

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.13
(259 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
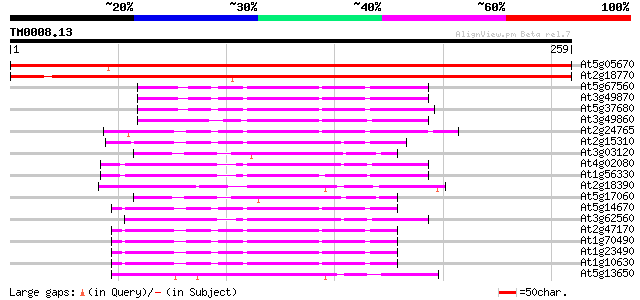
Score E
Sequences producing significant alignments: (bits) Value
At5g05670 signal recognition particle receptor beta subunit-like... 385 e-108
At2g18770 putative signal recognition particle receptor beta sub... 361 e-100
At5g67560 ADP-ribosylation factor-like protein 53 1e-07
At3g49870 ADP-RIBOSYLATION FACTOR like protein 53 2e-07
At5g37680 ADP-ribosylation factor-like protein (K12B20.15) 52 2e-07
At3g49860 putative protein 48 6e-06
At2g24765 ADP-ribosylation factor 3 like protein (At2g24765) 46 2e-05
At2g15310 putative ADP-ribosylation factor 45 5e-05
At3g03120 putative ADP-ribosylation factor 44 7e-05
At4g02080 SAR1/GTP-binding secretory factor 44 1e-04
At1g56330 unknown protein 44 1e-04
At2g18390 putative ADP-ribosylation factor 43 1e-04
At5g17060 ADP-ribosylation factor -like protein 43 2e-04
At5g14670 ADP-ribosylation factor - like protein 42 3e-04
At3g62560 Sar1-like GTP binding protein 42 3e-04
At2g47170 ADP-ribosylation factor 1 42 3e-04
At1g70490 ADP-ribosylation factor 1 like protein 42 3e-04
At1g23490 unknown protein (At1g23490) 42 3e-04
At1g10630 similar to ADP-ribosylation factor gb|AAD17207; simila... 42 3e-04
At5g13650 GTP-binding protein typA (tyrosine phosphorylated prot... 41 6e-04
>At5g05670 signal recognition particle receptor beta subunit-like
protein
Length = 260
Score = 385 bits (990), Expect = e-108
Identities = 186/260 (71%), Positives = 225/260 (86%), Gaps = 1/260 (0%)
Query: 1 MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLL-LSLRLFKRAKTNTVV 59
M+ LE K + +YL+++PP QLY A + +FTT+LL LS+RL +R K+NTV+
Sbjct: 1 MENLEDLKILAEQWSHQGIEYLQKIPPNQLYAAIGVLLFTTILLFLSIRLVRRTKSNTVL 60
Query: 60 LTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHS 119
L+GLTGSGKTVLFYQLRDGS+HQGTVTSMEPNEGTF+LHSE TKKGK+KPVH+VDVPGHS
Sbjct: 61 LSGLTGSGKTVLFYQLRDGSSHQGTVTSMEPNEGTFVLHSENTKKGKIKPVHLVDVPGHS 120
Query: 120 RLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
RLRPKL+E+LPQAA +VFVVDA++FLPNCRAASEYLY+ILT +VVKKKIP+L+ CNKTD
Sbjct: 121 RLRPKLEEFLPQAAAIVFVVDALEFLPNCRAASEYLYEILTNANVVKKKIPVLLCCNKTD 180
Query: 180 KVTAHTKEFIRRQLEKEIDKLRASRSAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADA 239
K+TAHTKEFIR+Q+EKEI+KLRASRSAVS AD+ N+FT+G+ GE FSFT C NKVT A+A
Sbjct: 181 KLTAHTKEFIRKQMEKEIEKLRASRSAVSTADIANDFTIGIEGEVFSFTHCSNKVTVAEA 240
Query: 240 SGLTGEISQLEEFIREYVKP 259
SGLTGE Q+E+FIREY+KP
Sbjct: 241 SGLTGETIQIEDFIREYIKP 260
>At2g18770 putative signal recognition particle receptor beta
subunit
Length = 260
Score = 361 bits (926), Expect = e-100
Identities = 176/260 (67%), Positives = 220/260 (83%), Gaps = 4/260 (1%)
Query: 1 MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVL 60
+++L+ E+ S+ L ++++++PP QLY A + + T+ LLS+RLF+R K+NTV+L
Sbjct: 4 LEDLKIVAEQWSKQGL---EFVQKIPPPQLYTAIGVLLLATIWLLSIRLFRRTKSNTVLL 60
Query: 61 TGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSET-TKKGKVKPVHIVDVPGHS 119
+GL+GSGKTVLFYQLRDGS+HQG VTSMEPNEGTF+LH+E TKKGKVKPVH++DVPGHS
Sbjct: 61 SGLSGSGKTVLFYQLRDGSSHQGAVTSMEPNEGTFVLHNENNTKKGKVKPVHLIDVPGHS 120
Query: 120 RLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
RL KL+EYLP+AA VVFVVDA++FLPN RAASEYLYDILT SV+K K P+L+ CNKTD
Sbjct: 121 RLISKLEEYLPRAAAVVFVVDALEFLPNIRAASEYLYDILTNASVIKNKTPVLLCCNKTD 180
Query: 180 KVTAHTKEFIRRQLEKEIDKLRASRSAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADA 239
KVTAHTKEFIR+Q+EKEIDKLR SRSA+S AD+ N+FTLG+ GE FSF+ C NKVT A+A
Sbjct: 181 KVTAHTKEFIRKQMEKEIDKLRVSRSAISTADIANDFTLGIEGEVFSFSHCHNKVTVAEA 240
Query: 240 SGLTGEISQLEEFIREYVKP 259
SGLTGE Q++EFIRE+VKP
Sbjct: 241 SGLTGETDQVQEFIREHVKP 260
>At5g67560 ADP-ribosylation factor-like protein
Length = 184
Score = 53.1 bits (126), Expect = 1e-07
Identities = 44/134 (32%), Positives = 64/134 (46%), Gaps = 11/134 (8%)
Query: 60 LTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHS 119
L GL +GKT L + G G M P G + KG V + + D+ G
Sbjct: 24 LIGLQNAGKTSLVNVVATG----GYSEDMIPTVG---FNMRKVTKGSVT-IKLWDLGGQP 75
Query: 120 RLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
R R + Y + +V+VVDA D N + L+D+L+K S+ IPLL+L NK D
Sbjct: 76 RFRSMWERYCRSVSAIVYVVDAAD-PDNLSVSKSELHDLLSKTSL--NGIPLLVLGNKID 132
Query: 180 KVTAHTKEFIRRQL 193
K A +KE + ++
Sbjct: 133 KPGALSKEALTDEM 146
>At3g49870 ADP-RIBOSYLATION FACTOR like protein
Length = 184
Score = 52.8 bits (125), Expect = 2e-07
Identities = 44/134 (32%), Positives = 64/134 (46%), Gaps = 11/134 (8%)
Query: 60 LTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHS 119
L GL +GKT L + G G M P G + KG V + + D+ G
Sbjct: 24 LIGLQNAGKTSLVNVVATG----GYSEDMIPTVG---FNMRKVTKGNVT-IKLWDLGGQP 75
Query: 120 RLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
R R + Y + +V+VVDA D N + L+D+L+K S+ IPLL+L NK D
Sbjct: 76 RFRSMWERYCRAVSAIVYVVDAAD-PDNLSVSKSELHDLLSKTSL--NGIPLLVLGNKID 132
Query: 180 KVTAHTKEFIRRQL 193
K A +KE + ++
Sbjct: 133 KPGALSKEALTDEM 146
>At5g37680 ADP-ribosylation factor-like protein (K12B20.15)
Length = 184
Score = 52.4 bits (124), Expect = 2e-07
Identities = 47/137 (34%), Positives = 64/137 (46%), Gaps = 11/137 (8%)
Query: 60 LTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHS 119
L GL +GKT L + G G M P G + KG V + I D+ G
Sbjct: 24 LVGLQNAGKTSLVNAIATG----GYSEDMIPTVG---FNMRKVTKGNVT-IKIWDLGGQR 75
Query: 120 RLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
R R + Y + +V+V+DA D + + L D+LTK S+ IPLLIL NK D
Sbjct: 76 RFRTMWERYCRGVSAIVYVIDAAD-RDSVPISRSELNDLLTKPSL--NGIPLLILGNKID 132
Query: 180 KVTAHTKEFIRRQLEKE 196
K A +K+ + QL E
Sbjct: 133 KSEALSKQALVDQLGLE 149
>At3g49860 putative protein
Length = 165
Score = 47.8 bits (112), Expect = 6e-06
Identities = 39/134 (29%), Positives = 64/134 (47%), Gaps = 11/134 (8%)
Query: 60 LTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHS 119
L GL SGKT L + G + + ++ N + TK+ + + D+ G
Sbjct: 5 LVGLQNSGKTSLVNVVATGEYSEDMIPTVGFNM------RKVTKENVA--IRLWDLGGQP 56
Query: 120 RLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
R R + Y + +V+VVDA D N + L+D+L+ S++ IPLL+L NK D
Sbjct: 57 RFRCMWERYCRAVSMIVYVVDAAD-TENLSVSRSELHDLLSNASLI--GIPLLVLGNKID 113
Query: 180 KVTAHTKEFIRRQL 193
A +KE + ++
Sbjct: 114 IHGALSKEALTEEM 127
>At2g24765 ADP-ribosylation factor 3 like protein (At2g24765)
Length = 182
Score = 46.2 bits (108), Expect = 2e-05
Identities = 45/167 (26%), Positives = 75/167 (43%), Gaps = 17/167 (10%)
Query: 44 LLSLRLFKRA---KTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
+L R+F K +++ GL +GKT + Y+L+ G V S P G + E
Sbjct: 3 ILFTRMFSSVFGNKEARILVLGLDNAGKTTILYRLQ-----MGEVVSTIPTIG---FNVE 54
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + +K + D+ G + +RP Y P V++VVD+ D A E + IL
Sbjct: 55 TVQYNNIK-FQVWDLGGQTSIRPYWRCYFPNTQAVIYVVDSSD-TDRIGVAKEEFHAILE 112
Query: 161 KGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASRSAV 207
+ + K +LI NK D A + L E+ K+++ + A+
Sbjct: 113 EDEL--KGAVVLIFANKQDLPGALDDAAVTEAL--ELHKIKSRQWAI 155
>At2g15310 putative ADP-ribosylation factor
Length = 205
Score = 44.7 bits (104), Expect = 5e-05
Identities = 39/139 (28%), Positives = 66/139 (47%), Gaps = 13/139 (9%)
Query: 45 LSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKK 104
++ R ++K +++ GL GSGKT + Y+L+ G V + P G + ET +
Sbjct: 8 IAKRFLPKSKVR-ILMVGLDGSGKTTILYKLK-----LGEVVTTVPTIG---FNLETVEY 58
Query: 105 GKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSV 164
+ + D+ G ++R Y A G++FVVD+ D A +E L+ ILT +
Sbjct: 59 KGIN-FTVWDIGGQEKIRKLWRHYFQNAQGLIFVVDSSDSERLSEARNE-LHRILTDNEL 116
Query: 165 VKKKIPLLILCNKTDKVTA 183
+ +L+ NK D A
Sbjct: 117 --EGACVLVFANKQDSRNA 133
>At3g03120 putative ADP-ribosylation factor
Length = 192
Score = 44.3 bits (103), Expect = 7e-05
Identities = 39/124 (31%), Positives = 59/124 (47%), Gaps = 16/124 (12%)
Query: 58 VVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPV--HIVDV 115
VV+ GL +GKT + Y+L H G V S P G + +K + K V + DV
Sbjct: 20 VVMLGLDAAGKTTILYKL-----HIGEVLSTVPTIGFNV------EKVQYKNVIFTVWDV 68
Query: 116 PGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILC 175
G +LRP Y G+++VVD++D +A E+ DI+ ++ I L+
Sbjct: 69 GGQEKLRPLWRHYFNNTDGLIYVVDSLDRERIGKAKQEF-QDIIRDPFMLNSVI--LVFA 125
Query: 176 NKTD 179
NK D
Sbjct: 126 NKQD 129
>At4g02080 SAR1/GTP-binding secretory factor
Length = 193
Score = 43.5 bits (101), Expect = 1e-04
Identities = 45/151 (29%), Positives = 69/151 (44%), Gaps = 14/151 (9%)
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 10 VLASLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQHPTSEELSI------ 61
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
GK+K D+ GH R +Y + VV++VDA D + E L +L+
Sbjct: 62 --GKIK-FKAFDLGGHQIARRVWKDYYAKVDAVVYLVDAYDKERFAESKKE-LDALLSDE 117
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
S+ +P LIL NK D A +++ +R L
Sbjct: 118 SLA--SVPFLILGNKIDIPYAASEDELRYHL 146
>At1g56330 unknown protein
Length = 193
Score = 43.5 bits (101), Expect = 1e-04
Identities = 45/151 (29%), Positives = 67/151 (43%), Gaps = 14/151 (9%)
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 10 ILASLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQHPTSEELSI------ 61
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
GK+K D+ GH R +Y + VV++VDA D A S+ D L
Sbjct: 62 --GKIK-FKAFDLGGHQIARRVWKDYYAKVDAVVYLVDAYD--KERFAESKRELDALLSD 116
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
+ +P LIL NK D A +++ +R L
Sbjct: 117 EAL-ATVPFLILGNKIDIPYAASEDELRYHL 146
>At2g18390 putative ADP-ribosylation factor
Length = 185
Score = 43.1 bits (100), Expect = 1e-04
Identities = 48/163 (29%), Positives = 75/163 (45%), Gaps = 17/163 (10%)
Query: 42 LLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSET 101
LL + ++ K+ K +++ GL SGKT + ++ T + T + N T I T
Sbjct: 3 LLSIIRKIKKKEKEMRILMVGLDNSGKTTIVLKINGEDTSVISPT-LGFNIKTIIYQKYT 61
Query: 102 TKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDF--LPNCRAASEYLYDIL 159
++I DV G +R Y Q G+V+VVD+ D L +C+ L ++L
Sbjct: 62 --------LNIWDVGGQKTIRSYWRNYFEQTDGLVWVVDSSDLRRLDDCKME---LDNLL 110
Query: 160 TKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKE-IDKLR 201
+ + LLIL NK D A T + I + L E +DK R
Sbjct: 111 KEERLAGSS--LLILANKQDIQGALTPDEIGKVLNLESMDKSR 151
>At5g17060 ADP-ribosylation factor -like protein
Length = 192
Score = 42.7 bits (99), Expect = 2e-04
Identities = 38/124 (30%), Positives = 57/124 (45%), Gaps = 16/124 (12%)
Query: 58 VVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIV--DV 115
VV+ GL +GKT + Y+L H G V S P G + +K + K V DV
Sbjct: 20 VVMLGLDAAGKTTILYKL-----HIGEVLSTVPTIGFNV------EKVQYKNVMFTVWDV 68
Query: 116 PGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILC 175
G +LRP Y G+++VVD++D +A E + + K + I +L+
Sbjct: 69 GGQEKLRPLWRHYFNNTDGLIYVVDSLDRERIGKAKQE--FQEIIKDPFMLNSI-ILVFA 125
Query: 176 NKTD 179
NK D
Sbjct: 126 NKQD 129
>At5g14670 ADP-ribosylation factor - like protein
Length = 188
Score = 42.4 bits (98), Expect = 3e-04
Identities = 36/132 (27%), Positives = 62/132 (46%), Gaps = 13/132 (9%)
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 11 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 61
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 62 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 117
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 118 DAVLLVFANKQD 129
>At3g62560 Sar1-like GTP binding protein
Length = 193
Score = 42.4 bits (98), Expect = 3e-04
Identities = 41/140 (29%), Positives = 62/140 (44%), Gaps = 12/140 (8%)
Query: 54 KTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIV 113
K ++ GL +GKT L + L+D Q T +E I GK+K
Sbjct: 19 KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQHPTSEELSI--------GKIK-FKAF 69
Query: 114 DVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLI 173
D+ GH R +Y + VV++VDA D + E L +L+ S+ +P LI
Sbjct: 70 DLGGHQIARRVWKDYYAKVDAVVYLVDAYDKERFAESKKE-LDALLSDESLA--NVPFLI 126
Query: 174 LCNKTDKVTAHTKEFIRRQL 193
L NK D A +++ +R L
Sbjct: 127 LGNKIDIPYAASEDELRYHL 146
>At2g47170 ADP-ribosylation factor 1
Length = 181
Score = 42.4 bits (98), Expect = 3e-04
Identities = 36/132 (27%), Positives = 62/132 (46%), Gaps = 13/132 (9%)
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 11 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 61
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 62 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 117
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 118 DAVLLVFANKQD 129
>At1g70490 ADP-ribosylation factor 1 like protein
Length = 181
Score = 42.4 bits (98), Expect = 3e-04
Identities = 36/132 (27%), Positives = 62/132 (46%), Gaps = 13/132 (9%)
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 11 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 61
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 62 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 117
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 118 DAVLLVFANKQD 129
>At1g23490 unknown protein (At1g23490)
Length = 181
Score = 42.4 bits (98), Expect = 3e-04
Identities = 36/132 (27%), Positives = 62/132 (46%), Gaps = 13/132 (9%)
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 11 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 61
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 62 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 117
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 118 DAVLLVFANKQD 129
>At1g10630 similar to ADP-ribosylation factor gb|AAD17207; similar
to ESTs gb|N65779, gb|AI996946.1, gb|AI725909.1,
gb|T45928, gb|T42202
Length = 181
Score = 42.4 bits (98), Expect = 3e-04
Identities = 36/132 (27%), Positives = 62/132 (46%), Gaps = 13/132 (9%)
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 11 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 61
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 62 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 117
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 118 DAVLLVFANKQD 129
>At5g13650 GTP-binding protein typA (tyrosine phosphorylated protein
A)
Length = 609
Score = 41.2 bits (95), Expect = 6e-04
Identities = 41/162 (25%), Positives = 66/162 (40%), Gaps = 20/162 (12%)
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQL-------RDGSTHQGTV---TSMEPNEGTFIL 97
+L +R + + GKT L + RD Q + +E G IL
Sbjct: 9 QLDRRDNVRNIAIVAHVDHGKTTLVDSMLRQAKVFRDNQVMQERIMDSNDLERERGITIL 68
Query: 98 HSETTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDF-LPNCRAASEYLY 156
T+ K V+I+D PGHS +++ L GV+ VVD+V+ +P R L
Sbjct: 69 SKNTSITYKNTKVNIIDTPGHSDFGGEVERVLNMVDGVLLVVDSVEGPMPQTRFV---LK 125
Query: 157 DILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEID 198
L G V +++ NK D+ +A + + E I+
Sbjct: 126 KALEFGHAV------VVVVNKIDRPSARPEFVVNSTFELFIE 161
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,370,449
Number of Sequences: 26719
Number of extensions: 211145
Number of successful extensions: 696
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 661
Number of HSP's gapped (non-prelim): 51
length of query: 259
length of database: 11,318,596
effective HSP length: 97
effective length of query: 162
effective length of database: 8,726,853
effective search space: 1413750186
effective search space used: 1413750186
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0008.13


