

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.15
(375 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
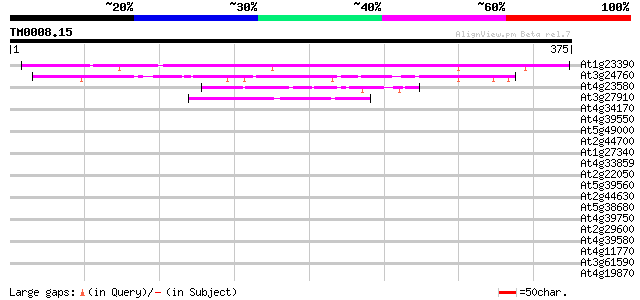
Score E
Sequences producing significant alignments: (bits) Value
At1g23390 unknown protein 280 1e-75
At3g24760 unknown protein 84 1e-16
At4g23580 putative protein 45 8e-05
At3g27910 hypothetical protein 42 4e-04
At4g34170 putative protein 40 0.002
At4g39550 unknown protein 39 0.006
At5g49000 unknown protein 38 0.010
At2g44700 unknown protein 38 0.010
At1g27340 unknown protein 38 0.010
At4g33859 putative protein 37 0.023
At2g22050 hypothetical protein 37 0.023
At5g39560 putative protein 36 0.030
At2g44630 unknown protein 36 0.039
At5g38680 putative protein 35 0.087
At4g39750 putative protein 34 0.15
At2g29600 hypothetical protein 33 0.19
At4g39580 putative protein 33 0.25
At4g11770 putative protein 33 0.25
At3g61590 unknown protein 33 0.25
At4g19870 unknown protein 33 0.33
>At1g23390 unknown protein
Length = 394
Score = 280 bits (715), Expect = 1e-75
Identities = 160/380 (42%), Positives = 221/380 (58%), Gaps = 18/380 (4%)
Query: 9 EEAPIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIILTQSTRAS 68
EE+ I GD+LE+ILS++PL+ L AC VS +W AVF SL +T+ PWL + Q
Sbjct: 16 EESSIDGDILESILSYLPLLDLDSACQVSKSWNRAVFYSLRRLKTM-PWLFVYNQRNSPP 74
Query: 69 HVIT--AHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALH 126
+ + A AYDP+S W+E+ H S RSSHSTLLY LSPA F+FS DA H
Sbjct: 75 YTMATMAMAYDPKSEAWIELNTASSPVEHVSVA---RSSHSTLLYALSPARFSFSTDAFH 131
Query: 127 LDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMES--NTWVMCQSMP 184
L W H PRVWR DPIVA VG+ +++AGG CDFE+D AVE++D+ES W C+SMP
Sbjct: 132 LTWQHVAPPRVWRIDPIVAVVGRSLIIAGGVCDFEEDRFAVELFDIESGDGAWERCESMP 191
Query: 185 AMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSVGYCVT-G 243
L S+ STWLSVAV+ E++ VTE+ SG+T F+ V+ W DLCP + Y + G
Sbjct: 192 DFLYESASSTWLSVAVSSEKMYVTEKRSGVTCSFNPVTRSWTKLLDLCPGECSLYSRSIG 251
Query: 244 MLGKRLMVAGVVGDGEDVKAVKLWAVKGGLGSGMT-EEVGEMPEELLEKFKGDSDE--LG 300
RL++AG++GD + ++LW V S + E +G MPE LEK +G + + L
Sbjct: 252 FSVNRLIMAGIIGDEYNPTGIELWEVIDSDESHLKFESIGSMPETYLEKLRGINSDWPLT 311
Query: 301 SLEVTWVGNFVYLRNTLEI-EELVMCEVVNGSRCEWRSVRNAAV-----DGGTRMVVCGS 354
S+ + VG+ VY+ E E+V E+ G C+WR++ NA R++V S
Sbjct: 312 SIVLNAVGDMVYVHGAAENGGEIVAAEIEGGKLCKWRTLPNADATWKKSHAAERVIVACS 371
Query: 355 EVRLEDLQSAMVSGIQTCRT 374
V DL+ A + + T
Sbjct: 372 NVGFSDLKLAFRNNLSFLST 391
>At3g24760 unknown protein
Length = 383
Score = 84.3 bits (207), Expect = 1e-16
Identities = 93/347 (26%), Positives = 152/347 (43%), Gaps = 45/347 (12%)
Query: 16 DVLETILSHVPLIHLVPACHVSNTWKHAVFS---SLAHTRTIKPWLIILTQSTRASHVIT 72
DV E+IL H+P+ LV VS W+ + S S + + + PWL + +S
Sbjct: 19 DVTESILYHLPIPSLVRFTLVSKQWRSLITSLPPSPSPSPSSPPWLFLFGIHNTSSFHNQ 78
Query: 73 AHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHST-LLYTLSPAEFAFSIDALHLDWHH 131
+ A+DP S+ W ++LP PS++++ S L+T +P F+FS L +W
Sbjct: 79 SFAFDPLSNTW--LRLP------PSSSSSDHLVGSNRFLFTTAP-RFSFS-PILKPNWRF 128
Query: 132 APAPRVWRTDPIV------ARVGQRVVVAG-----GACDFEDDPLAVEMYDMESNTWVMC 180
R R +P++ + + ++V G G D E+ LAV++YD ++W +C
Sbjct: 129 TSPVRFPRINPLLTVFTTLSNSSKLILVGGSSRIGGLVDIEER-LAVQIYDPVLDSWELC 187
Query: 181 QSMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGM--TFCFDTVSMKWDGPYDLCPDQSVG 238
+PA + L+ A+ V + S +FC D S W L P +
Sbjct: 188 SPLPADFRSGQDHQTLTSALFKRRFYVFDNYSCFISSFCLD--SYTWSDVQTLKP-PGLS 244
Query: 239 YCVTGMLGKRLMVAGVVGDGEDVKAVKLWAVKGGLGSGMTEEVGEMPEELLEKFKGDSDE 298
+ L++ G+ G + LW+++ GS E+ MPE+LL DE
Sbjct: 245 FAYLNSCNGMLVLGGMCG-----FSFNLWSIEE--GSMEFSEIAVMPEDLLFGLVDSDDE 297
Query: 299 ---LGSLEVTWVGNFVYLRNTLEIEEL--VMCEVVNGSR--CEWRSV 338
SL+ GN VY+ N ++ +CE+ G C WR V
Sbjct: 298 DDKFRSLKCAGSGNLVYVFNDDCHKKFPACVCEIGGGENGICSWRRV 344
>At4g23580 putative protein
Length = 383
Score = 44.7 bits (104), Expect = 8e-05
Identities = 46/156 (29%), Positives = 67/156 (42%), Gaps = 24/156 (15%)
Query: 129 WHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMPAMLK 188
WH AP+ RV R P + R+ V GG CD D +E++D ++ TW Q +
Sbjct: 159 WHEAPSMRVGRVFPSACTLDGRIYVTGG-CDNLDTMNWMEIFDTKTQTWEFLQIPSEEI- 216
Query: 189 GSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPD------QSVGYCVT 242
GS +LSV+ G V V + +T+ KW D+C + S YCV
Sbjct: 217 -CKGSEYLSVSYQG-TVYVKSDEKDVTYKMH--KGKW-READICMNNGWSLSSSSSYCVV 271
Query: 243 GMLGKRLMVAGVVGDGE----DVKAVKLWAVKGGLG 274
+ R +GE D+K + W + GLG
Sbjct: 272 ENVFYRYC------EGEIRWYDLKN-RAWTILKGLG 300
>At3g27910 hypothetical protein
Length = 294
Score = 42.4 bits (98), Expect = 4e-04
Identities = 32/122 (26%), Positives = 51/122 (41%), Gaps = 5/122 (4%)
Query: 120 FSIDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVM 179
F ID LH + P+ RV R V ++ V GG + VE++D+E TW
Sbjct: 65 FVIDCLHGTFQFLPSMRVPRGCAAFGIVDGKIYVIGGYNKADSLDNWVEVFDLEKQTW-- 122
Query: 180 CQSMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSVGY 239
+S + L V +++ + + +G+ FD W+ + L D VG
Sbjct: 123 -ESFSGLCNEELSKITLKSVVMNKKIYIMDRGNGIV--FDPKKGVWERDFLLDRDWVVGS 179
Query: 240 CV 241
CV
Sbjct: 180 CV 181
>At4g34170 putative protein
Length = 293
Score = 40.4 bits (93), Expect = 0.002
Identities = 30/95 (31%), Positives = 45/95 (46%), Gaps = 4/95 (4%)
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
+D+ W AP+ RV R P V + ++ V GG CD D +E++D ++ TW Q
Sbjct: 113 MDSRSHTWREAPSMRVPRMFPSVCTLDGKIYVMGG-CDNLDSTNWMEVFDTKTQTWEFLQ 171
Query: 182 SMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTF 216
+ G GS + SV G V V E +T+
Sbjct: 172 IPSEEIFG--GSAYESVRYEG-TVYVWSEKKDVTY 203
>At4g39550 unknown protein
Length = 392
Score = 38.5 bits (88), Expect = 0.006
Identities = 31/108 (28%), Positives = 44/108 (40%), Gaps = 7/108 (6%)
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
+D WH P RV R P V ++ V GG D + +E++D + TW
Sbjct: 167 LDCQSHTWHEGPGMRVERRYPAANVVEGKIYVTGGCKDCSNSSNWMEVFDPRTQTWESVS 226
Query: 182 SMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDT---VSMKWD 226
S A + G S S V GE ++ S G+ + MKWD
Sbjct: 227 SPGAEIGGC--SIHKSAVVEGE--ILIANSHGLIYKPKEGRWERMKWD 270
>At5g49000 unknown protein
Length = 328
Score = 37.7 bits (86), Expect = 0.010
Identities = 18/49 (36%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Query: 129 WHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTW 177
WH AP+ R+ R P V ++ VAGG +F D +E++D+++ TW
Sbjct: 113 WHEAPSMRMKRNYPAANVVDGKIYVAGGLEEF-DSSKWMEVFDIKTQTW 160
>At2g44700 unknown protein
Length = 368
Score = 37.7 bits (86), Expect = 0.010
Identities = 49/216 (22%), Positives = 90/216 (40%), Gaps = 19/216 (8%)
Query: 16 DVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAH-TRTI--KPWLII--LTQSTRASH- 69
+++ IL+ VP + C VS + V S H R++ K +L I + ++ R +
Sbjct: 34 EIISMILARVPKRYYPILCSVSKNMRSLVRSPEIHKARSLLGKDYLYIGFIDENYRPVYD 93
Query: 70 ---VITAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALH 126
+ +++ ++ P+P + + + AV + + +P+ D
Sbjct: 94 YWYTLRRIENSTTENLFESIEFPYPSEPNRFSMNAVGPKIFFISESCTPSSRLSIFDTRF 153
Query: 127 LDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMPAM 186
+ P V R V VG +V V GG ++DD +A E +D+ + TW ++ P
Sbjct: 154 GELRQGPCLLVKRGYNCVGLVGGKVYVIGG---YQDDEIAAESFDLNTQTW---EAAPIP 207
Query: 187 LKGSSGSTWLSVAVAG--EEVLVTEESSGMTFCFDT 220
+ S W+ A +V GMT C+DT
Sbjct: 208 DEKES-HRWICKANVSFDRKVCALRSREGMT-CYDT 241
>At1g27340 unknown protein
Length = 467
Score = 37.7 bits (86), Expect = 0.010
Identities = 82/366 (22%), Positives = 139/366 (37%), Gaps = 58/366 (15%)
Query: 16 DVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIK------PWLIILTQSTRASH 69
D+ E ++S +P+ V W +A+ S + +R PW +T +
Sbjct: 123 DLFEDVVSRLPMATFFQFRAVCRKW-NALIDSDSFSRCFTELPQTIPWFYTITHE----N 177
Query: 70 VITAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALHLDW 129
V + YDP W HP A + S +L S +D H ++
Sbjct: 178 VNSGQVYDPSLKKW----------HHPIIPALPKKS--IVLPMASAGGLVCFLDIGHRNF 225
Query: 130 HHA-PAPRVWRTDP-----IVARVGQRVVVAGGACDFEDDPLAV------EMYDMESNTW 177
+ + P + +R P + +RV + + G + L V E+YD SN W
Sbjct: 226 YVSNPLTKSFRELPARSFKVWSRVAVGMTLNGNSTSHGYKVLWVGCEGEYEVYDSLSNVW 285
Query: 178 VMCQSMPAMLKGSSGSTWLSVAVAGEEVL--VTEESSGMTFCFDTVSMKW-----DGPYD 230
++P+ +K + S VA L + + G+ +D VS KW GP D
Sbjct: 286 TKRGTIPSNIKLPVLLNFKSQPVAIHSTLYFMLTDPEGI-LSYDMVSGKWKQFIIPGPPD 344
Query: 231 LCPDQSVGYCVTGMLGKRLMVAGVVGDGEDVKAVKLWAVKGGLGSGMTEEVGEMPEELLE 290
L D ++ C G+RLM+ G++ V +W ++ + + +EV MP
Sbjct: 345 L-SDHTLAAC-----GERLMLVGLLTKNA-ATCVCIWELQ--KMTLLWKEVDRMPNIWCL 395
Query: 291 KFKGDSDELGSLEVTWVGNFVYLRNTLEIEELVMCEVVNGSRCEWRSVRNAAVDGGTR-- 348
+F G + L G + L +L ++ N EW V V G +
Sbjct: 396 EFYGKHIRMNCLGNK--GCLILL--SLRSRQMNRLITYNAVTREWTKVPGCTVPRGRKRL 451
Query: 349 MVVCGS 354
+ CG+
Sbjct: 452 WIACGT 457
>At4g33859 putative protein
Length = 379
Score = 36.6 bits (83), Expect = 0.023
Identities = 36/139 (25%), Positives = 62/139 (43%), Gaps = 10/139 (7%)
Query: 94 GHPSATAAVRSSHSTLLYTLSPAEFAFSIDALHLDWHHAPAPRVWRTDPIVARVGQRVVV 153
G + A++ ++ S LS +D+ W AP+ RV R P + R+ V
Sbjct: 123 GSENKNASINATGSKTYNALSSV---MVMDSRSHTWREAPSMRVARVFPSACTLDGRIYV 179
Query: 154 AGGACDFEDDPLAVEMYDMESNTWVMCQSMPAMLKGSSGSTWLSVAVAGEEVLVTEESSG 213
GG C+ + +E++D ++ TW Q +P+ + GS +LS++ V V
Sbjct: 180 TGG-CENLNSMNWMEIFDTKTQTWEFLQ-IPSE-EVCKGSEYLSISYQ-RTVYVGSREKD 235
Query: 214 MTFCFDTVSMKWDGPYDLC 232
+T+ KW G D+C
Sbjct: 236 VTYKMH--KGKWRGA-DIC 251
>At2g22050 hypothetical protein
Length = 349
Score = 36.6 bits (83), Expect = 0.023
Identities = 44/188 (23%), Positives = 79/188 (41%), Gaps = 21/188 (11%)
Query: 7 LPEEAPIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFS-SLAHTRTIKPW-LIILTQS 64
LP + +++ L VP + + C VS T + V S L+ RT+ P + ++ S
Sbjct: 28 LPSFTSLPDEIVLDCLQRVPRSYYLNLCRVSKTLRSLVRSPELSRLRTLLPKNSVYVSFS 87
Query: 65 TRASHVITAHAYD------------PRSHVWVEMKLPHPLKGHPSA--TAAVRSSHSTLL 110
+V Y ++ + +K+P P H S ++AV S +
Sbjct: 88 QNIINVPPDTIYRWFTLKKKTMKTAMKTFRYKLVKIPIPFPSHHSMYNSSAVGSEIYFVG 147
Query: 111 YTLSPAEFAFSIDALHLDWHHAPAPRVWRTDPI-VARVGQRVVVAGGACDFEDDPLAVEM 169
+ P + +D + P+ +V RTD V + ++ V GG +D + VE+
Sbjct: 148 GSFEPMSELWILDTRTGMFRQGPSMKVARTDEASVGVINGKIYVIGGC----EDKIQVEV 203
Query: 170 YDMESNTW 177
YD +S +W
Sbjct: 204 YDPKSRSW 211
>At5g39560 putative protein
Length = 395
Score = 36.2 bits (82), Expect = 0.030
Identities = 18/60 (30%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Query: 129 WHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMPAMLK 188
W P V R+DP+ + QR+ V GG + ++ E++D+++ TW S A L+
Sbjct: 154 WRDGPNMTVARSDPVAVLIDQRIYVLGGR-EMDESDDWFEVFDIKTQTWRALPSFRAGLE 212
>At2g44630 unknown protein
Length = 372
Score = 35.8 bits (81), Expect = 0.039
Identities = 40/181 (22%), Positives = 76/181 (41%), Gaps = 23/181 (12%)
Query: 16 DVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAH-TRTIKP----WLIILTQSTRAS-- 68
D++ IL+ VP + C VS + + + S H TR++ +L T++T +
Sbjct: 30 DIVLNILALVPKRYYPILCCVSKSLRSLIRSPEIHKTRSLHGKDSLYLCFSTRTTYPNRN 89
Query: 69 ----HVITAHAYDPR----SHVWVEMKLPHPLKGHPSATAAVRSSHSTLL----YTLSPA 116
H T D + +V+V + +P+ GH S + + + + Y +
Sbjct: 90 RTTFHWFTLRRNDNKMNTTENVFVSIDVPYR-PGHASYPLSNIAIDTEIYCIPGYNFPSS 148
Query: 117 EFAFSIDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNT 176
+ D W P+ +V R V VG ++ V GG + + E++D+++ T
Sbjct: 149 SIVWIFDTQSGQWRQGPSMQVERLSATVGLVGGKIYVIGGN---RGEEILAEVFDLKTQT 205
Query: 177 W 177
W
Sbjct: 206 W 206
>At5g38680 putative protein
Length = 357
Score = 34.7 bits (78), Expect = 0.087
Identities = 26/123 (21%), Positives = 49/123 (39%), Gaps = 2/123 (1%)
Query: 57 WLIILTQSTRASHVITAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPA 116
W + + +A T++ +V + +PHP S+ AV S+ + ++SP
Sbjct: 83 WFTLCLKPDQALSSETSNKKKSSGYVLATVSIPHPRLVQRSSLVAVGSNIYNIGRSISPY 142
Query: 117 EFAFSIDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPL--AVEMYDMES 174
D W AP+ V + + ++ VAG D + L E++D ++
Sbjct: 143 SSVSIFDCRSHTWREAPSLPVELVEVSAGVLDGKIYVAGSCKDGDSLNLKNTFEVFDTKT 202
Query: 175 NTW 177
W
Sbjct: 203 QVW 205
>At4g39750 putative protein
Length = 912
Score = 33.9 bits (76), Expect = 0.15
Identities = 25/98 (25%), Positives = 45/98 (45%), Gaps = 5/98 (5%)
Query: 129 WHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMPAMLK 188
W AP+ V R + + ++ V GG C ++ E++D ++ TW A L+
Sbjct: 698 WRKAPSMTVARANVFAYVINGKIYVMGG-CAADESKYWAEVFDPKTQTWKPLTDPGAELR 756
Query: 189 GSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWD 226
SS + +AV+ ++ V + S + +D KWD
Sbjct: 757 VSS---IIGMAVSEGKIYV-KNSYVKDYVYDPEEDKWD 790
Score = 30.0 bits (66), Expect = 2.2
Identities = 14/49 (28%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Query: 129 WHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTW 177
W +AP V R +P+ +++ V GG + E E++D ++ TW
Sbjct: 345 WRNAPDMTVARANPVAYVFDRKIYVMGGCAETESANWG-EVFDPKTQTW 392
>At2g29600 hypothetical protein
Length = 415
Score = 33.5 bits (75), Expect = 0.19
Identities = 21/72 (29%), Positives = 38/72 (52%), Gaps = 5/72 (6%)
Query: 116 AEFAFSIDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESN 175
A+ F ID W + +V R+ A + + V GG+ DD VE++++E+N
Sbjct: 175 AKTVFVIDCRFHTWRYLQEMQVARSYAASAVIDGMIYVVGGSTKRSDD--WVEVFNVETN 232
Query: 176 TWVMCQSMPAML 187
TW +++P++L
Sbjct: 233 TW---ENVPSVL 241
>At4g39580 putative protein
Length = 375
Score = 33.1 bits (74), Expect = 0.25
Identities = 18/64 (28%), Positives = 31/64 (48%), Gaps = 1/64 (1%)
Query: 114 SPAEFAFSIDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDME 173
+P+ +D W AP+ RV R P + ++ VAGG C+ +E++D +
Sbjct: 143 APSSSVSVLDCQCEKWREAPSMRVARNYPTATVLDGKIYVAGG-CEDCTSLDCIEVFDPK 201
Query: 174 SNTW 177
+ TW
Sbjct: 202 TQTW 205
>At4g11770 putative protein
Length = 396
Score = 33.1 bits (74), Expect = 0.25
Identities = 19/72 (26%), Positives = 35/72 (48%), Gaps = 5/72 (6%)
Query: 120 FSIDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGG--ACDFEDDPLAVEMYDMESNTW 177
F +D WH AP+ +V R P+V + ++ V G D+ + +E++D ++ W
Sbjct: 168 FFLDCRSHTWHEAPSMQVARKSPLVNVLDGKIYVVEGWRGSDYSN---LIEIFDPKTQKW 224
Query: 178 VMCQSMPAMLKG 189
S A ++G
Sbjct: 225 EHVPSPSAEMRG 236
>At3g61590 unknown protein
Length = 411
Score = 33.1 bits (74), Expect = 0.25
Identities = 23/80 (28%), Positives = 34/80 (41%), Gaps = 10/80 (12%)
Query: 16 DVLETILSHVPLIHLVPACHVSNTWKHAVFSS------LAHTRTIKPWLIILTQSTRASH 69
D+LE ILS +P+ + A V W V S ++ + +PW + T + S
Sbjct: 46 DLLERILSFLPIASIFRAGTVCKRWNEIVSSRRFLCNFSNNSVSQRPWYFMFTTTDDPS- 104
Query: 70 VITAHAYDPRSHVWVEMKLP 89
+AYDP W LP
Sbjct: 105 ---GYAYDPIIRKWYSFDLP 121
>At4g19870 unknown protein
Length = 400
Score = 32.7 bits (73), Expect = 0.33
Identities = 50/224 (22%), Positives = 80/224 (35%), Gaps = 42/224 (18%)
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
+D W AP+ V R P + ++ V GG + E++D+++ TW C
Sbjct: 165 LDCRSNTWRDAPSMTVARKRPFICLYDGKIYVIGGYNKLSESEPWAEVFDIKTQTW-ECL 223
Query: 182 SMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWD-------GPYDLCPD 234
S P G+ +A E + + T+ +D +W+ P C
Sbjct: 224 SDP----GTEIRNCTIYRIAEIEGKIHFGYTQKTYAYDPKQGEWECCEGEVAFPRSQCVM 279
Query: 235 QSVGYCVTGMLGKRLMVAGVVGDGEDVKAVKLWAVKGGLGSGMTEEVGEMPEELLEKFK- 293
+SV Y ED K W+ G E+V + E LLE K
Sbjct: 280 ESVLYTFANNY-----------TWEDDYGCKWWST-----DGYGEKVKGL-ESLLEIHKR 322
Query: 294 --GDSDEL-------GSLEVTWVGNFVYLRNTLEIEELVMCEVV 328
G SD G L + W G Y+++ + + C V+
Sbjct: 323 NGGSSDNTTKLVACGGKLLLLWEG---YMKHNPNNRKKIWCAVI 363
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.133 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,921,039
Number of Sequences: 26719
Number of extensions: 386755
Number of successful extensions: 1015
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 969
Number of HSP's gapped (non-prelim): 70
length of query: 375
length of database: 11,318,596
effective HSP length: 101
effective length of query: 274
effective length of database: 8,619,977
effective search space: 2361873698
effective search space used: 2361873698
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0008.15


