

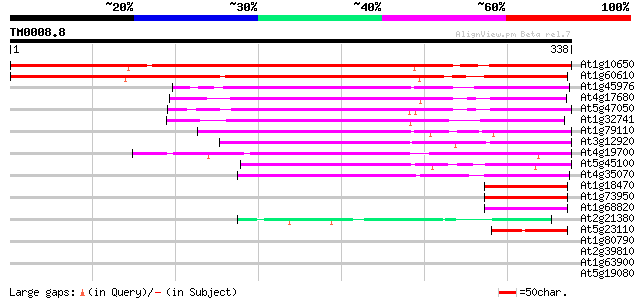
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.8
(338 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g10650 similar to putative inhibitor of apoptosis gi|3924605;... 382 e-106
At1g60610 unknown protein 363 e-101
At1g45976 S-ribonuclease binding like protein (SBP1) 164 8e-41
At4g17680 unknown protein 145 2e-35
At5g47050 unknown protein 135 3e-32
At1g32741 unknown protein 133 2e-31
At1g79110 unknown protein 122 3e-28
At3g12920 unknown protein 119 2e-27
At4g19700 unknown protein 109 2e-24
At5g45100 unknown protein 105 5e-23
At4g35070 unknown protein 102 3e-22
At1g18470 unknown protein 52 4e-07
At1g73950 unknown protein (At1g73950) 49 4e-06
At1g68820 unknown protein (At1g68820) 45 6e-05
At2g21380 putative kinesin heavy chain 44 1e-04
At5g23110 putative protein 42 5e-04
At1g80790 hypothetical protein 39 0.005
At2g39810 unknown protein 37 0.012
At1g63900 putative RING zinc finger protein 37 0.015
At5g19080 unknown protein 37 0.020
>At1g10650 similar to putative inhibitor of apoptosis gi|3924605;
similar to ESTs gb|AI994578.1, gb|T04171, gb|AA728525
Length = 339
Score = 382 bits (980), Expect = e-106
Identities = 197/351 (56%), Positives = 254/351 (72%), Gaps = 25/351 (7%)
Query: 1 MFGGNNGNRLLPVFLDENQFQCQTNA-TNQLQLFGNLQAGRNVDPVSYIGNEHISSMIQP 59
M GNNGN PVF++ENQ Q QTN +NQL L GN+ G VDPV+Y N+++ MI+P
Sbjct: 1 MLSGNNGNTAPPVFMNENQLQYQTNLRSNQLHLLGNMGGGCTVDPVNYFANDNLVPMIRP 60
Query: 60 NKRSREMEDIS-----KKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERN 114
NKR RE E IS ++Q+LQ+SLNYN ++ +P N +STGLRLSYDDDE N
Sbjct: 61 NKRGREAESISNNIQRQQQKLQMSLNYNY--NNTSVREEVPKENLVSTGLRLSYDDDEHN 118
Query: 115 SSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMAS 174
SSVTSASGS+ A I SL D++R +L RQ++E D +IK+Q Q+++GVRDMKQ+H+AS
Sbjct: 119 SSVTSASGSILAASPIFQSLDDSLRIDLHRQKDEFDQFIKIQAAQMAKGVRDMKQRHIAS 178
Query: 175 LLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRN 234
LT++EKG+ KKL+EK+ EI +MN+KN+EL ERIKQVA+EAQNWHYRAKYNESVVN L+
Sbjct: 179 FLTTLEKGVSKKLQEKDHEINDMNKKNKELVERIKQVAMEAQNWHYRAKYNESVVNVLKA 238
Query: 235 NLQQAISH------GVEQGKEGFGDSEVDDAA-SYIDPNNFLSIPGAPMKSIHPRYQGME 287
NLQQA+SH +QGKEGFGDSE+DDAA SYIDPNN IH R
Sbjct: 239 NLQQAMSHNNSVIAAADQGKEGFGDSEIDDAASSYIDPNN----NNNNNMGIHQR----- 289
Query: 288 NLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
+ C+ C K+VS+L++PCRHLSLCK+CD F +CPVC+ +K++ V+V+ S
Sbjct: 290 -MRCKMCNVKEVSVLIVPCRHLSLCKECDVFTKICPVCKSLKSSCVQVFFS 339
>At1g60610 unknown protein
Length = 340
Score = 363 bits (933), Expect = e-101
Identities = 189/354 (53%), Positives = 257/354 (72%), Gaps = 34/354 (9%)
Query: 1 MFGGNNGNRLLPVFLDENQFQCQTNAT-NQLQLFGNLQAGRNVDPVSYIGNEHISSMIQP 59
M GGNN N + V ++++QF+ Q+N + NQL L G ++AG +DPV+Y N++++ M++
Sbjct: 1 MLGGNNDNPVPQVLMNDSQFRYQSNTSLNQLHLLGTMRAGCTIDPVNYFANDNLAPMMRL 60
Query: 60 NK-RSREMED---ISKKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERNS 115
N R RE E+ + ++Q+LQISLNYN ++ +P N +STGLRLSYDDDERNS
Sbjct: 61 NSTRGRETENNNIMQRQQKLQISLNYNYNNNNTVVQDEVPKQNLVSTGLRLSYDDDERNS 120
Query: 116 SVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASL 175
SVTSA+GS+T P + SLGDNIR +L+RQ +ELD +IK + +Q+++GVRD+KQ+H+ S
Sbjct: 121 SVTSANGSITTP--VYQSLGDNIRLDLNRQNDELDQFIKFRADQMAKGVRDIKQRHVTSF 178
Query: 176 LTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNN 235
+T++EK + KKL+EK+ EIE+MN+KNREL ++IKQVAVEAQNWHY+AKYNESVVNAL+ N
Sbjct: 179 VTALEKDVSKKLQEKDHEIESMNKKNRELVDKIKQVAVEAQNWHYKAKYNESVVNALKVN 238
Query: 236 LQQAISHGVE------------QGKEGFGDSEVDD-AASYIDPNNFLSIPGAPMKSIHPR 282
LQQ +SHG + Q KEGFGDSE+DD AASY N+L+IPG P
Sbjct: 239 LQQVMSHGNDNNAVGGGVADHHQMKEGFGDSEIDDEAASY----NYLNIPGMP------- 287
Query: 283 YQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVY 336
+ C+ C K+VS+LL+PCRHLSLCKDCD F VCPVCQ +KT+SV+V+
Sbjct: 288 ---STGMRCKLCNVKNVSVLLVPCRHLSLCKDCDVFTGVCPVCQSLKTSSVQVF 338
>At1g45976 S-ribonuclease binding like protein (SBP1)
Length = 325
Score = 164 bits (414), Expect = 8e-41
Identities = 89/239 (37%), Positives = 142/239 (59%), Gaps = 22/239 (9%)
Query: 99 ALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKE 158
++STGL LS D N+ V S+ GS +++ +GD+I EL RQ ++D ++K+Q +
Sbjct: 109 SVSTGLGLSLD----NARVASSDGS-----ALLSLVGDDIDRELQRQDADIDRFLKIQGD 159
Query: 159 QLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNW 218
QL + D ++ ++ +E+ +V+KLREK+EE+E +NRKN+EL R++Q+ +EA+ W
Sbjct: 160 QLRHAILDKIKRGQQKTVSLMEEKVVQKLREKDEELERINRKNKELEVRMEQLTMEAEAW 219
Query: 219 HYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSEVDDAASYIDPNNFLSIPGAPMKS 278
RAKYNE+++ AL NL +A EG GDSEVDD AS + +
Sbjct: 220 QQRAKYNENMIAALNYNLDRAQGR-PRDSIEGCGDSEVDDTASCFNGRD----------- 267
Query: 279 IHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYL 337
+ + CR C +++ MLL+PC H+ LCK+C+ ++ CP+CQ K +EVY+
Sbjct: 268 -NSNNNTKTMMMCRFCGVREMCMLLLPCNHMCLCKECERKLSSCPLCQSSKFLGMEVYM 325
>At4g17680 unknown protein
Length = 314
Score = 145 bits (367), Expect = 2e-35
Identities = 85/248 (34%), Positives = 136/248 (54%), Gaps = 38/248 (15%)
Query: 97 PNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQ 156
PN +STGLRL +D + S SL ++ ++ RQ++ELD +I+ Q
Sbjct: 94 PNVVSTGLRLFHDQSQNQQQFFS-------------SLPGDVTGKIKRQRDELDRFIQTQ 140
Query: 157 KEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQ 216
E+L R + D +++ LL + E+ + +KLR+KE E+E R++ EL R+ + EA+
Sbjct: 141 GEELRRTLADNRERRYVELLCAAEEIVGRKLRKKEAELEKATRRHAELEARVAHIVEEAR 200
Query: 217 NWHYRAKYNESVVNALRNNLQQAISHGVEQ-------GKEGFGDSEVDDAAS-YIDPNNF 268
NW RA E+ V++L +LQQAI++ ++ G++G E +DA S Y+DP
Sbjct: 201 NWQLRAATREAEVSSLHAHLQQAIANRLDTAAKQSTFGEDGGDAEEAEDAESVYVDPER- 259
Query: 269 LSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCD-GFINVCPVCQM 327
++ I P SCR C+ K +++ +PC+HL LC CD G + VCP+C
Sbjct: 260 -------IELIGP--------SCRICRRKSATVMALPCQHLILCNGCDVGAVRVCPICLA 304
Query: 328 IKTASVEV 335
+KT+ VEV
Sbjct: 305 VKTSGVEV 312
>At5g47050 unknown protein
Length = 300
Score = 135 bits (340), Expect = 3e-32
Identities = 85/253 (33%), Positives = 134/253 (52%), Gaps = 37/253 (14%)
Query: 96 NPNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKL 155
NPN +STGLRLS R S ++ P + ++ E+ Q +EL+ ++++
Sbjct: 75 NPNVVSTGLRLS-----REQSQNQEQRFLSFP------ITGDVAGEIKSQTDELNRFLQI 123
Query: 156 QKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEA 215
Q EQL R + + +++ LL + E+ + ++LREKE EIE R++ EL R Q+ EA
Sbjct: 124 QGEQLKRMLAENSERNYRELLRTTEESVRRRLREKEAEIEKATRRHVELEARATQIETEA 183
Query: 216 QNWHYRAKYNESVVNALRNNLQQA--ISHG------VE-QGKEGFGDSEVDDAAS-YIDP 265
+ W RA E+ +L+ L QA ++HG VE Q G E +DA S Y+DP
Sbjct: 184 RAWQMRAAAREAEATSLQAQLHQAVVVAHGGGVITTVEPQSGSVDGVDEAEDAESAYVDP 243
Query: 266 NNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVC 325
+ ++ I P CR C+ + ++L +PCRHL +C +CDG + +CP+C
Sbjct: 244 DR--------VEMIGP--------GCRICRRRSATVLALPCRHLVMCTECDGSVRICPLC 287
Query: 326 QMIKTASVEVYLS 338
K +SVEV+ S
Sbjct: 288 LSTKNSSVEVFYS 300
>At1g32741 unknown protein
Length = 312
Score = 133 bits (334), Expect = 2e-31
Identities = 81/254 (31%), Positives = 130/254 (50%), Gaps = 47/254 (18%)
Query: 95 PNPNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNI-RTELDRQQEELDHYI 153
P N + TGLRL +D+ I L +++ ++RQ EELD ++
Sbjct: 88 PPSNMVHTGLRLFSGEDQAQK---------------ISHLSEDVFAAHINRQSEELDEFL 132
Query: 154 KLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAV 213
Q E+L R + + ++ H +LL ++E+ +V+KLREKE EIE R++ EL R Q+
Sbjct: 133 HAQAEELRRTLAEKRKMHYKALLGAVEESLVRKLREKEVEIERATRRHNELVARDSQLRA 192
Query: 214 EAQNWHYRAKYNESVVNALRNNLQQAI------------SHGVEQGKEGFGDSEVDDAAS 261
E Q W RAK +E +L++ LQQA+ S E+G S VDDA S
Sbjct: 193 EVQVWQERAKAHEDAAASLQSQLQQAVNQCAGGCVSAQDSRAAEEGLLCTTISGVDDAES 252
Query: 262 -YIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFIN 320
Y+DP + ++ +C+AC+ ++ +++++PCRHLS+C CD
Sbjct: 253 VYVDP------------------ERVKRPNCKACREREATVVVLPCRHLSICPGCDRTAL 294
Query: 321 VCPVCQMIKTASVE 334
CP+C ++ +SVE
Sbjct: 295 ACPLCLTLRNSSVE 308
>At1g79110 unknown protein
Length = 355
Score = 122 bits (306), Expect = 3e-28
Identities = 75/240 (31%), Positives = 127/240 (52%), Gaps = 21/240 (8%)
Query: 114 NSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMA 173
+S V +A+ + T L +I + +++QQ E+D ++ L E++ + + +++
Sbjct: 121 SSCVNAATTTTTTTLFSFLGQDIDISSHMNQQQHEIDRFVSLHMERVKYEIEEKRKRQAR 180
Query: 174 SLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALR 233
+++ +IE+G+VK+LR KEEE E + + N L ER+K +++E Q W A+ NE+ N LR
Sbjct: 181 TIMEAIEQGLVKRLRVKEEERERIGKVNHALEERVKSLSIENQIWRDLAQTNEATANHLR 240
Query: 234 NNLQQAISHGVEQGKEGFG----DSEVDDAASYIDPNNFLSIPGAPMKSIHPRYQGMENL 289
NL+ ++ V+ G G +E DDA S S G +++ R G+E
Sbjct: 241 TNLEHVLAQ-VKDVSRGAGLEKNMNEEDDAESCCGS----SCGGGGEETVRRRV-GLERE 294
Query: 290 S-----------CRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
+ CR C ++ +LL+PCRHL LC C ++ CP+C K ASV V +S
Sbjct: 295 AQDKAERRRRRMCRNCGEEESCVLLLPCRHLCLCGVCGSSVHTCPICTSPKNASVHVNMS 354
>At3g12920 unknown protein
Length = 335
Score = 119 bits (298), Expect = 2e-27
Identities = 68/219 (31%), Positives = 118/219 (53%), Gaps = 10/219 (4%)
Query: 127 PPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKK 186
P ++ LG ++ + + + ++D I E++ + + ++ ++ ++E+G++K
Sbjct: 119 PTDPLMFLGQDLSSNVQQHHFDIDRLISNHVERMRMEIEEKRKTQGRRIVEAVEQGLMKT 178
Query: 187 LREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQ 246
LR K++EI ++ + N L E++K + VE Q W A+ NE+ VNALR+NLQQ ++ VE+
Sbjct: 179 LRAKDDEINHIGKLNLFLEEKVKSLCVENQIWRDVAQSNEATVNALRSNLQQVLA-AVER 237
Query: 247 GKEGFGDSEVDDAASYIDPNN-------FLSIPGAPMKSIHPRYQGMENLSCRACKAKDV 299
+ + DDA S N+ + G + GM CR+C +
Sbjct: 238 NRWEEPPTVADDAQSCCGSNDEGDSEEERWKLAGEAQDTKKMCRVGMS--MCRSCGKGEA 295
Query: 300 SMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
S+LL+PCRH+ LC C +N CP+C+ KTAS+ V LS
Sbjct: 296 SVLLLPCRHMCLCSVCGSSLNTCPICKSPKTASLHVNLS 334
>At4g19700 unknown protein
Length = 304
Score = 109 bits (272), Expect = 2e-24
Identities = 77/268 (28%), Positives = 126/268 (46%), Gaps = 21/268 (7%)
Query: 75 LQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERNSSVT--SASGSMTAPPSIIL 132
L N+N + ++ ++P LST + + + N+ V S + ++P I +
Sbjct: 53 LAAKANFNKAESGLSYNFTVP---PLSTKRQRDFQFSDSNAPVKRRSVAFDSSSPSLINV 109
Query: 133 SLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEE 192
L I+ + QQ E+D ++ Q E+L + +Q L ++++ I KKL+EK++
Sbjct: 110 ELVSQIQNQ---QQSEIDRFVAQQTEKLRIEIEARQQTQTRMLASAVQNVIAKKLKEKDD 166
Query: 193 EIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFG 252
EI + N L ER+K + VE Q W A+ NE+ N LR NL Q ++
Sbjct: 167 EIVRIRNLNWVLQERVKSLYVENQIWRDIAQTNEANANTLRTNLDQVLA----------- 215
Query: 253 DSEVDDAASYIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLC 312
E AS + ++ S G+ C+ C ++ S+L++PCRHL LC
Sbjct: 216 QLETFPTASAVVEDDAESSCGSCCGDGGGEAVTAVGGGCKRCGEREASVLVLPCRHLCLC 275
Query: 313 KDCDG--FINVCPVCQMIKTASVEVYLS 338
C G + CPVC M+ ASV V +S
Sbjct: 276 TVCGGSALLRTCPVCDMVMNASVHVNMS 303
>At5g45100 unknown protein
Length = 294
Score = 105 bits (261), Expect = 5e-23
Identities = 66/204 (32%), Positives = 103/204 (50%), Gaps = 19/204 (9%)
Query: 140 TELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNR 199
+++ +Q E+D ++ Q E L + ++ L ++++ I+KKL+ K+EEI M +
Sbjct: 104 SQIQQQNSEIDRFVAQQTETLRIELEARQRTQTRMLASAVQNAILKKLKAKDEEIIRMGK 163
Query: 200 KNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGD---SEV 256
N L ER+K + VE Q W A+ NE+ N LR+NL+Q ++ + F E
Sbjct: 164 LNWVLQERVKNLYVENQIWRDLAQTNEATANNLRSNLEQVLAQ--VDDLDAFRRPLVEEA 221
Query: 257 DDAASYIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDC- 315
DDA S S G + ++ N C+ C S+L++PCRHL LC C
Sbjct: 222 DDAESSCG-----SCDGGDVTAV-------VNGGCKRCGELTASVLVLPCRHLCLCTVCG 269
Query: 316 -DGFINVCPVCQMIKTASVEVYLS 338
+ CPVC M+ TASV V +S
Sbjct: 270 SSALLRTCPVCDMVMTASVHVNMS 293
>At4g35070 unknown protein
Length = 265
Score = 102 bits (254), Expect = 3e-22
Identities = 54/201 (26%), Positives = 103/201 (50%), Gaps = 11/201 (5%)
Query: 138 IRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENM 197
+ ++++Q++E+D +IK+Q E+L +++ +++ M +L +E + + +KEEE+
Sbjct: 74 LAAQMEKQKQEIDQFIKIQNERLRYVLQEQRKREMEMILRKMESKALLLMSQKEEEMSKA 133
Query: 198 NRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSEVD 257
KN EL + ++++ +E Q W A+ NE++V L L+Q G++EV+
Sbjct: 134 LNKNMELEDLLRKMEMENQTWQRMARENEAIVQTLNTTLEQVRERAAT--CYDAGEAEVE 191
Query: 258 DAASYI-DPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCD 316
D S+ + S+P MK M + C +L +PCRHL C DC+
Sbjct: 192 DEGSFCGGEGDGNSLPAKKMK--------MSSCCCNCGSNGVTRVLFLPCRHLCCCMDCE 243
Query: 317 GFINVCPVCQMIKTASVEVYL 337
+ +CP+C K + +E +
Sbjct: 244 EGLLLCPICNTPKKSRIEALI 264
>At1g18470 unknown protein
Length = 467
Score = 52.4 bits (124), Expect = 4e-07
Identities = 20/50 (40%), Positives = 30/50 (60%)
Query: 287 ENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVY 336
E + CR C KD+S++L+PCRH LC+ C CP+C++ + VY
Sbjct: 416 EKVLCRVCFEKDISLVLLPCRHRVLCRTCADKCTTCPICRIDIEKRLSVY 465
>At1g73950 unknown protein (At1g73950)
Length = 466
Score = 48.9 bits (115), Expect = 4e-06
Identities = 18/50 (36%), Positives = 30/50 (60%)
Query: 287 ENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVY 336
E + CR C +++S++L+PCRH LC++C CP C++ + VY
Sbjct: 415 EKVLCRVCFEREISVVLLPCRHRVLCRNCSDKCKKCPFCRITIEERLPVY 464
>At1g68820 unknown protein (At1g68820)
Length = 468
Score = 45.1 bits (105), Expect = 6e-05
Identities = 17/50 (34%), Positives = 28/50 (56%)
Query: 287 ENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVY 336
E + CR C ++++L+PCRH LC C CP+C+++ + VY
Sbjct: 417 EKILCRVCFEDPINVVLLPCRHHVLCSTCCEKCKKCPICRVLIEERMPVY 466
>At2g21380 putative kinesin heavy chain
Length = 1058
Score = 43.9 bits (102), Expect = 1e-04
Identities = 46/196 (23%), Positives = 78/196 (39%), Gaps = 39/196 (19%)
Query: 138 IRTELDRQQEELDHYIKLQKEQLSRGVRDM-----KQKHMASLLTSIEKGIVKKLREKEE 192
I E ++ EE K ++E L + +M K K S SI+K + +++E
Sbjct: 883 IEEEFRKKAEEA----KRREEALENDLANMWVLVAKLKKANSGALSIQKSDEAEPAKEDE 938
Query: 193 --EIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEG 250
E++N N +N L ER + N H ++ L + +++ KE
Sbjct: 939 VTELDNKNEQNAILKER------QLVNGHEEVIVAKAEETPKEEPLVARLKARMQEMKEK 992
Query: 251 FGDSEVDDAASYIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLS 310
S+ AA+ D N+ + C+ C + +L+PCRH
Sbjct: 993 EMKSQAAAAAN-ADANSHI---------------------CKVCFESPTATILLPCRHFC 1030
Query: 311 LCKDCDGFINVCPVCQ 326
LCK C + CP+C+
Sbjct: 1031 LCKSCSLACSECPICR 1046
>At5g23110 putative protein
Length = 4706
Score = 42.0 bits (97), Expect = 5e-04
Identities = 13/46 (28%), Positives = 29/46 (62%), Gaps = 1/46 (2%)
Query: 291 CRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVY 336
C+ C+ K+V + ++PC H+ LC+ C ++ CP C++ ++ ++
Sbjct: 4660 CQICQTKEVEVTIVPCGHV-LCRHCSTSVSRCPFCRLQVNRTIRIF 4704
>At1g80790 hypothetical protein
Length = 634
Score = 38.5 bits (88), Expect = 0.005
Identities = 29/133 (21%), Positives = 66/133 (48%), Gaps = 16/133 (12%)
Query: 90 WSASIPNPNALSTGLRLSYDDDERNSSVTSASGSMTA-----PPSIILSLGDNIRTELDR 144
WS + AL+ R D+D++ S V ++S + + +L L D + +
Sbjct: 337 WSKQLDKKQALTELERQKLDEDKKKSDVMNSSLQLASLEQKKTDDRVLRLVDEHKR---K 393
Query: 145 QQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNREL 204
++E L+ ++L+KE S+ M+ + + KG +K ++ ++E+ E + +K +++
Sbjct: 394 KEETLNKILQLEKELDSKQKLQMEIQEL--------KGKLKVMKHEDEDDEGIKKKMKKM 445
Query: 205 AERIKQVAVEAQN 217
E +++ E Q+
Sbjct: 446 KEELEEKCSELQD 458
>At2g39810 unknown protein
Length = 927
Score = 37.4 bits (85), Expect = 0.012
Identities = 22/78 (28%), Positives = 37/78 (47%), Gaps = 20/78 (25%)
Query: 269 LSIPGAPMKSIHPRYQGMENLS---------------CRACK-----AKDVSMLLIPCRH 308
+S+P P S P + +++L+ CRA + + V+ +L PC H
Sbjct: 16 ISLPTQPNYSSKPVQEALKHLASINLRELCNEAKVERCRATRDLASCGRFVNYVLNPCGH 75
Query: 309 LSLCKDCDGFINVCPVCQ 326
SLC +C +VCP+C+
Sbjct: 76 ASLCTECCQRCDVCPICR 93
>At1g63900 putative RING zinc finger protein
Length = 115
Score = 37.0 bits (84), Expect = 0.015
Identities = 12/46 (26%), Positives = 23/46 (49%)
Query: 291 CRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVY 336
C C ++ + + +PC H+ C C + CP+C+ +V+ Y
Sbjct: 68 CVICLEQEYNAVFVPCGHMCCCTACSSHLTSCPLCRRRIDLAVKTY 113
>At5g19080 unknown protein
Length = 378
Score = 36.6 bits (83), Expect = 0.020
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Query: 291 CRACKAKDVSMLLIPCRHLSLCKDCDGFI----NVCPVCQ 326
C C + ++PCRHL LC DC + N CP+C+
Sbjct: 321 CVICLTEPKDTAVMPCRHLCLCSDCAEELRFQTNKCPICR 360
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.131 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,661,575
Number of Sequences: 26719
Number of extensions: 328347
Number of successful extensions: 1725
Number of sequences better than 10.0: 158
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 117
Number of HSP's that attempted gapping in prelim test: 1598
Number of HSP's gapped (non-prelim): 220
length of query: 338
length of database: 11,318,596
effective HSP length: 100
effective length of query: 238
effective length of database: 8,646,696
effective search space: 2057913648
effective search space used: 2057913648
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0008.8


