

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.11
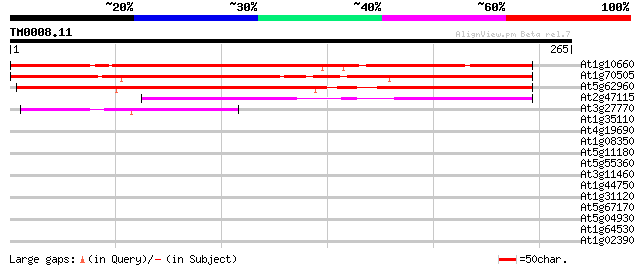
(265 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g10660 unknown protein 266 8e-72
At1g70505 putative protein 251 4e-67
At5g62960 unknown protein 220 7e-58
At2g47115 putative protein 111 5e-25
At3g27770 unknown protein 83 1e-16
At1g35110 unknown protein 38 0.005
At4g19690 Fe(II) transport protein (IRT1) 30 1.7
At1g08350 endosomal like protein 30 1.7
At5g11180 putative protein (AtGLR2.6) 29 2.3
At5g55360 wax synthase-like protein 29 3.0
At3g11460 hypothetical protein 28 5.1
At1g44750 Unknown protein (At1g44750) 28 5.1
At1g31120 putative potassium transporter 28 5.1
At5g67170 unknown protein 28 6.6
At5g04930 ATPase 27 8.7
At1g64530 unknown protein 27 8.7
At1g02390 putative protein 27 8.7
>At1g10660 unknown protein
Length = 320
Score = 266 bits (680), Expect = 8e-72
Identities = 134/249 (53%), Positives = 173/249 (68%), Gaps = 10/249 (4%)
Query: 1 METDTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAGLLYE 60
M DTTA SYWLNWR +CAL ++ + L + LIWKYE K R +R E Q++ G L++
Sbjct: 1 MAADTTASSYWLNWRVLLCALILLAPIVLAAVLIWKYE--GKRRRQR-ESQRELPGTLFQ 57
Query: 61 DELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGL 120
DE W TC KRIHP+WLLA+R+ SF+ +L LLI N+V DG GI YFYTQWTFTLVT+YFG
Sbjct: 58 DEAWTTCFKRIHPLWLLAFRVFSFVAMLTLLISNVVRDGAGIFYFYTQWTFTLVTLYFGY 117
Query: 121 ASFFSIYGCFFKHDKFEGNTVGSACL-NTERGTYVAP-TIDGDTDIPHMYKSTDAYQEPH 178
AS S+YGC + + GN + +TE+GTY P +DG+ + K+++ E
Sbjct: 118 ASVLSVYGCCIYNKEASGNMESYTSIGDTEQGTYRPPIALDGEGNTS---KASNRPSEAP 174
Query: 179 TQNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFL 238
+ TAG W YIFQI+FQTCAGAVVLTD+VFW ++YPF K ++L VCMHS+NAVFL
Sbjct: 175 ARKTAGFWVYIFQILFQTCAGAVVLTDIVFWAIIYPF--TKGYKLSFLDVCMHSLNAVFL 232
Query: 239 LGETSLNGM 247
LG+TSLN +
Sbjct: 233 LGDTSLNSL 241
>At1g70505 putative protein
Length = 338
Score = 251 bits (640), Expect = 4e-67
Identities = 129/253 (50%), Positives = 171/253 (66%), Gaps = 14/253 (5%)
Query: 1 METDTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQ----QQRAG 56
M ++T SYWLNWRF +CA++++ ++ L S+LIW+YE K R +RG+ Q +Q G
Sbjct: 27 MTSETNIPSYWLNWRFFVCAIFVLTSLFLSSYLIWRYEGPIK-RKKRGDDQSLELEQLTG 85
Query: 57 LLYEDELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTI 116
++Y+DE WNT +K IHP WLL +R+ F+VLLGL+ GN +ADG GI FYTQWTFTLVTI
Sbjct: 86 VVYDDESWNTSVKEIHPNWLLGFRVFGFVVLLGLISGNAIADGTGIFIFYTQWTFTLVTI 145
Query: 117 YFGLASFFSIYGCFFKHDKFEGNTVGSACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQE 176
YFGL S SIY F D E + ++ E+GTY P G+ + +++KS+ +
Sbjct: 146 YFGLGSLVSIYR-FRSPDNGENRV---SIVDEEQGTYRPP---GNAENSNVFKSSSGHDR 198
Query: 177 PH--TQNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVN 234
+ T+ A GYI QI+FQTCAGAV+LTD VFW ++YPFL KDF L F V MHSVN
Sbjct: 199 ENMSTRQVATTLGYIHQILFQTCAGAVLLTDGVFWFIIYPFLTAKDFNLDFFIVIMHSVN 258
Query: 235 AVFLLGETSLNGM 247
A+FLLGET LN +
Sbjct: 259 AIFLLGETFLNSL 271
>At5g62960 unknown protein
Length = 347
Score = 220 bits (560), Expect = 7e-58
Identities = 109/248 (43%), Positives = 157/248 (62%), Gaps = 17/248 (6%)
Query: 4 DTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGE---RQQQRAGLLYE 60
+TT SYW NWR IC +W+ + +FLI+KYE F + R + GE +++ +G +YE
Sbjct: 28 NTTESSYWFNWRVMICCIWMAIATVITAFLIFKYEGFRRKRSDVGEVDGGEKEWSGNVYE 87
Query: 61 DELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGL 120
DE W CL+ IHP WLLA+R+V+F VLL +LI + DG I ++YTQWTF L+T+YFGL
Sbjct: 88 DETWRPCLRNIHPAWLLAFRVVAFFVLLVMLIVIGLVDGPTIFFYYTQWTFGLITLYFGL 147
Query: 121 ASFFSIYGCFFKHDKFEGNTVGS-ACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHT 179
S S++GC+ + + G+ V S +++ER A + D I Q ++
Sbjct: 148 GSLLSLHGCYQYNKRAAGDRVDSIEAIDSER----ARSKGADNTI---------QQSQYS 194
Query: 180 QNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLL 239
N AG WGY+FQI+FQ AGAV+LTD VFW ++ PFL D+ L + + MHS+NA+FLL
Sbjct: 195 SNPAGFWGYVFQIIFQMNAGAVLLTDCVFWFIIVPFLEIHDYSLNVLVINMHSLNAIFLL 254
Query: 240 GETSLNGM 247
G+ +LN +
Sbjct: 255 GDAALNSL 262
>At2g47115 putative protein
Length = 289
Score = 111 bits (277), Expect = 5e-25
Identities = 65/185 (35%), Positives = 88/185 (47%), Gaps = 37/185 (20%)
Query: 63 LWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLAS 122
LW +C R+HP WLL R SF+ + LL +++ I +YT+WTF LV IYF +
Sbjct: 59 LWASCWTRLHPGWLLFTRSTSFLSMAALLAWDVIKWDASIFVYYTEWTFMLVIIYFAMGI 118
Query: 123 FFSIYGCFFKHDKFEGNTVGSACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHTQNT 182
S+YGC + T++ D D+
Sbjct: 119 VASVYGCLIHLKEL--------------------TLETDEDV-----------------V 141
Query: 183 AGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGET 242
G F+ +T AGAVVLTD+VFWLV+ PFL+ F L +CMH+ NA FLL ET
Sbjct: 142 VEKVGDEFRRRLETSAGAVVLTDIVFWLVIVPFLSTTRFGLNTLTICMHTANAGFLLLET 201
Query: 243 SLNGM 247
LN +
Sbjct: 202 LLNSL 206
>At3g27770 unknown protein
Length = 108
Score = 83.2 bits (204), Expect = 1e-16
Identities = 40/105 (38%), Positives = 56/105 (53%), Gaps = 8/105 (7%)
Query: 6 TALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAG--LLYEDEL 63
T+ YW NWR +CA+W+I M + ++WKYE D + Q G +L D++
Sbjct: 10 TSFDYWFNWRVLLCAIWVIVPMIVSLLVLWKYE------DSSVQTQPSLNGNDVLCIDDV 63
Query: 64 WNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQ 108
W C +RIHP WLL +R++ F LL I G I Y+YTQ
Sbjct: 64 WRPCFERIHPGWLLGFRVLGFCFLLANNIARFANRGWRIYYYYTQ 108
>At1g35110 unknown protein
Length = 1311
Score = 38.1 bits (87), Expect = 0.005
Identities = 32/119 (26%), Positives = 53/119 (43%), Gaps = 22/119 (18%)
Query: 75 WLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQ------------WTFTLVTIYFGLAS 122
WL +++ ++L L+ +VA GIL F W V + FG+ S
Sbjct: 39 WLWTLGVIAVVILSARLLVVVVATRGGILAFVLPDRRGLVTVEVRGWWLLAVRVRFGVLS 98
Query: 123 FFSIYGCFFKHDKFEGNTVGSACLNTERGTYVA-------PTIDGDTDIPHMYKSTDAY 174
FS+ + K G T GS L+ +RG +V+ P+ D+++P+ +TD Y
Sbjct: 99 SFSV--LIYSERKLLGLT-GSRFLSCKRGVFVSAGSTSQEPSSQWDSNLPNRLFATDHY 154
>At4g19690 Fe(II) transport protein (IRT1)
Length = 339
Score = 29.6 bits (65), Expect = 1.7
Identities = 21/74 (28%), Positives = 36/74 (48%), Gaps = 13/74 (17%)
Query: 76 LLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASFFSIYGCFFKHDK 135
LL YR+++ ++ LG+++ ++V +G+ T T T+ + L CF H
Sbjct: 180 LLRYRVIAMVLELGIIVHSVV---IGLSLGATSDTCTIKGLIAAL--------CF--HQM 226
Query: 136 FEGNTVGSACLNTE 149
FEG +G L E
Sbjct: 227 FEGMGLGGCILQAE 240
>At1g08350 endosomal like protein
Length = 589
Score = 29.6 bits (65), Expect = 1.7
Identities = 34/121 (28%), Positives = 54/121 (44%), Gaps = 9/121 (7%)
Query: 23 IIFTMGLGSFLIWKY---EEFNKSRDERGERQQQRAGLLYEDELWNTCLKRIHPVWLLAY 79
++ +GL SFL ++ E + S + ER++ L++ D C + I WL A
Sbjct: 238 VVLLIGLISFLFMRHLKNELRSYSIGDEEERKEAGWKLVHSDVF--RCPRNIS--WLCAI 293
Query: 80 RIVSFIVLLGLLIGNMVADGVGILYFYTQ-WTFTLVTIYFGLASFFSIYGCFFKHDKFEG 138
+ + LL L+I G LY Y + T + I + L S + Y H +FEG
Sbjct: 294 -LGTGTQLLILIIALFALAFTGFLYPYNRGMLLTSLVIMYTLTSIVAGYTSTSFHSQFEG 352
Query: 139 N 139
N
Sbjct: 353 N 353
>At5g11180 putative protein (AtGLR2.6)
Length = 906
Score = 29.3 bits (64), Expect = 2.3
Identities = 22/70 (31%), Positives = 36/70 (51%), Gaps = 4/70 (5%)
Query: 63 LWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILY--FYTQWTFTLVTIYFGL 120
L+N L R P+WLL + ++F+VLLG ++ VGI+ T +L I L
Sbjct: 3 LFNHLLSRALPLWLLFF--INFLVLLGKSQQEVLQVQVGIVLDTNATLAALSLRAINMSL 60
Query: 121 ASFFSIYGCF 130
+ F++ + F
Sbjct: 61 SEFYNTHNGF 70
>At5g55360 wax synthase-like protein
Length = 342
Score = 28.9 bits (63), Expect = 3.0
Identities = 19/59 (32%), Positives = 27/59 (45%), Gaps = 2/59 (3%)
Query: 73 PVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGL--ASFFSIYGC 129
P+WL A ++V F+VLL + +L F + F + I F L A F GC
Sbjct: 116 PIWLFAIKVVIFVVLLQMYEYKQYLSPALLLVFNSLHIFLELEIVFMLVKALVFITLGC 174
>At3g11460 hypothetical protein
Length = 623
Score = 28.1 bits (61), Expect = 5.1
Identities = 13/30 (43%), Positives = 18/30 (59%)
Query: 232 SVNAVFLLGETSLNGMVKYLWLSRLFPGSC 261
SV++V +LG L + +YLWL R G C
Sbjct: 153 SVDSVTMLGLVPLCTVPEYLWLGRSLHGQC 182
>At1g44750 Unknown protein (At1g44750)
Length = 379
Score = 28.1 bits (61), Expect = 5.1
Identities = 20/86 (23%), Positives = 40/86 (46%), Gaps = 3/86 (3%)
Query: 77 LAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASFFSIYGCFFKHDKF 136
L Y ++ +++L ++ G+ + VG+LY T++L+ +F +++ F KF
Sbjct: 111 LKYIVLIYVLLGVIIAGDNMLYSVGLLYLSAS-TYSLICA--TQLAFNAVFSYFINAQKF 167
Query: 137 EGNTVGSACLNTERGTYVAPTIDGDT 162
+ S L + +A D DT
Sbjct: 168 TALILNSVVLLSFSAALIALNDDADT 193
>At1g31120 putative potassium transporter
Length = 796
Score = 28.1 bits (61), Expect = 5.1
Identities = 11/28 (39%), Positives = 17/28 (60%), Gaps = 1/28 (3%)
Query: 12 LNWRFS-ICALWIIFTMGLGSFLIWKYE 38
+ W F+ I LW +F +G F IWK++
Sbjct: 248 VGWLFAPIVFLWFLFIASIGMFNIWKHD 275
>At5g67170 unknown protein
Length = 375
Score = 27.7 bits (60), Expect = 6.6
Identities = 14/40 (35%), Positives = 19/40 (47%)
Query: 142 GSACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHTQN 181
GS C NTE+ V ++GD D + D E T+N
Sbjct: 214 GSCCDNTEKAEQVLLQVNGDVDAAIEFLIADQGMESLTEN 253
>At5g04930 ATPase
Length = 1158
Score = 27.3 bits (59), Expect = 8.7
Identities = 18/63 (28%), Positives = 25/63 (39%), Gaps = 6/63 (9%)
Query: 6 TALSYWL------NWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAGLLY 59
T YW W F C L I+ T L F I E+ + D R R+ ++ G
Sbjct: 1081 TLPGYWAIFQVGKTWMFWFCLLAIVVTSLLPRFAIKFLVEYYRPSDVRIAREAEKLGTFR 1140
Query: 60 EDE 62
E +
Sbjct: 1141 ESQ 1143
>At1g64530 unknown protein
Length = 841
Score = 27.3 bits (59), Expect = 8.7
Identities = 17/57 (29%), Positives = 25/57 (43%), Gaps = 9/57 (15%)
Query: 136 FEGNTVGSACLNT-ERGTYVAPTIDGDTDIPHMYKSTDAYQEPHTQNTAGAWGYIFQ 191
F+G+ +G C++T + YV H++ DA E H Q G G FQ
Sbjct: 328 FDGSCMGKVCMSTSDLAVYVVDA--------HVWGFRDACAEHHLQKGQGVAGRAFQ 376
>At1g02390 putative protein
Length = 530
Score = 27.3 bits (59), Expect = 8.7
Identities = 16/45 (35%), Positives = 26/45 (57%), Gaps = 4/45 (8%)
Query: 188 YIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHS 232
Y + F+ AG V+ + +F LVLYPF++ + +GL + M S
Sbjct: 70 YFMVVAFE--AGGVIRS--LFLLVLYPFISLMSYEMGLKTMVMLS 110
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.143 0.473
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,294,471
Number of Sequences: 26719
Number of extensions: 262541
Number of successful extensions: 708
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 690
Number of HSP's gapped (non-prelim): 19
length of query: 265
length of database: 11,318,596
effective HSP length: 97
effective length of query: 168
effective length of database: 8,726,853
effective search space: 1466111304
effective search space used: 1466111304
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0008.11


