

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.14
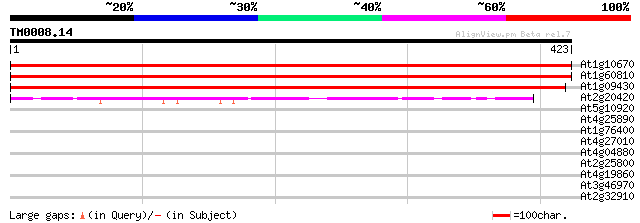
(423 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g10670 unknown protein 767 0.0
At1g60810 unknown protein 766 0.0
At1g09430 unknown protein 694 0.0
At2g20420 succinyl-CoA ligase beta subunit 64 2e-10
At5g10920 argininosuccinate lyase (AtArgH) 31 1.5
At4g25890 putative acidic ribosomal protein 31 1.5
At1g76400 putative ribophorin I (dolichyl-diphosphooligosacchari... 31 1.5
At4g27010 putative protein 30 1.9
At4g04880 adenosine deaminase like protein 30 1.9
At2g25800 unknown protein 29 5.6
At4g19860 unknown protein 28 7.3
At3g46970 starch phosphorylase H (cytosolic form) - like protein 28 7.3
At2g32910 unknown protein 28 7.3
>At1g10670 unknown protein
Length = 423
Score = 767 bits (1980), Expect = 0.0
Identities = 378/423 (89%), Positives = 402/423 (94%)
Query: 1 MARKKIREYDSKRLVKEHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPD 60
MARKKIREYDSKRLVKEHFKRLSGKELPI+S Q+ +TD +EL +KEPWLSS KLVVKPD
Sbjct: 1 MARKKIREYDSKRLVKEHFKRLSGKELPIRSVQINETTDLNELVEKEPWLSSEKLVVKPD 60
Query: 61 MLFGKRGKSGLVALNLDFAQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN 120
MLFGKRGKSGLVAL LDFA VA+FVKERLGKEVEM GCKGPITTFIVEPF+PHNEE+YLN
Sbjct: 61 MLFGKRGKSGLVALKLDFADVATFVKERLGKEVEMSGCKGPITTFIVEPFVPHNEEYYLN 120
Query: 121 IVSERLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEIVAPLVATLPLEIKGEIEE 180
+VS+RLG SISFSECGGI+IEENWDKVKT+F+PTG SLT EI APLVATLPLEIK EIEE
Sbjct: 121 VVSDRLGCSISFSECGGIEIEENWDKVKTIFLPTGASLTPEICAPLVATLPLEIKAEIEE 180
Query: 181 FLKVIFTLFQDLDFTFLEMNPFTLVDGKPCPLDMRGELDDTAAFKNFKKWGNIEFPLPFG 240
F+KVIFTLFQDLDFTFLEMNPFTLVDG P PLDMRGELDDTAAFKNFKKWG+IEFPLPFG
Sbjct: 181 FIKVIFTLFQDLDFTFLEMNPFTLVDGSPYPLDMRGELDDTAAFKNFKKWGDIEFPLPFG 240
Query: 241 RVMSATESFVHELDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFASELGN 300
RVMS TESF+H LDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLG+ASELGN
Sbjct: 241 RVMSPTESFIHGLDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGYASELGN 300
Query: 301 YAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGIIRALKE 360
YAEYSGAPKEDEVLQYARVVIDCATANPDG+ RALVIGGGIANFTDVAATF+GIIRALKE
Sbjct: 301 YAEYSGAPKEDEVLQYARVVIDCATANPDGKSRALVIGGGIANFTDVAATFNGIIRALKE 360
Query: 361 KESKLKAARMHIYVRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATMTGICKEAIQCIT 420
KE+KLKAARMHI+VRRGGPNYQKGLA MRALG++IG+PIEVYGPEATMTGICKEAIQ IT
Sbjct: 361 KEAKLKAARMHIFVRRGGPNYQKGLAKMRALGDDIGVPIEVYGPEATMTGICKEAIQYIT 420
Query: 421 AAA 423
AAA
Sbjct: 421 AAA 423
>At1g60810 unknown protein
Length = 423
Score = 766 bits (1978), Expect = 0.0
Identities = 377/423 (89%), Positives = 404/423 (95%)
Query: 1 MARKKIREYDSKRLVKEHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPD 60
MARKKIREYDSKRLVKEHFKRLSG+ELPI+S Q+ TD +EL ++EPWLSS KLVVKPD
Sbjct: 1 MARKKIREYDSKRLVKEHFKRLSGQELPIRSVQINQETDLNELVEREPWLSSEKLVVKPD 60
Query: 61 MLFGKRGKSGLVALNLDFAQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN 120
MLFGKRGKSGLVALNLDFA VA+FVKERLGKEVEM GCKGPITTFIVEPF+PHNEEFYLN
Sbjct: 61 MLFGKRGKSGLVALNLDFADVATFVKERLGKEVEMSGCKGPITTFIVEPFVPHNEEFYLN 120
Query: 121 IVSERLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEIVAPLVATLPLEIKGEIEE 180
IVS+RLG SISFSECGGIDIEENWDKVKT+ IPTG SLT EI APLVATLPLEIKGE+E+
Sbjct: 121 IVSDRLGCSISFSECGGIDIEENWDKVKTITIPTGASLTFEICAPLVATLPLEIKGELED 180
Query: 181 FLKVIFTLFQDLDFTFLEMNPFTLVDGKPCPLDMRGELDDTAAFKNFKKWGNIEFPLPFG 240
F++VIFTLF+DLDFTFLEMNPFTLVDGKP PLDMRGELDDTAAFKNFKKWG+IEFP+PFG
Sbjct: 181 FIQVIFTLFEDLDFTFLEMNPFTLVDGKPYPLDMRGELDDTAAFKNFKKWGDIEFPMPFG 240
Query: 241 RVMSATESFVHELDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFASELGN 300
RVMS+TESF+H LDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLG+ASELGN
Sbjct: 241 RVMSSTESFIHGLDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGYASELGN 300
Query: 301 YAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGIIRALKE 360
YAEYSGAPKEDEVLQYARVVIDCATANPDG+ RALVIGGGIANFTDVAATF+GIIRALKE
Sbjct: 301 YAEYSGAPKEDEVLQYARVVIDCATANPDGKSRALVIGGGIANFTDVAATFNGIIRALKE 360
Query: 361 KESKLKAARMHIYVRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATMTGICKEAIQCIT 420
KE+KLKAARMHI+VRRGGPNYQKGLA MR+LG+EIG+PIEVYGPEATMTGICKEAIQ IT
Sbjct: 361 KEAKLKAARMHIFVRRGGPNYQKGLAKMRSLGDEIGVPIEVYGPEATMTGICKEAIQYIT 420
Query: 421 AAA 423
AAA
Sbjct: 421 AAA 423
>At1g09430 unknown protein
Length = 424
Score = 694 bits (1790), Expect = 0.0
Identities = 335/419 (79%), Positives = 383/419 (90%)
Query: 1 MARKKIREYDSKRLVKEHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPD 60
MARKKIREYDSKRL+KEH KRL+ +L I+SAQVT STDF+EL ++E WLSS+KLVVKPD
Sbjct: 1 MARKKIREYDSKRLLKEHLKRLANIDLQIRSAQVTESTDFTELTNQESWLSSTKLVVKPD 60
Query: 61 MLFGKRGKSGLVALNLDFAQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN 120
MLFGKRGKSGLVAL LD A+VA FVK RLG EVEM GCK PITTFIVEPF+PH++E+YL+
Sbjct: 61 MLFGKRGKSGLVALKLDLAEVADFVKARLGTEVEMEGCKAPITTFIVEPFVPHDQEYYLS 120
Query: 121 IVSERLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEIVAPLVATLPLEIKGEIEE 180
IVS+RLG +ISFSECGGI+IEENWDKVKT+F+P S+T E+ APL+ATLPLE++ +I
Sbjct: 121 IVSDRLGCTISFSECGGIEIEENWDKVKTIFLPAEKSMTLEVCAPLIATLPLEVRAKIGN 180
Query: 181 FLKVIFTLFQDLDFTFLEMNPFTLVDGKPCPLDMRGELDDTAAFKNFKKWGNIEFPLPFG 240
F+ F +FQDLDF+F+EMNPFTLVDG+P PLDMRGELDDTAAFKNF KWG+IEFPLPFG
Sbjct: 181 FIMGAFAVFQDLDFSFMEMNPFTLVDGEPFPLDMRGELDDTAAFKNFNKWGDIEFPLPFG 240
Query: 241 RVMSATESFVHELDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFASELGN 300
RV+S+TE+F+H LDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLG+ASELGN
Sbjct: 241 RVLSSTENFIHGLDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGYASELGN 300
Query: 301 YAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGIIRALKE 360
YAEYSGAP E+EVLQYARVVIDCAT +PDG+KRAL+IGGGIANFTDVAATF+GIIRAL+E
Sbjct: 301 YAEYSGAPNEEEVLQYARVVIDCATTDPDGRKRALLIGGGIANFTDVAATFNGIIRALRE 360
Query: 361 KESKLKAARMHIYVRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATMTGICKEAIQCI 419
KE++LKA+RMHIYVRRGGPNYQ GLA MRALGEE+G+P+EVYGPEATMTGICK AI CI
Sbjct: 361 KETRLKASRMHIYVRRGGPNYQTGLARMRALGEELGVPLEVYGPEATMTGICKRAIDCI 419
>At2g20420 succinyl-CoA ligase beta subunit
Length = 421
Score = 63.9 bits (154), Expect = 2e-10
Identities = 100/415 (24%), Positives = 172/415 (41%), Gaps = 56/415 (13%)
Query: 1 MARKKIREYDSKRLVKEHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPD 60
+ R I EY L+ ++ G +P A + +QD P + S+LVVK
Sbjct: 24 LRRLNIHEYQGAELMGKY-----GVNVPKGVAASSLEEVKKAIQDVFP--NESELVVKSQ 76
Query: 61 MLFGKRG----KSGLVA-LNLDFAQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPHN- 114
+L G RG KSGL +++ A + ++ +V + GP + + ++
Sbjct: 77 ILAGGRGLGTFKSGLKGGVHIVKRDEAEEIAGKMLGQVLVTKQTGPQGKVVSKVYLCEKL 136
Query: 115 ---EEFYLNIVSER--LGNSISFSECGGIDIEENWDKVKTVFIPTGVS----LTSEIVAP 165
E Y +I+ +R G I + GG IE+ +K + I + +T E A
Sbjct: 137 SLVNEMYFSIILDRKSAGPLIIACKKGGTSIEDLAEKFPDMIIKVPIDVFAGITDEDAAK 196
Query: 166 LV---ATLPLEIKGEIEEFLKVIFTLFQDLDFTFLEMNPFTLVD-GKPCPLDMRGELDDT 221
+V A + K IE+ +K ++ LF+ D T LE+NP + D + DD
Sbjct: 197 VVDGLAPKAADRKDSIEQ-VKKLYELFRKTDCTMLEINPLAETSTNQLVAADAKLNFDDN 255
Query: 222 AAFKNFKKWGNIEFPLPFGRVMSATESFVHELDEKTSASLKFTVLNPKGRIWTMVAGGGA 281
AAF+ V + + + E +A + + G I MV G G
Sbjct: 256 AAFRQ-------------KEVFAMRDPTQEDPREVAAAKVDLNYIGLDGEIGCMVNGAGL 302
Query: 282 SVIYADTVGDLGFASELGNYAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALV-IGGG 340
++ D + G N+ + G E +V++ +++ + D K LV I GG
Sbjct: 303 AMATMDIIKLHG--GTPANFLDVGGNASEHQVVEAFKIL-----TSDDKVKAILVNIFGG 355
Query: 341 IANFTDVAATFSGIIRALKEKESKLKAARMHIYVRRGGPNYQKGLALMRALGEEI 395
I +A SGI+ A KE A ++ + VR G N ++G +++ G ++
Sbjct: 356 IMKCDVIA---SGIVNAAKE-----VALKVPVVVRLEGTNVEQGKRILKESGMKL 402
>At5g10920 argininosuccinate lyase (AtArgH)
Length = 517
Score = 30.8 bits (68), Expect = 1.5
Identities = 35/148 (23%), Positives = 66/148 (43%), Gaps = 23/148 (15%)
Query: 10 DSKRLVKEHFKRLSGKELPI----KSAQVTGSTDFSELQDKEPWLSSSKLVVKPDMLFGK 65
D LV+ R+ G + + K + + DF E DKEP S+K ++
Sbjct: 343 DPMELVRGKSARVIGDLVTVLTLCKGLPLAYNRDFQE--DKEPMFDSTKTIM-------- 392
Query: 66 RGKSGLVALNLDFAQVASFVKERLGKEVEMGGCKG-PITTFIVEPFIPHNEEFYLNIVSE 124
G++ ++ +FAQ +F ++R+ K + G + ++V+ +P +IV +
Sbjct: 393 ----GMIDVSAEFAQNVTFNEDRIKKSLPAGHLDATTLADYLVKKGMPFRSSH--DIVGK 446
Query: 125 RLGNSISFS-ECGGIDIEENWDKVKTVF 151
+G +S E + +EE K+ VF
Sbjct: 447 LVGVCVSKGCELQNLSLEE-MKKLSPVF 473
>At4g25890 putative acidic ribosomal protein
Length = 119
Score = 30.8 bits (68), Expect = 1.5
Identities = 17/59 (28%), Positives = 28/59 (46%), Gaps = 4/59 (6%)
Query: 254 DEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFASELGNYAEYSGAPKEDE 312
D F++++P ++ ++ GGG ++ G AS G E + APKEDE
Sbjct: 46 DSSGGVQSSFSLVSPTSAVFQVIVGGGGGGGFSAG----GAASSGGGAGEAAAAPKEDE 100
>At1g76400 putative ribophorin I
(dolichyl-diphosphooligosaccharide-protein
glycosyltransferase)
Length = 614
Score = 30.8 bits (68), Expect = 1.5
Identities = 21/84 (25%), Positives = 41/84 (48%), Gaps = 5/84 (5%)
Query: 68 KSGLVALNLDFAQVASFVK----ERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLNIVS 123
+S +++ L A++ S+ K + G E++ G K + ++ P + H E V+
Sbjct: 168 ESQSLSIKLPNARIESYTKFENTKLQGSELKYGPYKN-LQSYSYSPIVVHFESKAAFAVA 226
Query: 124 ERLGNSISFSECGGIDIEENWDKV 147
E+L I S G + + EN++ V
Sbjct: 227 EKLVREIEVSHWGNVQVTENYNVV 250
>At4g27010 putative protein
Length = 2535
Score = 30.4 bits (67), Expect = 1.9
Identities = 31/105 (29%), Positives = 48/105 (45%), Gaps = 10/105 (9%)
Query: 283 VIYADTVGDLG--FASELGNYAEYSGAPK---EDEVLQYARVVIDCATANPDGQKRALVI 337
++ +T GDL + N+AE A K E + L Y +I ++P+G+ R+ I
Sbjct: 53 LLKGETGGDLLRLYFQSSPNFAELLEAWKLRHEKQGLSYIFSLIQTILSHPEGKDRSTDI 112
Query: 338 GGGIANFTD--VAATFSGIIRALKEKESKLKAARMHI---YVRRG 377
G I F V I + L KE K ++A + + VRRG
Sbjct: 113 GRAIDQFGRLLVEEKLDDIYKELNSKEGKQQSAALSLLASIVRRG 157
>At4g04880 adenosine deaminase like protein
Length = 355
Score = 30.4 bits (67), Expect = 1.9
Identities = 16/61 (26%), Positives = 31/61 (50%), Gaps = 7/61 (11%)
Query: 112 PHNEEFYLNIVSERLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEIVAPLVATLP 171
P + L+ R+G++ F + E+W K+K+ IP + LTS IV ++++
Sbjct: 224 PKEIQAMLDFKPHRIGHACFFKD-------EDWTKLKSFRIPVEICLTSNIVTKSISSID 276
Query: 172 L 172
+
Sbjct: 277 I 277
>At2g25800 unknown protein
Length = 993
Score = 28.9 bits (63), Expect = 5.6
Identities = 13/47 (27%), Positives = 26/47 (54%)
Query: 20 KRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPDMLFGKR 66
+++ LP++ Q S+DF++ Q+ + WL S V++ +L R
Sbjct: 177 RKIESVVLPLELLQQLKSSDFTDQQEYDAWLKRSLKVLEAGLLLHPR 223
>At4g19860 unknown protein
Length = 535
Score = 28.5 bits (62), Expect = 7.3
Identities = 13/29 (44%), Positives = 17/29 (57%)
Query: 211 PLDMRGELDDTAAFKNFKKWGNIEFPLPF 239
P + RG L+D F+ KKW N+ P PF
Sbjct: 413 PGEHRGILNDHRVFRMLKKWLNVGEPDPF 441
>At3g46970 starch phosphorylase H (cytosolic form) - like protein
Length = 841
Score = 28.5 bits (62), Expect = 7.3
Identities = 36/158 (22%), Positives = 64/158 (39%), Gaps = 13/158 (8%)
Query: 37 STDFSELQDKEPWLSSSKLVVKPDMLFGKRGKSGLVALNLDFAQVASFVKERLGKEVE-- 94
S + S++ K WL + K + D+L G R + L ++A + K+RL + +E
Sbjct: 499 SPELSDIITK--WLKTDKWITDLDLLTGLRQFADNEELQSEWASAKTANKKRLAQYIERV 556
Query: 95 MGGCKGPITTFIVEPFIPHNEEFYLNIVSERLGNSISFSECGGIDIEENWDKVKTVFIPT 154
G P + F ++ H Y + LG F + + EE V +
Sbjct: 557 TGVSIDPTSLFDIQVKRIHE---YKRQLMNILGVVYRFKKLKEMKPEERKKTVPRTVMIG 613
Query: 155 GVSLTSEIVAPLVATLPLEI------KGEIEEFLKVIF 186
G + + A + L ++ E+ E+LKV+F
Sbjct: 614 GKAFATYTNAKRIVKLVNDVGDVVNSDPEVNEYLKVVF 651
>At2g32910 unknown protein
Length = 691
Score = 28.5 bits (62), Expect = 7.3
Identities = 13/61 (21%), Positives = 33/61 (53%), Gaps = 3/61 (4%)
Query: 4 KKIREYDSKRLVKEHFKRLSGKELPIKSAQVTGSTDFSELQ---DKEPWLSSSKLVVKPD 60
KK+++ ++++K +++G+E IK + +G E + D EP+L + + + +
Sbjct: 117 KKVKKLVKRKIIKGTAAQVAGEENVIKEGEQSGEPSLGESEKDKDSEPYLGGNDMEFQKE 176
Query: 61 M 61
+
Sbjct: 177 L 177
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.138 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,442,697
Number of Sequences: 26719
Number of extensions: 411240
Number of successful extensions: 896
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 886
Number of HSP's gapped (non-prelim): 14
length of query: 423
length of database: 11,318,596
effective HSP length: 102
effective length of query: 321
effective length of database: 8,593,258
effective search space: 2758435818
effective search space used: 2758435818
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0008.14


