%), Gaps = 45/289 (15%)
Query: 5 ETLVSACAGGSIDRKITCETLANKTDPPERHP----------DSPPESFWLSKDEEHDWF 54
+ + SAC GGS DRKI+CETLA+ + + D PPES+ LSK+ + +W
Sbjct: 3 QVVASACTGGS-DRKISCETLADDNEDSPHNSKIRPVSISAVDFPPESYSLSKEAQLEWL 61
Query: 55 DRNALYERKESTKGSTSSSTTN--------SQRFSLNLKSRIIGLPKPQKPSFTDAKNRR 106
+ NA +ERKES KG++S+ +N S R SL K+ II LPKPQK F +AK RR
Sbjct: 62 NDNAFFERKESQKGNSSAPISNPNTNPNSSSHRISLKSKASIIRLPKPQKTCFNEAKKRR 121
Query: 107 NHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSS 166
N + + + PKR S KS + +EP SPKVSC+GRVRSKRDRSR R++ +
Sbjct: 122 NCRIARTL-MIPKRIGSRLKSDPTLSEPCSPKVSCIGRVRSKRDRSR----RMQRQKSGR 176
Query: 167 IDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRETDSAA--EDSTVTNSVVKARDST- 223
++ ++ P + +K GFF FRAIFR+G K + + A D V+ V R ST
Sbjct: 177 TNSFKDKPVPV-KKPGFFASFRAIFRTGGGCKDLSASGAHAPRRDVVVSPPRVSVRRSTD 235
Query: 224 --------------ASVNDASFAESI-SSEP--PGLGGMMRFASGRRSE 255
N SI EP PGLGGM RF SGRR +
Sbjct: 236 IRGRLPPGDVGKSSPQRNSTGSRRSIDGGEPVLPGLGGMTRFTSGRRPD 284
>At1g20310 unknown protein
Length = 240
Score = 43.1 bits (100), Expect = 2e-04
Identities = 40/136 (29%), Positives = 61/136 (44%), Gaps = 28/136 (20%)
Query: 125 TKSVTSHAEPSSPKVSCMGRVRSKRDR------SRSRSLRLRNSRKSSIDAGEENPGRIG 178
TK AEP+SPKVSC+G+V+ R + ++L+ +S S + E+N
Sbjct: 66 TKYEAVFAEPTSPKVSCIGQVKLARPKCPEKKNKAPKNLKTASSLSSCVIKEEDN----- 120
Query: 179 RKHGFFERFRAIFRSGRRKKAHRETDSAAEDSTVTNSVVKARDSTASVNDASFAESISSE 238
G F + + IF R R+++S A + AR+ + DA A
Sbjct: 121 ---GSFSKLKRIF--SMRSYPSRKSNSTA------FAAAAAREHPIAEVDAVTA------ 163
Query: 239 PPGLGGMMRFASGRRS 254
P LG M +FAS R +
Sbjct: 164 APSLGAMKKFASSREA 179
>At1g76070 unknown protein
Length = 272
Score = 42.4 bits (98), Expect = 3e-04
Identities = 40/128 (31%), Positives = 61/128 (47%), Gaps = 28/128 (21%)
Query: 133 EPSSPKVSCMGRVR--------SKRDRSRSRSLRLRNSRKSSIDAGEENPGRIGRKHGFF 184
EP+SPKVSC+G+++ K++++ S + + +S E+ GR+ + F
Sbjct: 82 EPTSPKVSCIGQIKLGKSKCPTGKKNKAPSSLIPKISKTSTSSLTKEDEKGRLSKIKSIF 141
Query: 185 ERFRAIFRSGRRKKAHRETDSAAEDSTVTNSVVKARDSTASVNDASFAESISSEPPGLGG 244
A R+ R K+H SAA++ VT VV STA+V P LG
Sbjct: 142 SFSPASGRNTSR-KSHPTAVSAADEHPVT--VV----STAAV-------------PSLGQ 181
Query: 245 MMRFASGR 252
M +FAS R
Sbjct: 182 MKKFASSR 189
>At3g15095 unknown protein, 3' partial
Length = 417
Score = 38.1 bits (87), Expect = 0.005
Identities = 34/109 (31%), Positives = 49/109 (44%), Gaps = 21/109 (19%)
Query: 56 RNALYERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIK 115
R+A S + TS S N+ LN S + G +K S + +N N+ NNI
Sbjct: 53 RSACLTTSLSRRLRTSGSLKNASAGVLN--SPMFGANGGRKRSGSGYENSNNNNN-NNI- 108
Query: 116 LFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRK 164
EPSSPKV+C+G+VR K + + +R R+ RK
Sbjct: 109 -----------------EPSSPKVTCIGQVRVKTRKHVKKKMRARSRRK 140
>At1g78120 hypothetical protein
Length = 530
Score = 36.2 bits (82), Expect = 0.019
Identities = 32/88 (36%), Positives = 38/88 (42%), Gaps = 16/88 (18%)
Query: 28 KTDPPERHPDSPPESFWLSKDEEHDWFDRNALYERKESTKGSTSSSTTNSQRFSLN-LKS 86
K P E HP ES S D RK + GS S T S+RFSLN +
Sbjct: 82 KPKPDENHPRKSSESARKSSDSA-----------RKSISSGS---SRTESKRFSLNGVMG 127
Query: 87 RIIGLPKPQ-KPSFTDAKNRRNHKPCNN 113
II P+P K T K+R KP N+
Sbjct: 128 NIIVKPQPAVKTDVTQTKSRWEGKPVNH 155
>At1g32810 hypothetical protein
Length = 654
Score = 35.4 bits (80), Expect = 0.032
Identities = 31/120 (25%), Positives = 52/120 (42%), Gaps = 3/120 (2%)
Query: 30 DPPERHPDSPPESFWLSKDEEHDWFDRNALYERKESTKGSTSSSTT--NSQRFSLNLKSR 87
+P E+ P P S K +H+ +++ + +ES+ S +SS T N K
Sbjct: 208 EPSEKAPKHPKFSITSKKSMQHNRTSHSSVSKTRESSSSSKTSSATRINGGSSEAPSKHS 267
Query: 88 IIG-LPKPQKPSFTDAKNRRNHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVR 146
+ G PK +KP + ++ + + I L P S H SSP+V + R+R
Sbjct: 268 LSGTFPKNEKPGQSIFQSSTKNPVQSIISLAPNLSDEELALRLHHQLNSSPRVPRVPRMR 327
>At1g20970 hypothetical protein
Length = 1498
Score = 33.1 bits (74), Expect = 0.16
Identities = 37/155 (23%), Positives = 69/155 (43%), Gaps = 15/155 (9%)
Query: 3 QAETLVSACAGGSIDRKITC--ETLANKTDPPERHPDSPPESFWLSKDEEHDWFDRNALY 60
+A S G IDR++ C E KT P D PP+ +S+ EE D +R+++
Sbjct: 641 EAHVAPSIIEDGEIDREVNCGSEVNVTKTTPVAVREDIPPKE--VSEMEESDVKERSSIN 698
Query: 61 ERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIK--LFP 118
+E +T+S + + + +L+S+++ T AK+ + +P N +
Sbjct: 699 TDEEV---ATASVASEIKTCAQDLESKVV---TSTDTIHTGAKDCVDSQPAENKEGNKLI 752
Query: 119 KRSVSVTKSVTSHAE---PSSPKVSCMGRVRSKRD 150
K + + S+ + + S K+ C G V + D
Sbjct: 753 KNEIRLCTSLVENQKDGVDSIYKLLCSGNVVDRTD 787
>At5g03110 putative protein
Length = 283
Score = 32.7 bits (73), Expect = 0.21
Identities = 13/21 (61%), Positives = 17/21 (80%)
Query: 133 EPSSPKVSCMGRVRSKRDRSR 153
EPSSPKV+CMG+VR R + +
Sbjct: 64 EPSSPKVTCMGQVRVNRSKPK 84
>At2g37100 unknown protein
Length = 297
Score = 32.7 bits (73), Expect = 0.21
Identities = 18/53 (33%), Positives = 29/53 (53%), Gaps = 2/53 (3%)
Query: 99 FTDAKNRRNHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDR 151
F K+R + + +F +R + + T EP+SPKV+CMG+VR R +
Sbjct: 24 FLRTKSRSRSRSRSRRPIFFRRKNASAAAETQ--EPTSPKVTCMGQVRINRSK 74
>At5g32630 putative protein
Length = 856
Score = 32.3 bits (72), Expect = 0.27
Identities = 22/64 (34%), Positives = 29/64 (44%), Gaps = 4/64 (6%)
Query: 20 ITCETLANKTDPPERHPDSPPESFWLSKDEEHDWFDRNALYERKESTKGSTSSSTTNSQR 79
I E +N+ PP S KDE DWFD + L E + STS+S +S R
Sbjct: 379 INVECQSNRIKQKAATLGEPPNSTEKKKDEIKDWFDCSCL----EDLQISTSASIYSSSR 434
Query: 80 FSLN 83
S +
Sbjct: 435 LSFH 438
>At1g70620 unknown protein
Length = 897
Score = 32.3 bits (72), Expect = 0.27
Identities = 30/102 (29%), Positives = 43/102 (41%), Gaps = 23/102 (22%)
Query: 62 RKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKRS 121
RK S+ S SS + ++ S K+R P P K+RR H
Sbjct: 819 RKRSSPSSDESSDDSKRKSSSKRKNRS---PSP-------GKSRRRH------------- 855
Query: 122 VSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSR 163
VS + H++ + S + RSKR RSRSRS R+ +
Sbjct: 856 VSSRSPHSKHSQHKNTLYSSHDKSRSKRSRSRSRSPHRRHRK 897
>At5g37010 serine-rich protein - like
Length = 637
Score = 32.0 bits (71), Expect = 0.35
Identities = 42/166 (25%), Positives = 65/166 (38%), Gaps = 13/166 (7%)
Query: 93 KPQKPSFTDAKNRRNHKPCNNIKLFPK-RSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDR 151
KP+K F K+RR+H+ + P+ SVS KS S+ P + R R
Sbjct: 68 KPRKEVFV-IKHRRSHEKSSKTTTDPEDSSVSDVKSTGSNHNPVDVDAILIQCGRLSRSN 126
Query: 152 SRSRSLRLRNSRKSSIDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRETDSAAEDST 211
S + R + K S D ++N G + G + ++ HR+ E
Sbjct: 127 SAAAKTRRYSGSKRSFDF-DQNGGDADAEDGGED-------EEAERRIHRQRQRGGESPR 178
Query: 212 VTNSVVKARDSTASVNDASFAESISSEPPGLGGMMRF--ASGRRSE 255
+R+ S ++ S + S G GG R + GRRSE
Sbjct: 179 ERRRRTPSRERDDSKSNRSGSRERGSSGNG-GGSRRVSRSPGRRSE 223
>At1g16610 arginine/serine-rich protein
Length = 414
Score = 31.6 bits (70), Expect = 0.46
Identities = 22/62 (35%), Positives = 30/62 (47%), Gaps = 6/62 (9%)
Query: 118 PKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSSIDAGEENPGRI 177
P S S ++S + SSP S R RSRSRSL +S S+ +G +P R
Sbjct: 11 PSVSGSSSRSSSRSRSGSSPSRSI------SRSRSRSRSLSSSSSPSRSVSSGSRSPPRR 64
Query: 178 GR 179
G+
Sbjct: 65 GK 66
>At3g49600 putative protein
Length = 1672
Score = 30.4 bits (67), Expect = 1.0
Identities = 50/220 (22%), Positives = 75/220 (33%), Gaps = 29/220 (13%)
Query: 13 GGSIDRKITCETLANKTDPPERHPDSPPESFWLSKDEEHDWFDRNALYERKESTKGSTSS 72
GG D + K RH DS S+ +EH R+ K+ KG
Sbjct: 201 GGDDDDVDVVKRHKKKESKKRRHDDS-------SESDEHG---RDRRRRSKKKAKGRKQE 250
Query: 73 STTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKRSVSVTKSVTSHA 132
S ++S S D K R KP K +R SV+
Sbjct: 251 SESDSSSSDSESDS-----------DSDDGKKRGRKKPTKTTKKRSRRKRSVSSESEEVE 299
Query: 133 EPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSSI-----DAGEENPGRIGRKHGFFERF 187
S K+ + +RS S+ LR ++ +S D+ P K ++
Sbjct: 300 SDDSKKLRKSHKKSLPSNRSGSKELRDKHDEQSRAGRKRHDSDVSEPESEDNKQPLRKKE 359
Query: 188 RAIFRSGRRKKAHRETDSA--AEDSTVTNSVVKARDSTAS 225
A +R G+++K E A +D + ARDS S
Sbjct: 360 EA-YRGGQKQKRDDEDVEADHLKDRYTRDDKKAARDSDDS 398
>At3g42800 putative protein
Length = 341
Score = 30.4 bits (67), Expect = 1.0
Identities = 30/131 (22%), Positives = 57/131 (42%), Gaps = 14/131 (10%)
Query: 14 GSIDRKITCETLANKTDPPERHPDSPPESFWLSKDEEHDWFDRNALYERKESTKGSTSSS 73
GSI+ ++ ++ + P + D DEEH + R++ +T S SSS
Sbjct: 36 GSINENVSS---SSSSPSPNKKDDKLTTLRRAIMDEEHWLYARSS----TTTTNSSDSSS 88
Query: 74 TTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPK-------RSVSVTK 126
++S+ S K R+ L + K S + + + N+ +LF K ++V + +
Sbjct: 89 FSSSEAESYRTKRRLRKLAEQGKRSGDERQRTKRTVMDNDSRLFSKSDDDKKPKAVKIIE 148
Query: 127 SVTSHAEPSSP 137
+ +P SP
Sbjct: 149 ELKRSKQPVSP 159
>At1g78110 unknown protein
Length = 342
Score = 30.4 bits (67), Expect = 1.0
Identities = 24/105 (22%), Positives = 45/105 (42%), Gaps = 16/105 (15%)
Query: 67 KGSTSSSTTNSQRFSLNLKSRIIGLPKP----QKPSFTDAKNRRNHKPC----------- 111
+GS+SS + ++ + PKP +PS + R +H+
Sbjct: 9 RGSSSSGYSADLLVCFPSRTHLALTPKPICSPSRPSDSSTNRRPHHRRQLSKLSGGGGGG 68
Query: 112 -NNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSR 155
+ L+ K++ S AEP+SPKV+C G+++ + + R
Sbjct: 69 HGSPVLWAKQASSKNMGGDEIAEPTSPKVTCAGQIKVRPSKCGGR 113
>At1g27650 U2 snRNP auxiliary factor, putative
Length = 296
Score = 30.4 bits (67), Expect = 1.0
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 6/66 (9%)
Query: 146 RSKRDRSRSRSLRLRNSRKSSIDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRETDS 205
R R RSRSRS+ RN R D +P R+ +R R +R G K++ ++
Sbjct: 194 RGSRSRSRSRSISPRNKR----DNDRRDPSH--REFSHRDRDREFYRHGSGKRSSERSER 247
Query: 206 AAEDST 211
D +
Sbjct: 248 QERDGS 253
>At4g02720 unknown protein
Length = 422
Score = 30.0 bits (66), Expect = 1.3
Identities = 41/184 (22%), Positives = 70/184 (37%), Gaps = 17/184 (9%)
Query: 38 SPPESFWLSKDEEHDWFDRNALYERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKP 97
SPP DE D ER +++KG ++S+ S L+SR +K
Sbjct: 65 SPPRDQNEDSDENADEIQDKNGGERDDNSKGKERKGKSDSESESDGLRSR------KRKS 118
Query: 98 SFTDAKNRRNHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSL 157
+ +K RR ++ + S S + + SS + ++ RS R
Sbjct: 119 KSSRSKRRRKRSYDSDSESEGSESDSEEEDRRRRRKSSSKRKKSRSSRSFRKKRSHRRKT 178
Query: 158 RLRNSRKSSID----------AGEENPGRIGRKHGFFERFRAIFRS-GRRKKAHRETDSA 206
+ +S +SS + +GEE + K + RS G + K+ ++D
Sbjct: 179 KYSDSDESSDEDSKAEISASSSGEEEDTKSKSKRRKKSSDSSSKRSKGEKTKSGSDSDGT 238
Query: 207 AEDS 210
EDS
Sbjct: 239 EEDS 242
Score = 29.3 bits (64), Expect = 2.3
Identities = 24/96 (25%), Positives = 41/96 (42%), Gaps = 10/96 (10%)
Query: 151 RSRSRSLRLRNSRKSSIDAGEENPG----------RIGRKHGFFERFRAIFRSGRRKKAH 200
+ +S+S R + RK S D+ E+ G R RK + RS R+K++H
Sbjct: 115 KRKSKSSRSKRRRKRSYDSDSESEGSESDSEEEDRRRRRKSSSKRKKSRSSRSFRKKRSH 174
Query: 201 RETDSAAEDSTVTNSVVKARDSTASVNDASFAESIS 236
R ++ ++ KA S +S + +S S
Sbjct: 175 RRKTKYSDSDESSDEDSKAEISASSSGEEEDTKSKS 210
>At3g16260 unknown protein
Length = 942
Score = 29.6 bits (65), Expect = 1.8
Identities = 25/81 (30%), Positives = 28/81 (33%), Gaps = 8/81 (9%)
Query: 34 RHPDSPPESFWLSKDEEHDWFDRNALYERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPK 93
R SPP W S F+ R K S +S F N K R GL K
Sbjct: 56 RSGPSPPRRKWSS-------FEEQKRKGRSPMEKDKAISFNHSSDSFEFN-KRRAEGLDK 107
Query: 94 PQKPSFTDAKNRRNHKPCNNI 114
KP +N R P N I
Sbjct: 108 VDKPKKNLKRNTRTLNPTNTI 128
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.126 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,140,251
Number of Sequences: 26719
Number of extensions: 255795
Number of successful extensions: 855
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 809
Number of HSP's gapped (non-prelim): 71
length of query: 268
length of database: 11,318,596
effective HSP length: 98
effective length of query: 170
effective length of database: 8,700,134
effective search space: 1479022780
effective search space used: 1479022780
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0010.15


