

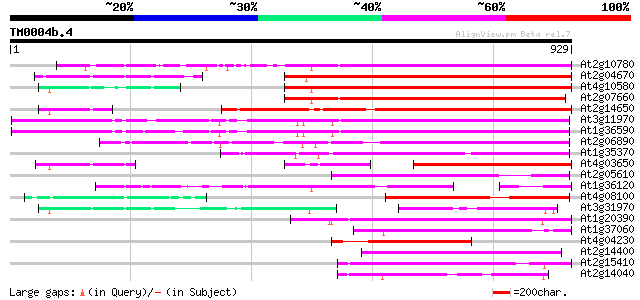
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0004b.4
(929 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 613 e-175
At2g04670 putative retroelement pol polyprotein 573 e-163
At4g10580 putative reverse-transcriptase -like protein 572 e-163
At2g07660 putative retroelement pol polyprotein 550 e-156
At2g14650 putative retroelement pol polyprotein 542 e-154
At3g11970 hypothetical protein 431 e-120
At1g36590 hypothetical protein 431 e-120
At2g06890 putative retroelement integrase 404 e-112
At1g35370 hypothetical protein 377 e-104
At4g03650 putative reverse transcriptase 355 5e-98
At2g05610 putative retroelement pol polyprotein 316 3e-86
At1g36120 putative reverse transcriptase gb|AAD22339.1 286 5e-77
At4g08100 putative polyprotein 259 6e-69
At3g31970 hypothetical protein 218 1e-56
At1g20390 hypothetical protein 213 4e-55
At1g37060 Athila retroelment ORF 1, putative 211 2e-54
At4g04230 putative transposon protein 201 2e-51
At2g14400 putative retroelement pol polyprotein 194 2e-49
At2g15410 putative retroelement pol polyprotein 184 2e-46
At2g14040 putative retroelement pol polyprotein 184 3e-46
>At2g10780 pseudogene
Length = 1611
Score = 613 bits (1580), Expect = e-175
Identities = 381/900 (42%), Positives = 490/900 (54%), Gaps = 108/900 (12%)
Query: 78 FSGGFDPEEADLWLKELEKIFTFLRTTAEMKVDYETYLLTGEAEYWW------RGARAMM 131
F G DP AD W L++ F R + + D + L G+A WW RG
Sbjct: 182 FMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKRRGDEVRS 241
Query: 132 EADHQAITWECFRGAFLDKYFPRSARAAKEAQFLRLRQGGMTVAEYAAKLESLAKHFRYF 191
AD F F KYFP A E +L L QG TV EY + L ++
Sbjct: 242 FAD--------FEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRYVG-- 291
Query: 192 RGQIDEGYMCERFIEGLCYELQRAVQPLGLNRYQVLVEKTKGIE-AIDNARGKFQGLNKP 250
R +E RFI GL E++ N LVE+ IE I+ R LN+
Sbjct: 292 RELEEEQAQLRRFIRGLRIEIRNHCLVRTFNSVSELVERAAMIEEGIEEERY----LNR- 346
Query: 251 YQGSGGPARTNQGR--GDKGRHFQKKPYVRPQGRGTTSGSFYPTGGNAIALRTPSGN--R 306
P R NQ DK R F K + + TG + SG +
Sbjct: 347 ---EKAPIRNNQSTKPADKKRKFDKVDNTKSDAK---------TGECVTCGKNHSGTCWK 394
Query: 307 EDVTCFRCNKKGHYANHC-----------SESLAACWNCNKPGHTAAECRIPKVEAAANV 355
C RC K H C E C+ C K GH EC PK+ A
Sbjct: 395 AIGACGRCGSKDHAIQSCPKMEPGQSKVLGEETRTCFYCGKTGHLKREC--PKLTAEKQ- 451
Query: 356 AGAR-------------RPTAGGRVYSISGTEAEEDDGLIRSTCEIAGNSLIALFDSGAT 402
AG R R RVY +S E D G R+ I G F
Sbjct: 452 AGQRDNRGGNGLPPPPKRQAVASRVYELS--EEANDAGNFRA---ITGG-----FRKEPN 501
Query: 403 HSFIDIACAARLKLEVSKLPFELTVSTPASKSLVTNTACLECPWMYLDKKFVANLICLPL 462
+ + A + + L ++V V N + A+LI +PL
Sbjct: 502 TDYGMVRAAGGQAMYPTGLVRGISV--------VVNGVNMP-----------ADLIIVPL 542
Query: 463 KGLDVIIGMDWLSHHHVLLDCANKVVIFP-DAGLAEF-----------LNSYFSKLSLRK 510
K DVI+GMDWL + +DC + F D G+ +F +++ ++ L K
Sbjct: 543 KKHDVILGMDWLGKYKAHIDCHRGRIQFERDEGMLKFQGIRTTSGSLVISAIQAERMLGK 602
Query: 511 GALSSLMSTTVVEAKENG-VHGIAVVQDFEDVFPEDVPGIPPVRDMEFTIDIVPGTGPIS 569
G + L + T E + + I +V +F DVF V G+PP R FTI++ PGT PIS
Sbjct: 603 GCEAYLATITTKEVGASAELKDIPIVNEFSDVFAA-VSGVPPDRSDPFTIELEPGTTPIS 661
Query: 570 IAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRSRLCVDYRQLNKV 629
APYRMAPAE+ +LK QLE+L KGFIRPS SPWGAPVL VKKKDG RLC+DYR LNKV
Sbjct: 662 KAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKDGSFRLCIDYRGLNKV 721
Query: 630 TIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVM 689
T+KN+YPLPRID+LMDQL GA FSKIDL SGYHQI ++ D++KTAFRTRY H+E++VM
Sbjct: 722 TVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVM 781
Query: 690 PFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVL 749
PFG+TNAPA FM MN +F FLD FV++FI+DIL+YS++ E H+EHLR VL+ LR+ L
Sbjct: 782 PFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLREHEL 841
Query: 750 YANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRR 809
+A +KC FW V FLGHVIS +G++VDP K+ ++ W RP+ T+IRSF+GLAGYYRR
Sbjct: 842 FAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRR 901
Query: 810 FIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDA 869
F+ FA +A PLT+LT K+ F W+++CE+SF ++K LT APVL LP+E EPY VY DA
Sbjct: 902 FVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLPEEGEPYTVYTDA 961
Query: 870 SYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYLYGSTFTVFS 929
S GLGCVLMQ +AY+SRQL+ HE+NYPTHDLE+AAVVF LKIWR YLYG+ +++
Sbjct: 962 SIVGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYT 1021
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 573 bits (1477), Expect = e-163
Identities = 278/489 (56%), Positives = 361/489 (72%), Gaps = 16/489 (3%)
Query: 455 ANLICLPLKGLDVIIGMDWLSHHHVLLDCANKVVIF-------------PDAGLAEFLNS 501
A+LI P++ DVI+GMDWL H+ V LDC V F P +G + +++
Sbjct: 393 ADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPTSG-SLVISA 451
Query: 502 YFSKLSLRKGALSSLMSTTVVEAK-ENGVHGIAVVQDFEDVFPEDVPGIPPVRDMEFTID 560
+K + KG + L++ ++ E+ + V I V+Q+FEDVF + + G+PP R FTI+
Sbjct: 452 VQAKKMIEKGCEAYLVTISMPESLGQVAVSDIRVIQEFEDVF-QSLQGLPPSRSDPFTIE 510
Query: 561 IVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRSRLC 620
+ PGT P+S APYRMAPAE+TELK QLEDL KGFIRPS SPWGAPVL VKKKDG RLC
Sbjct: 511 LEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLC 570
Query: 621 VDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTR 680
+DYR LN VT+KN+YPLPRID+L+DQL+GA FSKIDL SGYHQI + + D++KTAFRTR
Sbjct: 571 IDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTR 630
Query: 681 YGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQV 740
YGH+E++VMPF +TNAPA FM MN +F FLD FV++FIDDIL+YS++ EEHE HLR+V
Sbjct: 631 YGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRV 690
Query: 741 LQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSF 800
++ LR++ L+A +KC FW E+ FLGH++S EG++VDP K+EA+ W RP T+IRSF
Sbjct: 691 MEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSF 750
Query: 801 IGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEK 860
+ L GYYRRF++GFA +A P+TKLT K+ PF W+ +CE+ F +K+ LT+ PVL LP+
Sbjct: 751 LRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHG 810
Query: 861 EPYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYL 920
+PY VY DAS GLGCVLMQ K +AY+SRQL+ HE NYPTHDLE+A V+FALKIWR YL
Sbjct: 811 QPYMVYTDASRVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYL 870
Query: 921 YGSTFTVFS 929
YG VF+
Sbjct: 871 YGGKVQVFT 879
Score = 92.8 bits (229), Expect = 8e-19
Identities = 80/282 (28%), Positives = 117/282 (41%), Gaps = 26/282 (9%)
Query: 41 AQNQAEENARRVQREERELAAAQTRGLNDFKRQDPPKFSGGFDPEEADLWLKELEKIFTF 100
A AE R EE + R + +R FS G PEEAD W +++ F
Sbjct: 104 APRMAEVQQRAAVAEE---VPSYLRMMEQLQRIGTWYFSSGTSPEEADSWRSRVQRNFGS 160
Query: 101 LRTTAEMKVDYETYLLTGEAEYWWRGARA-MMEADHQAITWECFRGAFLDKYFPRSARAA 159
R AE +VD + L G+A WWR A +AD ++W F F KYFP+ A
Sbjct: 161 SRCPAEYRVDLAVHFLEGDAHLWWRSVTARRRQAD---MSWADFVAEFNAKYFPQEALDR 217
Query: 160 KEAQFLRLRQGGMTVAEYAAKLESLAKHFRYFRGQIDEGYMCERFIEGLCYELQRAVQPL 219
EA+FL L QG +V EY + L + RG D+ RF+ GL +L+ +
Sbjct: 218 MEARFLELTQGEWSVREYDREFNRLLAYAG--RGMEDDQAQMRRFLRGLRPDLRVQCRVS 275
Query: 220 GLNRYQVLVEKTKGIEAIDNARGKFQGLNKPYQGSGGPARTNQGRGDKGRHFQKKPYVRP 279
LVE +E ++ + + G++ Q + +G K QK+ + P
Sbjct: 276 QYATKAALVETAAEVE--EDLQRQVVGVSPAVQTKNTQQQVTPSKGGKPAQGQKRKWDHP 333
Query: 280 Q--GRGTTSGSFYPTGGNAIALRTPSGNREDVTCFRCNKKGH 319
G+G +G F S D T C ++GH
Sbjct: 334 SRAGQGGRAGYF-------------SCGSLDHTVADCTERGH 362
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 572 bits (1473), Expect = e-163
Identities = 277/488 (56%), Positives = 364/488 (73%), Gaps = 14/488 (2%)
Query: 455 ANLICLPLKGLDVIIGMDWLSHHHVLLDCANKVVIF--PDAGLAE----------FLNSY 502
A+LI P++ DVI+GMDWL ++ V LD V F P+ L +++
Sbjct: 367 ADLIISPVELYDVILGMDWLDYYRVHLDWHRGRVFFERPEGRLVYQGVRPISGSLVISAV 426
Query: 503 FSKLSLRKGALSSLMSTTVVEAK-ENGVHGIAVVQDFEDVFPEDVPGIPPVRDMEFTIDI 561
++ + KG + L++ ++ E+ + V I VVQ+F+DVF + + G+PP + FTI++
Sbjct: 427 QAEKMIEKGCEAYLVTISMPESVGQVAVSDIRVVQEFQDVF-QSLQGLPPSQSDPFTIEL 485
Query: 562 VPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRSRLCV 621
PGT P+S APYRMAPAE+ ELK QL+DL KGFIRPS SPWGAPVL VKKKDG RLC+
Sbjct: 486 EPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLCI 545
Query: 622 DYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRY 681
DYR+LN+VT+KNRYPLPRID+L+DQL+GA FSKIDL SGYHQI + + D++KTAFRTRY
Sbjct: 546 DYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRY 605
Query: 682 GHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVL 741
GH+E++VMPFG+TNAPAVFM MN +F FLD FV++FIDDIL+YS++ EE E HLR+V+
Sbjct: 606 GHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVM 665
Query: 742 QVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFI 801
+ LR++ L+A +KC FW E+ FLGH++S EG++VDP K+EA+ W RP T+IRSF+
Sbjct: 666 EKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFL 725
Query: 802 GLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKE 861
G AGYYRRF++GFA +A P+TKLT K+ PF W+++CE+ F +K+ LT+ PVL LP+ +
Sbjct: 726 GWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTSTPVLALPEHGQ 785
Query: 862 PYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYLY 921
PY VY DAS GLGCVLMQH K +AY+SRQL HE NYPTHDLE+AAV+FALKIWR YLY
Sbjct: 786 PYMVYTDASRVGLGCVLMQHGKVIAYASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLY 845
Query: 922 GSTFTVFS 929
G VF+
Sbjct: 846 GGKVQVFT 853
Score = 64.7 bits (156), Expect = 2e-10
Identities = 60/240 (25%), Positives = 91/240 (37%), Gaps = 38/240 (15%)
Query: 49 ARRVQREERELAAAQT-----RGLNDFKRQDPPKFSGGFDPEEADLWLKELEKIFTFLRT 103
A RV ++ A A+ R + +R D FSGG PEEAD W + + F R
Sbjct: 118 APRVAEVQQRAAVAEEVLSYLRMMEQLQRIDTGYFSGGTSPEEADSWRSRVGRNFGSSRC 177
Query: 104 TAEMKVDYETYLLTGEAEYWWRGARAMMEADHQAITWECFRGAFLDKYFPRSARAAKEAQ 163
AE +VD + L G+A WWR A ++W F F KYFP+ A
Sbjct: 178 PAEYRVDLAVHFLEGDAHLWWRSVTARRR--QTDMSWADFVAEFKAKYFPQEA------- 228
Query: 164 FLRLRQGGMTVAEYAAKLESLAKHFRYFRGQIDEGYMCERFIEGLCYELQRAVQPLGLNR 223
+ YA +G D+ RF+ GL +L+ +
Sbjct: 229 ----------LDPYAG------------QGMEDDQAQMRRFLRGLRPDLRVRCRVSQYAT 266
Query: 224 YQVLVEKTKGIEAIDNARGKFQGLNKPYQGSGGPARTNQGRGDKGRHFQKKPYVRPQGRG 283
LVE +E ++ + + G++ Q + +G K QK+ + P G
Sbjct: 267 KAALVETAAEVE--EDFQRQVVGVSPVVQPKKTQQQVTPSKGGKPAQGQKRKWDHPSRAG 324
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 550 bits (1418), Expect = e-156
Identities = 275/479 (57%), Positives = 349/479 (72%), Gaps = 14/479 (2%)
Query: 455 ANLICLPLKGLDVIIGMDWLSHHHVLLDCANKVVIFP-DAGLAEF-----------LNSY 502
A+LI +PLK DVI+GMDWL + +DC + + F D G+ +F +++
Sbjct: 30 ADLIIVPLKKHDVILGMDWLGKYKGHIDCHRERIQFERDEGMLKFQGIRTTSGSLVISAI 89
Query: 503 FSKLSLRKGALSSLMSTTVVEAKENG-VHGIAVVQDFEDVFPEDVPGIPPVRDMEFTIDI 561
++ L KG + L + T E + + I +V +F DVF V G+PP R FTI++
Sbjct: 90 QAERMLGKGCEAYLATITTKEVGASAELKDILIVNEFSDVFAA-VSGVPPDRSDPFTIEL 148
Query: 562 VPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRSRLCV 621
PGT PIS APYRMAPAE+ ELK QLE+L KGFIRPS SPWGAPVL VKKKDG RLC+
Sbjct: 149 EPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKDGSFRLCI 208
Query: 622 DYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRY 681
DYR LNKVT+KN+YPLPRID+LMDQL GA FSKIDL SGYHQI ++ D++KTAFRTRY
Sbjct: 209 DYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRY 268
Query: 682 GHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVL 741
GH+E++VMPFG+TNAPA FM MN +F FLD FV++FIDDIL++S++ E H+EHLR VL
Sbjct: 269 GHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVL 328
Query: 742 QVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFI 801
+ LR+ L+A +K FW V FLGHVIS +G++VDP K+ ++ W RP+ T+IRSF+
Sbjct: 329 ERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFL 388
Query: 802 GLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKE 861
GLAGYYRRF+ FA +A PLT+LT K+ F W+++CE+SF ++K L APVL LP+E E
Sbjct: 389 GLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLPEEGE 448
Query: 862 PYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYL 920
PY VY DAS GLGCVLMQ +AY+SRQL+ HE+NYPTHDLE+AAVVFALKIWR YL
Sbjct: 449 PYTVYTDASIVGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFALKIWRSYL 507
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 542 bits (1396), Expect = e-154
Identities = 288/581 (49%), Positives = 373/581 (63%), Gaps = 28/581 (4%)
Query: 351 AAANVAGARRPTAGGRVYSISGTEAEEDDGLIRSTCEIAGNSLIALFDSGATHSFIDIAC 410
AAA R G +I+G A E + T + G LFDSGA+H FI
Sbjct: 274 AAAPQGCTTREIGGTSNRAITGFLAHE---VCVGTLLVGGVEAHVLFDSGASHCFITPES 330
Query: 411 AARLKLEVSKLPFELTVSTPASKSLVTNTACLECPWMYLDKKFVANLICLPLKGLDVIIG 470
A+R + V + + + A+LI P++ DVI+G
Sbjct: 331 ASRGNIRGDPGEQLGAVKVAGGQFVAVLGRTKGVDIQIAGESMPADLIISPVELYDVILG 390
Query: 471 MDWLSHHHVLLDCANKVVIF--PDAGLAEFLNSYFSKLSLRKGALSSLMSTTVVEAKENG 528
MDWL H+ V LDC V F P+ L + + G+L + V+A++
Sbjct: 391 MDWLDHYRVHLDCHRGRVSFERPEGSLV------YQGVRPTSGSLV----ISAVQAEKMI 440
Query: 529 VHGIAVVQDFEDVFPEDVPGIPPVRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLE 588
G +FEDVF + + G+PP R FTI++ PGT P+S APYRMAPAE+ ELK QLE
Sbjct: 441 EKGCEAYLEFEDVF-QSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLE 499
Query: 589 DLTKKGFIRPSVSPWGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLK 648
DL GAPVL VKKKDG RLC+DYR LN VT+KN+YPLPRID+L+DQL+
Sbjct: 500 DLL------------GAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLR 547
Query: 649 GAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIF 708
GA FSKIDL SGYH I + + D++KTAFRTRYGH+E++VMPFG+TNAPA FM MN +F
Sbjct: 548 GATCFSKIDLTSGYHLIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVF 607
Query: 709 HPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGH 768
LD FV++FIDDIL+YS++ EEHE HLR+V++ LR++ L+A +KC FW E+ FLGH
Sbjct: 608 QEVLDEFVIIFIDDILVYSKSLEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGH 667
Query: 769 VIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKN 828
++S EG++VDP K+EA+ W P T+IRSF+GLAGYYRRF++GFA +A P+TKLT K+
Sbjct: 668 IVSAEGVSVDPEKIEAIRDWHTPTNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKD 727
Query: 829 QPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHRKAVAYS 888
PF W+ +CE+ F +K+ LT+ PVL LP+ EPY VY DAS GLGCVLMQ K +AY+
Sbjct: 728 VPFVWSPECEEGFVSLKEMLTSTPVLALPEHGEPYMVYTDASGVGLGCVLMQRGKVIAYA 787
Query: 889 SRQLKIHERNYPTHDLELAAVVFALKIWRHYLYGSTFTVFS 929
SRQL+ HE NYPTHDLE+AAV+FALKIWR YLYG VF+
Sbjct: 788 SRQLRKHEGNYPTHDLEMAAVIFALKIWRSYLYGGNVQVFT 828
Score = 73.2 bits (178), Expect = 6e-13
Identities = 47/128 (36%), Positives = 61/128 (46%), Gaps = 9/128 (7%)
Query: 49 ARRVQREERELAAAQT-----RGLNDFKRQDPPKFSGGFDPEEADLWLKELEKIFTFLRT 103
A RV ++ A A+ R + +R FSGG PE AD W +E+ F R
Sbjct: 122 APRVAEVQQRAAVAEEVPSYLRMMEQLQRIGTGYFSGGTSPEVADSWRSRVERNFGSSRC 181
Query: 104 TAEMKVDYETYLLTGEAEYWWRGARA-MMEADHQAITWECFRGAFLDKYFPRSARAAKEA 162
AE ++D + L G+A WWR A +AD I+W F F KYFP+ A EA
Sbjct: 182 PAEYRIDLAVHFLEGDAHLWWRSVTARRRQAD---ISWADFVAEFNAKYFPQEALDRMEA 238
Query: 163 QFLRLRQG 170
FL L QG
Sbjct: 239 HFLELTQG 246
>At3g11970 hypothetical protein
Length = 1499
Score = 431 bits (1107), Expect = e-120
Identities = 305/967 (31%), Positives = 470/967 (48%), Gaps = 89/967 (9%)
Query: 4 MEQAIANLTALMAQQVTNTAAREEAEAQRAATAALVAAQNQAEENARRVQREERELAAAQ 63
M + A+L A++++ E++ R+A + + +N++ +++ +
Sbjct: 40 MMKQFADLYAVLSRSTAGKMVDEQSTLDRSAPRSSQSMENRSGYPDPYRDARHQQVRSDH 99
Query: 64 TRGLNDFKRQDPPKFSGGFDPEEADLWLKELEKIFTFLRTTAEMKVDYETYLLTGEAEYW 123
N+ R F FD WL ++E+ F T +MKV A W
Sbjct: 100 FNAYNNLTRLGKIDFPR-FDGTRLKEWLFKVEEFFGVDSTPEDMKVKMAAIHFDSHASTW 158
Query: 124 WRGARAMMEADHQAITWECFRGAFLDKYFPRSARAAKEAQFLRLRQGGMTVAEYAAKLES 183
+ W+ + +++ E + L+ G + +Y K E
Sbjct: 159 HQSFIQSGVGLEVLYDWKGYVKLLKERFEDDCDDPMAELKHLQETDG---IIDYHQKFEL 215
Query: 184 LAKHFRYFRGQIDEGYMCERFIEGLCYELQ---RAVQPLGLNRYQVLVEKTKGIEAIDNA 240
+ R + E Y+ ++ GL + Q R QP + L
Sbjct: 216 IKT-----RVNLSEEYLVSVYLAGLRTDTQMHVRMFQPQTVRHCLFL------------- 257
Query: 241 RGKFQGLNKPYQGSGGPARTNQGRGDKGRHFQKKPYVRPQGRGTTSGSFYPTGGNA--IA 298
GK P + + TN+ G + +K G+F P ++
Sbjct: 258 -GKTYEKAHPKKPANTTWSTNRSAPTGGYNKYQKEGESKTDHYGNKGNFKPVSQQPKKMS 316
Query: 299 LRTPSGNREDVTCFRCNKK---GHYANHCSESLAACWNCNKPGHTAAECRI-------PK 348
+ S R C+ C++K HY H L + ++ A E + P+
Sbjct: 317 QQEMSDRRSKGLCYFCDEKYTPEHYLVHKKTQLFRM-DVDEEFEDAREELVNDDDEHMPQ 375
Query: 349 VEAAANVAGARRPTAGGRVYSISGTEAEEDDGLIRSTCEIAGNSLIALFDSGATHSFIDI 408
+ A V+G AG + + GT ++ + L DSG+TH+F+D
Sbjct: 376 ISVNA-VSGI----AGYKTMRVKGTYDKK--------------IIFILIDSGSTHNFLDP 416
Query: 409 ACAARLKLEVSKLPFELTVSTPASKSLVTNTACLECPWMYLDKKFVANLICLPLKGLDVI 468
AA+L +V VS + L + W F ++++ +PL+G+D++
Sbjct: 417 NTAAKLGCKVDTAGLT-RVSVADGRKLRVEGKVTDFSWKLQTTTFQSDILLIPLQGIDMV 475
Query: 469 IGMDWL------SHHHVLLDCA-----NKVVI--FPDAGLAEFLNSYFSKLSLRKGALSS 515
+G+ WL S L+ KV++ + E KL + L+
Sbjct: 476 LGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVREVKAQKLQKLQEDQVQLAM 535
Query: 516 LMSTTVVEAKENGVHGI--------------AVVQDFEDVFPEDVPGIPPVRDME-FTID 560
L V E+ E + I V+ ++ D+F E +PP R+ I
Sbjct: 536 LCVQEVSESTEGELCTINALTSELGEESVVEEVLNEYPDIFIEPT-ALPPFREKHNHKIK 594
Query: 561 IVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRSRLC 620
++ G+ P++ PYR + + E+ +EDL G ++ S SP+ +PV+LVKKKDG RLC
Sbjct: 595 LLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPYASPVVLVKKKDGTWRLC 654
Query: 621 VDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTR 680
VDYR+LN +T+K+ +P+P I+DLMD+L GA IFSKIDLR+GYHQ+R+ +DIQKTAF+T
Sbjct: 655 VDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTH 714
Query: 681 YGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQV 740
GH+EYLVMPFG+TNAPA F MN IF PFL +FV+VF DDIL+YS + EEH +HL+QV
Sbjct: 715 SGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQV 774
Query: 741 LQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSF 800
+V+R L+A +KC F + +V++LGH IS +GI DP+K++AV W +P T+ +R F
Sbjct: 775 FEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRGF 834
Query: 801 IGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEK 860
+GLAGYYRRF+ F IAGPL LT K F WT +Q+F+D+K L APVL+LP
Sbjct: 835 LGLAGYYRRFVRSFGVIAGPLHALT-KTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFD 893
Query: 861 EPYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYL 920
+ + V DA QG+G VLMQ +AY SRQLK + + ++ EL AV+FA++ WRHYL
Sbjct: 894 KQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYL 953
Query: 921 YGSTFTV 927
S F +
Sbjct: 954 LQSHFII 960
>At1g36590 hypothetical protein
Length = 1499
Score = 431 bits (1107), Expect = e-120
Identities = 306/967 (31%), Positives = 473/967 (48%), Gaps = 89/967 (9%)
Query: 4 MEQAIANLTALMAQQVTNTAAREEAEAQRAATAALVAAQNQAEENARRVQREERELAAAQ 63
M + A+L A++++ E++ R+A + + +N++ +++ +
Sbjct: 40 MMKQFADLYAVLSRSTAGKMVDEQSTLDRSAPRSSQSMENRSGYLDPYRDARHQQVRSDH 99
Query: 64 TRGLNDFKRQDPPKFSGGFDPEEADLWLKELEKIFTFLRTTAEMKVDYETYLLTGEAEYW 123
N+ R F FD WL ++E+ F T +MKV A W
Sbjct: 100 FNAYNNLTRLGKIDFPR-FDGTRLKEWLFKVEEFFGVDSTPEDMKVKMAAIHFDSHASTW 158
Query: 124 WRGARAMMEADHQAITWECFRGAFLDKYFPRSARAAKEAQFLRLRQGGMTVAEYAAKLES 183
+ W+ + +++ E + L+ G + +Y K E
Sbjct: 159 HQSFIQSGVGLEVLYDWKGYVKLLKERFEDDCDDPMAELKHLQETDG---IIDYHQKFEL 215
Query: 184 LAKHFRYFRGQIDEGYMCERFIEGLCYELQ---RAVQPLGLNRYQVLVEKTKGIEAIDNA 240
+ R + E Y+ ++ GL + Q R QP + R+ + + KT +
Sbjct: 216 IKT-----RVNLSEEYLVSVYLAGLRTDTQMHVRMFQPQTV-RHCLFLGKTYEKAHLKKP 269
Query: 241 RGKFQGLNKPYQGSGGPARTNQGRGDKGRHFQKKPYVRPQGRGTTSGSFYPTGGNA--IA 298
N+ G +G K H+ K G+F P ++
Sbjct: 270 ANTTWSTNRSAPTGGYNKYQKEGES-KTDHYGNK------------GNFKPVSQQPKKMS 316
Query: 299 LRTPSGNREDVTCFRCNKK---GHYANHCSESLAACWNCNKPGHTAAECRI-------PK 348
+ S R C+ C++K HY H L + ++ A E + P+
Sbjct: 317 QQEMSDRRSKGLCYFCDEKYTPEHYLVHKKTQLFRM-DVDEEFEDAREELVNDDDEHMPQ 375
Query: 349 VEAAANVAGARRPTAGGRVYSISGTEAEEDDGLIRSTCEIAGNSLIALFDSGATHSFIDI 408
+ A V+G AG + + GT ++ + L DSG+TH+F+D
Sbjct: 376 ISVNA-VSGI----AGYKTMRVKGTYDKK--------------IIFILIDSGSTHNFLDP 416
Query: 409 ACAARLKLEVSKLPFELTVSTPASKSLVTNTACLECPWMYLDKKFVANLICLPLKGLDVI 468
AA+L +V VS + L + W F ++++ +PL+G+D++
Sbjct: 417 NTAAKLGCKVDTAGLT-RVSVADGRKLRVEGKVTDFSWKLQTTTFQSDILLIPLQGIDMV 475
Query: 469 IGMDWL------SHHHVLLDCA-----NKVVI--FPDAGLAEFLNSYFSKLSLRKGALSS 515
+G+ WL S L+ KV++ + E KL + L+
Sbjct: 476 LGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVREVKAQKLQKLQEDQVQLAM 535
Query: 516 LMSTTVVEAKENGVHGI--------------AVVQDFEDVFPEDVPGIPPVRDME-FTID 560
L V E+ E + I V+ ++ D+F E +PP R+ I
Sbjct: 536 LCVQEVSESTEGELCTINALTSELGEESVVEEVLNEYPDIFIEPT-ALPPFREKHNHKIK 594
Query: 561 IVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRSRLC 620
++ G+ P++ PYR + + E+ +EDL G ++ S SP+ +PV+LVKKKDG RLC
Sbjct: 595 LLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPYASPVVLVKKKDGTWRLC 654
Query: 621 VDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTR 680
VDYR+LN +T+K+ +P+P I+DLMD+L GA IFSKIDLR+GYHQ+R+ +DIQKTAF+T
Sbjct: 655 VDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTH 714
Query: 681 YGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQV 740
GH+EYLVMPFG+TNAPA F MN IF PFL +FV+VF DDIL+YS + EEH +HL+QV
Sbjct: 715 SGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQV 774
Query: 741 LQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSF 800
+V+R L+A +KC F + +V++LGH IS +GI DP+K++AV W +P T+ +R F
Sbjct: 775 FEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRGF 834
Query: 801 IGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEK 860
+GLAGYYRRF+ F IAGPL LT K F WT +Q+F+D+K L APVL+LP
Sbjct: 835 LGLAGYYRRFVRSFGVIAGPLHALT-KTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFD 893
Query: 861 EPYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYL 920
+ + V DA QG+G VLMQ +AY SRQLK + + ++ EL AV+FA++ WRHYL
Sbjct: 894 KQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYL 953
Query: 921 YGSTFTV 927
S F +
Sbjct: 954 LQSHFII 960
>At2g06890 putative retroelement integrase
Length = 1215
Score = 404 bits (1038), Expect = e-112
Identities = 272/831 (32%), Positives = 404/831 (47%), Gaps = 112/831 (13%)
Query: 150 KYFPRSARAAKEAQFLRLRQGGMTVAEYAAKLESLAKHFRYFRGQIDEGYMCERFIEGLC 209
++ P + L QG +V EY ++E+L R I E E +
Sbjct: 8 RFVPSHYHRELHLKLRNLTQGNRSVEEYYKEMETLM-----LRADISEDR--EATLSRFL 60
Query: 210 YELQRAVQPLGLNRYQVLVEKTKGIEAIDNARGKFQGLNKPYQGSGGPARTNQGRGDKGR 269
+L R +Q +Y V +E+ + + K + ++ GSG A+ R ++
Sbjct: 61 GDLNRDIQDRLETQYYVQIEEMLHKAILFEQQVKRKSSSRSSYGSGTIAKPTYQREERTS 120
Query: 270 HFQKKPYVRPQGRGTTSGSFYPTGGNAIALRTPSGNREDVTCFRCNKKGHYANHCSESLA 329
+ KP V P+ + G A + DV C++C KGHYAN C
Sbjct: 121 SYHNKPIVSPRAESKPYAAVQDHKGKA---EISTSRVRDVRCYKCQGKGHYANECPNKRV 177
Query: 330 ACWNCNKPGHTAAECRIP----------KVEAAANVAGARRPTAGGRVYSISGTEAEEDD 379
N G E IP ++ A + ARR + E E+
Sbjct: 178 MILLDN--GEIEPEEEIPDSPSSLKENEELPAQGELLVARRTLS----VQTKTDEQEQRK 231
Query: 380 GLIRSTCEIAGNSLIALFDSGATHSFIDIACAARLKLEVSKLPFELTVSTPASKSLVTNT 439
L + C + G + D G+ + +L L+ ++ V +V
Sbjct: 232 NLFHTRCHVHGKVCSLIIDGGSCTNVASETMVKKLGLKWLNDSGKMRVKNQVVVPIVIG- 290
Query: 440 ACLECPWMYLDKKFVANLIC--LPLKGLDVIIGMDWLSHHHVLLDC-------------- 483
K+ ++C LP++ +++G W S V+ D
Sbjct: 291 ------------KYEDEILCDVLPMEAGHILLGRPWQSDRKVMHDGFTNRHSFEFKGGKT 338
Query: 484 --------------------ANKVVIFPD--AGLAEFLNSYFSK----LSLRKGALSSLM 517
+VV P+ A E ++Y SK L + K AL+SL
Sbjct: 339 ILVSMTPHEVYQDQIHLKQKKEQVVKQPNFFAKSGEVKSAYSSKQPMLLFVFKEALTSLT 398
Query: 518 STTVVEAKENGVHGIAVVQDFEDVFPEDVP-GIPPVRDMEFTIDIVPGTGPISIAPYRMA 576
+ V E +++QD++DVFPED P G+PP+R +E ID VPG + YR
Sbjct: 399 NFAPVLPSEM----TSLLQDYKDVFPEDNPKGLPPIRGIEHQIDFVPGASLPNRPAYRTN 454
Query: 577 PAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNRYP 636
P E EL+ Q KDG R+C D R +N VT+K +P
Sbjct: 455 PVETKELQRQ--------------------------KDGSWRMCFDCRAINNVTVKYCHP 488
Query: 637 LPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNA 696
+PR+DD++D+L G++IFSKIDL+SGYHQIR+ + D KTAF+T++G YE+LVMPFG+T+A
Sbjct: 489 IPRLDDMLDELHGSSIFSKIDLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHA 548
Query: 697 PAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKC 756
P+ FM MN + F+ FV+V+ DDIL+YS + EH EHL VL VLR + LYAN KC
Sbjct: 549 PSTFMRLMNHVLRAFIGIFVIVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKC 608
Query: 757 EFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAK 816
F + + FLG V+S +G+ VD KV+A+ W PKTV ++RSF GLAG+YRRF + F+
Sbjct: 609 TFCTDNLVFLGFVVSADGVKVDEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFST 668
Query: 817 IAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGC 876
I PLT++ +K+ F W + E++FQ +K +LT APVL L + + +E+ CDAS G+G
Sbjct: 669 IVAPLTEVMKKDVGFKWEKAQEEAFQSLKDKLTNAPVLILSEFLKTFEIECDASGIGIGA 728
Query: 877 VLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYLYGSTFTV 927
VLMQ +K +A+ S +L NYPT+D EL A+V AL+ W+HYL+ F +
Sbjct: 729 VLMQDQKLIAFFSEKLGGATLNYPTYDKELYALVRALQRWQHYLWPKVFVI 779
>At1g35370 hypothetical protein
Length = 1447
Score = 377 bits (969), Expect = e-104
Identities = 229/615 (37%), Positives = 339/615 (54%), Gaps = 58/615 (9%)
Query: 349 VEAAANVAGARRPTAGGRVYSISGTEAEEDDGLIRSTCEIAGNSLIALFDSGATHSFIDI 408
VE ++ ++P V ++SG + G ++ T + L L DSG+TH+FID
Sbjct: 339 VEVLSDDDHEQKPMPQISVNAVSGISGYKTMG-VKGTVD--KRDLFILIDSGSTHNFIDS 395
Query: 409 ACAARLKLEVSKLPFELTVSTPASKSLVTNTACLECPWMYLDKKFVANLICLPLKGLDVI 468
AA+L V V+ + L + W F ++++ +PL+G+D++
Sbjct: 396 TVAAKLGCHVESAGLT-KVAVADGRKLNVDGQIKGFTWKLQSTTFQSDILLIPLQGVDMV 454
Query: 469 IGMD--------------------------WLSHHHVLLDCANKVVIFPDAGLAEFLNSY 502
+G+ WL H ++ + + + +
Sbjct: 455 LGVQWLETLGRISWEFKKLEMQFFYKNQRVWL---HGIITGSVRDIKAHKLQKTQADQIQ 511
Query: 503 FSKLSLRK---------GALSSLMSTTVVEAKENGVHGIAVVQDFEDVFPEDVPGIPPVR 553
+ + +R+ G++S+L S V E+ + V++F DVF E +PP R
Sbjct: 512 LAMVCVREVVSDEEQEIGSISALTSDVVEESVVQNI-----VEEFPDVFAEPTD-LPPFR 565
Query: 554 DM-EFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKK 612
+ + I ++ G P++ PYR + E+ ++D+ K G I+ S SP+ +PV+LVKK
Sbjct: 566 EKHDHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSPFASPVVLVKK 625
Query: 613 KDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDI 672
KDG RLCVDY +LN +T+K+R+ +P I+DLMD+L G+ +FSKIDLR+GYHQ+R+ +DI
Sbjct: 626 KDGTWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDI 685
Query: 673 QKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREE 732
QKTAF+T GH+EYLVM FG+TNAPA F MN +F FL +FV+VF DDILIYS + EE
Sbjct: 686 QKTAFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEE 745
Query: 733 HEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPK 792
H+EHLR V +V+R L+A +K + LGH IS I DP+K++AV W P
Sbjct: 746 HKEHLRLVFEVMRLHKLFAKGSK--------EHLGHFISAREIETDPAKIQAVKEWPTPT 797
Query: 793 TVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAP 852
TV +R F+G AGYYRRF+ F IAGPL LT K F W+ + + +F +K L AP
Sbjct: 798 TVKQVRGFLGFAGYYRRFVRNFGVIAGPLHALT-KTDGFCWSLEAQSAFDTLKAVLCNAP 856
Query: 853 VLTLPQEKEPYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFA 912
VL LP + + V DA QG+ VLMQ +AY SRQLK + + ++ EL A +FA
Sbjct: 857 VLALPVFDKQFMVETDACGQGIRAVLMQKGHPLAYISRQLKGKQLHLSIYEKELLAFIFA 916
Query: 913 LKIWRHYLYGSTFTV 927
++ WRHYL S F +
Sbjct: 917 VRKWRHYLLPSHFII 931
>At4g03650 putative reverse transcriptase
Length = 839
Score = 355 bits (912), Expect = 5e-98
Identities = 160/261 (61%), Positives = 206/261 (78%)
Query: 669 DEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSR 728
+ D++KTAFRTRYGH+E++VMPFG+TNAPA FM MN +F FLD FV++FIDDIL+YS+
Sbjct: 518 EADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSK 577
Query: 729 NREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAW 788
+ EEHE HLR+V++ LR++ L+A +KC FW E+ FLGH++S EG++VDP K+EA+ W
Sbjct: 578 SPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDW 637
Query: 789 ERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRL 848
RP T+IRSF+GLAGYYRRFI+GFA +A P+TKLT K+ PF W+ +CE+ F +K+ L
Sbjct: 638 PRPTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEML 697
Query: 849 TTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAA 908
T+ PVL LP+ EPY VY DAS GLGC LMQ K +AY+SRQL+ HE NYPTHDLE+AA
Sbjct: 698 TSTPVLALPEHGEPYMVYTDASGVGLGCALMQRGKVIAYASRQLRKHEGNYPTHDLEMAA 757
Query: 909 VVFALKIWRHYLYGSTFTVFS 929
V+FALKIWR YLYG VF+
Sbjct: 758 VIFALKIWRSYLYGGKVQVFT 778
Score = 92.4 bits (228), Expect = 1e-18
Identities = 62/170 (36%), Positives = 82/170 (47%), Gaps = 10/170 (5%)
Query: 44 QAEENARRVQREERELAAAQT----RGLNDFKRQDPPKFSGGFDPEEADLWLKELEKIFT 99
QA R V+ ++R A + R + +R FSGG PEEAD W ++E+ F
Sbjct: 118 QAPVAPRVVEVQQRAAVAEEVPSYLRMMKQLQRIGTEYFSGGTSPEEADSWRSQVERNFG 177
Query: 100 FLRTTAEMKVDYETYLLTGEAEYWWRGARA-MMEADHQAITWECFRGAFLDKYFPRSARA 158
R AE +VD + L G+A WWR A +AD ++W F F KYFPR A
Sbjct: 178 SSRCPAEYRVDLTVHFLEGDAHLWWRSVTARRRQAD---MSWADFMAEFNAKYFPREALD 234
Query: 159 AKEAQFLRLRQGGMTVAEYAAKLESLAKHFRYFRGQIDEGYMCERFIEGL 208
EA+FL L QG +V EY K L + RG D+ RF+ GL
Sbjct: 235 RMEARFLELTQGVRSVREYDRKFNRLLVYAG--RGMEDDQAQMRRFLRGL 282
Score = 88.2 bits (217), Expect = 2e-17
Identities = 59/145 (40%), Positives = 81/145 (55%), Gaps = 10/145 (6%)
Query: 455 ANLICLPLKGLDVIIGMDWLSHHHVLLDCANKVVIFPDAGLAEFLNSYFSKLSLRKGALS 514
A+LI ++ DVI+GMDWL H+ V LDC V F E + + R+G L
Sbjct: 386 ADLIISHVELYDVILGMDWLDHYRVHLDCHRGRVSF------ERPEGSAGRENDREG-LR 438
Query: 515 SLMSTTVVEAK--ENGVHGIAVVQDFEDVFPEDVPGIPPVRDMEFTIDIVPGTGPISIAP 572
L + + V I VVQ+F+ VF + + G+PP R FTI++ P T P+S AP
Sbjct: 439 GLPGDDIYAGVCGQVAVSDIRVVQEFQYVF-QSLQGLPPSRSDPFTIELEPKTAPLSKAP 497
Query: 573 YRMAPAELTELKSQLEDLTKKGFIR 597
YRMAPA++ ELK QLEDL + +R
Sbjct: 498 YRMAPAKMAELKKQLEDLLAEADVR 522
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 316 bits (810), Expect = 3e-86
Identities = 169/395 (42%), Positives = 234/395 (58%), Gaps = 54/395 (13%)
Query: 534 VVQDFEDVFPEDVPGIPPVR-DMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTK 592
VV +F D+F E +PP R + I+++ + P++ PYR + E+ ++D+
Sbjct: 10 VVTEFPDIFVEPTD-LPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKIVDDMLA 68
Query: 593 KGFIRPSVSPWGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAI 652
G I+ S SP+ +PV+LVKKKDG RLCVDYR+LN +T+K+R+P+P I+DLMD+L G+ +
Sbjct: 69 SGTIQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNV 128
Query: 653 FSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFL 712
+SKIDLR+GYHQ+R+ DI KTAF+T GHYEYLVMPFG+TNAPA F MN F PFL
Sbjct: 129 YSKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKPFL 188
Query: 713 DRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS* 772
+FV+VF DDILIYS + EEH++HL V +V+R L+A +KC F + V++LGH IS
Sbjct: 189 RKFVLVFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEYLGHFISG 248
Query: 773 EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFA 832
EGIA DP+K++AV W P + + F+GL GYYRRF
Sbjct: 249 EGIATDPAKIKAVQDWPVPVNLKQLCGFLGLTGYYRRF---------------------- 286
Query: 833 WTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHRKAVAYSSRQL 892
+ V DA G+G VLMQ +A+ SRQL
Sbjct: 287 ------------------------------FVVEMDACGHGIGAVLMQEGHPLAFISRQL 316
Query: 893 KIHERNYPTHDLELAAVVFALKIWRHYLYGSTFTV 927
K + + ++ EL AV+F ++ WRHYL S F +
Sbjct: 317 KGKQLHLSIYEKELLAVIFVVRKWRHYLISSHFII 351
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 286 bits (731), Expect = 5e-77
Identities = 213/607 (35%), Positives = 291/607 (47%), Gaps = 94/607 (15%)
Query: 143 FRGAFLDKYFPRSARAAKEAQFLRLRQGGMTVAEYAAKLESLAKHFRYFRGQIDEGYMCE 202
F F KYFP+ A EA+FL L +G ++V EY + + L + RG D+
Sbjct: 157 FVAEFNAKYFPQEALDRMEARFLELTKGELSVREYGREFKRLLVYAG--RGMEDDQAQMR 214
Query: 203 RFIEGLCYELQRAVQPLGLNRYQVLVEKTKGIEAIDNARGKFQGLNKPYQGSGGPARTNQ 262
RF+ GL +L+ + +++Y T E ++ + + ++ Q +
Sbjct: 215 RFLRGLRPDLRVRCR---VSQYATKAALTAA-EVEEDLQRQVVAVSPSVQPKKIQQQVAP 270
Query: 263 GRGDKGRHFQKKPYVRPQGRGTTSGSFYPTGGNAIALRTPSGNREDVTCFRCNKKGHYAN 322
+G K QK+ + P G GG A G A+
Sbjct: 271 SKGGKPAQVQKRKWDHPFRAGQ--------GGRA---------------------GSAAD 301
Query: 323 HCSESLAACWNCNKPGHTAAECRIPKVEAAANVAGARRPTAGGRVYSI--SGTEAEEDDG 380
S S A AA AGA R A S SGT
Sbjct: 302 DSSTSTAG--------------------GAAQSAGAARSAAECAAVSTYCSGTTWLYYAD 341
Query: 381 LIRSTCEIAGNSLIALFDSGATHSFIDIACAARLKLEVSKLPFELTVSTPASKSLVTNTA 440
LI C G+ LFDSGA+H FI A+R + V + L
Sbjct: 342 LISGRCR--GH---VLFDSGASHCFITPESASRDNIRGEPGEQFGAVKIAGGQFLTVLGR 396
Query: 441 CLECPWMYLDKKFVANLICLPLKGLDVIIGMDWLSHHHVLLDCANKVVIF--PDAGLAE- 497
++ A+LI P+ D I+GMDWL H+ V LDC V F P+ L
Sbjct: 397 AKGVNIQIEEESMPADLIINPVVLYDAILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQ 456
Query: 498 ---------FLNSYFSKLSLRKGALSSLMSTTVVEAK-ENGVHGIAVVQDFEDVFPEDVP 547
++ + + KG + L++ ++ E+ + V I VVQ+FEDVF + +
Sbjct: 457 GVRPTSRSLVISEVQVEKMIEKGCEAYLVTISMSESVGQVAVSDIWVVQEFEDVF-QSLQ 515
Query: 548 GIPPVRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPV 607
G+PP R FTI++ GT P+S PYRM PAE+ ELK QLEDL KGFIRPS S WGAP
Sbjct: 516 GLPPSRSDPFTIELELGTAPLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPSTSRWGAP- 574
Query: 608 LLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRV 667
LN+VT+KN+YPLPRID+L+DQL+GA FSKIDL GYHQ +
Sbjct: 575 -----------------GLNRVTLKNKYPLPRIDELLDQLRGATCFSKIDLTPGYHQFPI 617
Query: 668 KDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYS 727
+ D++KTAFRTRYGH+E++VMPFG+TNAP M MN +F FLD FV++FIDDIL+Y
Sbjct: 618 AEADVRKTAFRTRYGHFEFVVMPFGLTNAPTALMRLMNSVFQEFLDEFVIIFIDDILVYF 677
Query: 728 RNREEHE 734
++ EEHE
Sbjct: 678 KSPEEHE 684
Score = 89.4 bits (220), Expect = 9e-18
Identities = 46/118 (38%), Positives = 61/118 (50%), Gaps = 39/118 (33%)
Query: 812 EGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASY 871
+GFA +A P+TKLT K+ PF W+ +CE+ F+
Sbjct: 691 KGFASMAQPMTKLTEKDIPFVWSPECEEGFRG---------------------------- 722
Query: 872 QGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYLYGSTFTVFS 929
K + Y+SRQL+ HE NYPTHDLE+AA++FALKIW YLYG VF+
Sbjct: 723 -----------KVIVYASRQLRKHEGNYPTHDLEMAAIIFALKIWGSYLYGGKVQVFT 769
>At4g08100 putative polyprotein
Length = 1054
Score = 259 bits (661), Expect = 6e-69
Identities = 134/306 (43%), Positives = 187/306 (60%), Gaps = 30/306 (9%)
Query: 622 DYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRY 681
D R +N +T+K R+P+PR+DD +D+L G++IFSKIDL+SGYHQ R+K+ D KTA +T+
Sbjct: 527 DCRAINNITVKYRHPIPRLDDTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEWKTAIKTKQ 586
Query: 682 GHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVL 741
YE+LVMPFG+TNAP FM MN + + FV+V+ DDIL+YS+N E H HL+ VL
Sbjct: 587 RLYEWLVMPFGLTNAPNTFMRLMNHVLRKHIGVFVIVYFDDILVYSKNLEYHVMHLKLVL 646
Query: 742 QVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFI 801
+LR + LYAN KC F + + FLG V+S +GI VD KV+A+ W PK V++
Sbjct: 647 DLLRKEKLYANLKKCTFCTDNLVFLGFVVSADGIKVDEEKVKAIREWPNPKNVSE----- 701
Query: 802 GLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKE 861
K+ F W + E +FQ +K++LT + V LP +
Sbjct: 702 -------------------------KDIGFKWEDAQENAFQALKEKLTNSSVPILPNFMK 736
Query: 862 PYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYLY 921
+E+ CDAS G+G VLMQ K +AY S +L NYPT+D EL A+V AL+ W+HYL+
Sbjct: 737 SFEIECDASGLGIGAVLMQDHKPIAYFSEKLGGATLNYPTYDKELYALVRALQTWQHYLW 796
Query: 922 GSTFTV 927
F +
Sbjct: 797 PKKFVI 802
Score = 51.2 bits (121), Expect = 3e-06
Identities = 67/304 (22%), Positives = 108/304 (35%), Gaps = 29/304 (9%)
Query: 25 REEAEAQRAATAALVAAQNQAEENARRVQREERELAAAQTRGLNDFKRQDPPKFSGGFDP 84
RE+ EA+ + A + +++R R ER GL + P F G +P
Sbjct: 57 REDEEARNYYSHA-------SSHSSQRRHRRERPPPRDPLGGL----KLKIPAFHGTDNP 105
Query: 85 EEADLWLKELEKIFTFLRTTAEMKVDYETYLLTGEAEYWWRG--ARAMMEADHQAITWEC 142
+ W +++E +F KV A WW D+ TW
Sbjct: 106 DTYLEWEQKIELVFLCQECLQSNKVKIAATKFYNYALSWWDQLVTSRRRTRDYPIKTWNQ 165
Query: 143 FRGAFLDKYFPRSARAAKEAQFLRLRQGGMTVAEYAAKLESLAKHFRYFRGQIDEGYMCE 202
+ ++ P + L QG TV EY ++E+L Q D +
Sbjct: 166 LKFVMRKRFVPSYYHRELHQRLRNLVQGSKTVEEYFLEMETLMLRADL---QEDGEAVMS 222
Query: 203 RFIEGLCYELQRAVQPLGLNRYQVLVEKTKGIEAIDNARGKFQGLNKPYQGSGGPARTNQ 262
RF+ GL E+Q ++ + ++ K E + K SG P+ +
Sbjct: 223 RFMGGLNREIQDRLETQHYVELEEMLHKAVMFEQQIKRKNARSSHTKTNYSSGKPSYQKE 282
Query: 263 GRGDKGRHFQKKPYVRPQGRGTTSGSFYPTG-GNAIALRTPSGNREDVTCFRCNKKGHYA 321
+ G KP+V+P+ P G G + R D+ CF+ HYA
Sbjct: 283 EK--FGYQNDSKPFVKPK-----PVDLDPKGKGKEVITRA-----RDIRCFKSQGLRHYA 330
Query: 322 NHCS 325
+ CS
Sbjct: 331 SKCS 334
>At3g31970 hypothetical protein
Length = 1329
Score = 218 bits (556), Expect = 1e-56
Identities = 114/276 (41%), Positives = 163/276 (58%), Gaps = 36/276 (13%)
Query: 644 MDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDY 703
M + G S I + + I + + +++KTAFRTRYGH+E++VMPFG+TN P FM
Sbjct: 552 MPESVGQVAVSDIRVVHEFEDIPIAEANVRKTAFRTRYGHFEFVVMPFGLTNIPTAFMRL 611
Query: 704 MNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEV 763
MN +F FLD FV++FIDDIL+YS++ EEHE ++ + A E
Sbjct: 612 MNSVFQEFLDEFVIIFIDDILVYSKSPEEHE---------VQSEESDGEAAGAEV----- 657
Query: 764 KFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTK 823
G++VD K+EA+ W RP T+IRSF+GLAGYY+RF++ FA +A P+TK
Sbjct: 658 ----------GVSVDLEKIEAIRDWPRPTNATEIRSFLGLAGYYKRFVKVFASMAQPMTK 707
Query: 824 LTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHRK 883
LT K+ PF W+ +C++ F +K+ LT+ PVL LP+ EPY VY DAS GL CVLMQ K
Sbjct: 708 LTGKDVPFVWSPECDEGFMSLKEMLTSTPVLALPEHGEPYMVYTDASRVGLDCVLMQRGK 767
Query: 884 AVA--------YSSRQLKIHERNY----PTHDLELA 907
++ +L + +R + +DLE+A
Sbjct: 768 VFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIA 803
Score = 113 bits (283), Expect = 4e-25
Identities = 126/511 (24%), Positives = 196/511 (37%), Gaps = 79/511 (15%)
Query: 49 ARRVQREERELAAAQT-----RGLNDFKRQDPPKFSGGFDPEEADLWLKELEKIFTFLRT 103
A RV ++ A A+ R + +R F GG PEEAD W +E F R
Sbjct: 124 APRVAEVQQRAAVAEEVPSYLRMMEQLQRIGTRYFFGGTSPEEADSWKSRVEHNFGSSRC 183
Query: 104 TAEMKVDYETYLLTGEAEYWWRGARAMMEADHQAITWECFRGAFLDKYFPRSARAAKEAQ 163
AE +VD + L G+A WWR A H ++W F F KYFP+ A EA+
Sbjct: 184 PAEYRVDLAVHFLEGDAHLWWRSVTARRRQAH--MSWADFVAEFNAKYFPQEALDRMEAR 241
Query: 164 FLRLRQGGMTVAEYAAKLESLAKHFRYFRGQIDEGYMCERFIEGLCYELQRAVQPLGLNR 223
FL L QG +V EY + L + RG D+ RF+ GL +L+ + L
Sbjct: 242 FLELTQGERSVREYEREFNRLLVYAG--RGMQDDQAQMRRFLRGLRPDLRVRCRVLQYAT 299
Query: 224 YQVLVEKTKGIEAIDNARGKFQGLNKPYQGSGGPARTNQGRGDKGRHFQKKPYVRPQGRG 283
LVE +E ++ + + G++ + + +G K QK+
Sbjct: 300 KAALVETAAEVE--EDLQRQVVGVSPAVKPKKTQQQVAPSKGGKPAQGQKR--------- 348
Query: 284 TTSGSFYPTGGNAIALRTPSGNREDVTCFRCNKKGHYANHCSESLAACWNCNKPGHTAAE 343
++GH +C + +P
Sbjct: 349 -------------------------------KERGHLRPNCPKLQRMAVAVVQPAVQHGA 377
Query: 344 CRIPKVEAAANVAGARRPTAGGRVYSISGTEAEEDDGLIRSTCEIAGNSLIALFDSGATH 403
+V+ A++A A + G E I ST + A LFDSGA+H
Sbjct: 378 QVQQRVQQLAHIAAAPQ-----------GYTTRE----IGSTSKRAITEAHVLFDSGASH 422
Query: 404 SFIDIACAARLKLEVSKLPFELTVSTPASKSLVTNTACLECPWMYLDKKFVANLICLPLK 463
FI A+R + V + L + +LI P++
Sbjct: 423 CFITPESASRGNIRGDPGEQLGAVKVAGGQFLAVLGRAKGVDIQIAGESMPVDLIISPVE 482
Query: 464 GLDVIIGMDWLSHHHVLLDCANKVVIF--PDAGL----------AEFLNSYFSKLSLRKG 511
DVI+GMDWL H+ V LDC V F P+ L + +++ ++ + KG
Sbjct: 483 LYDVILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQRVRPTSRSLVISAVQAEKMIEKG 542
Query: 512 ALSSLMSTTVVEA-KENGVHGIAVVQDFEDV 541
+ L++ ++ E+ + V I VV +FED+
Sbjct: 543 CEAYLVTISMPESVGQVAVSDIRVVHEFEDI 573
>At1g20390 hypothetical protein
Length = 1791
Score = 213 bits (542), Expect = 4e-55
Identities = 147/490 (30%), Positives = 233/490 (47%), Gaps = 28/490 (5%)
Query: 466 DVIIGMDWLSHHHVLLDCANKVVIFPDA-GLAEFLNSYFSKLSLRKGALSSLMSTTVVEA 524
+VI+G W+ + ++ V FP G+ +K R S L T +V
Sbjct: 657 NVILGTPWIHQMQAIPSTYHQCVKFPTHNGIFTLRAPKEAKTPSRSYEESELCRTEMVNI 716
Query: 525 KEN------GVHG----------IAVVQDFEDVFP---EDVPGIPP-VRDMEFTIDIVPG 564
E+ GV IA+++ F ED+ GI P + E +D P
Sbjct: 717 DESDPTRCVGVGAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNVD--PT 774
Query: 565 TGPISIAPYRMAPAELTELKSQLEDLTKKG-FIRPSVSPWGAPVLLVKKKDGRSRLCVDY 623
P+ ++ P + ++E L K G I W A ++VKKK+G+ R+CVDY
Sbjct: 775 FKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDY 834
Query: 624 RQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGH 683
LNK K+ YPLP ID L++ G + S +D SGY+QI + +D +KT+F T G
Sbjct: 835 TDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGT 894
Query: 684 YEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQV 743
Y Y VM FG+ NA A + ++N++ + R V V+IDD+L+ S E+H EHL + V
Sbjct: 895 YCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDV 954
Query: 744 LRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGL 803
L + N TKC F + +FLG+V++ GI +P ++ A+L P+ +++ G
Sbjct: 955 LNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGR 1014
Query: 804 AGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPY 863
RFI P L ++ F W +D E++F+ +K L+T P+L P+ E
Sbjct: 1015 IAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETL 1074
Query: 864 EVYCDASYQGLGCVLMQ----HRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHY 919
+Y S + VL++ ++ + Y+S+ L E YP + AVV + + R Y
Sbjct: 1075 YLYIAVSDHAVSSVLVREDRGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPY 1134
Query: 920 LYGSTFTVFS 929
T V +
Sbjct: 1135 FQSHTIAVLT 1144
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 211 bits (536), Expect = 2e-54
Identities = 134/381 (35%), Positives = 197/381 (51%), Gaps = 44/381 (11%)
Query: 569 SIAPYRMAPAELTEL-KSQLEDLTKKGFIRP-SVSPWGAPVLLVKKKDGRS--------- 617
SI P R L E+ K ++ L G I P S S W +PV V KK G +
Sbjct: 900 SIEPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDEL 959
Query: 618 ---------RLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVK 668
R+C++YR+LN + K +PLP ID ++++L + +D SG+ QI +
Sbjct: 960 IPTRTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIH 1019
Query: 669 DEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSR 728
D KT F YG + Y MPFG+ NAPA F M IF ++ V VF+DD +Y
Sbjct: 1020 PNDQGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGS 1079
Query: 729 NREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAW 788
+ +L +VL+ + L N KC F + E LG IS EGI VD +K++ ++
Sbjct: 1080 SFSSCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQL 1139
Query: 789 ERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRL 848
+ PKTV DIRSF+G AG+YR FI+ F+K+A PLT+L K FA+ ++C +F+ +K+ L
Sbjct: 1140 QPPKTVKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFDDECLTAFKLIKEAL 1199
Query: 849 TTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAA 908
TAP++ P P+E+ + Q++ Y T + EL A
Sbjct: 1200 ITAPIVQAPNWDFPFEI-------------------ITMDDAQVR-----YATTEKELLA 1235
Query: 909 VVFALKIWRHYLYGSTFTVFS 929
VVFA + +R YL GS T+++
Sbjct: 1236 VVFAFEKFRSYLVGSKVTIYT 1256
>At4g04230 putative transposon protein
Length = 315
Score = 201 bits (510), Expect = 2e-51
Identities = 102/233 (43%), Positives = 146/233 (61%), Gaps = 40/233 (17%)
Query: 534 VVQDFEDVFPEDVP-GIPPVRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTK 592
V+ ++D+FPE++P G+PP+
Sbjct: 106 VLHWYKDLFPEEIPPGLPPIH--------------------------------------- 126
Query: 593 KGFIRPSVSPWGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAI 652
KG+IR S+SP PVLLV KKDG R+C+D R +N +TIK R+P+PR+ D++D+L GA I
Sbjct: 127 KGYIRESLSPCAVPVLLVPKKDGTWRMCLDCRAINNITIKYRHPIPRLYDMLDELSGAII 186
Query: 653 FSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFL 712
FSK+DLRSGYHQ+R+++ D KTAF+T+ G YE LVMPFG+TNAP+ FM MN++ F+
Sbjct: 187 FSKVDLRSGYHQVRMREGDEWKTAFKTKQGLYECLVMPFGLTNAPSTFMRLMNQVLRSFI 246
Query: 713 DRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKF 765
+FVVV+ DDILIY+++ +H + L +L+ LR + LYAN KC F ++ F
Sbjct: 247 GKFVVVYFDDILIYNKSYSDHIQPLELILKTLRKEGLYANLKKCTFCSDKFFF 299
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 194 bits (493), Expect = 2e-49
Identities = 108/335 (32%), Positives = 179/335 (53%), Gaps = 3/335 (0%)
Query: 583 LKSQLEDLTKKGFIRPSVSP-WGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRID 641
+ +++ L K G IR P W A ++VKKK+G+ R+C+D+ LNK K+ +PLP ID
Sbjct: 480 VNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHID 539
Query: 642 DLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFM 701
L++ G + S +D SGY+QI + ED +KT F T G Y Y VMPFG+ NA A +
Sbjct: 540 RLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPFGLRNAGATYP 599
Query: 702 DYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLE 761
+N++F + + + V+IDD+LI S +E+H +HL + +L + N KC F +
Sbjct: 600 RLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKLNPAKCTFGVP 659
Query: 762 EVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPL 821
+FLG++++ GI +P+++ A L PK +++ G RFI + P
Sbjct: 660 SGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRFISRSTDKSLPF 719
Query: 822 TKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQH 881
++ + N+ F W E CE++F +K LT+ PVL P+ E +Y S + VL++
Sbjct: 720 YQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVSNHAVSGVLIRE 779
Query: 882 RKAVAYSSRQLKIHERN--YPTHDLELAAVVFALK 914
+ + +++ N Y D + A + +K
Sbjct: 780 DRGEQKPIYYIIVNQFNGDYEAKDSRMEAYLHVVK 814
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 184 bits (468), Expect = 2e-46
Identities = 124/392 (31%), Positives = 189/392 (47%), Gaps = 28/392 (7%)
Query: 544 EDVPGIPP-VRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSP 602
ED+PGI V E +D P P+ ++ P ++ +++ L G I P
Sbjct: 760 EDMPGIDSAVTCHELNVD--PTYKPLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVEVRYP 817
Query: 603 -WGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSG 661
W ++VKKK+G+ R+C+D+ LNK K+ +PLP ID L++ G + S +D SG
Sbjct: 818 DWLRNPVVVKKKNGKWRVCIDFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMDAFSG 877
Query: 662 YHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFID 721
Y+QI + D +KT F T G Y Y VMPFG+ NA A + +N++F LD + V+ID
Sbjct: 878 YNQILMHQNDREKTVFITDQGTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSMEVYID 937
Query: 722 DILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSK 781
D+L+ S EEH HLRQ QVL + N +KC F + +FLG++++ GI +P +
Sbjct: 938 DMLVKSLRAEEHITHLRQCFQVLNRYNMKLNPSKCTFGVTSGEFLGYLVTRRGIEANPKQ 997
Query: 782 VEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSF 841
+ A++ P+ +++ IG RFI P +L R N+ F W E CE
Sbjct: 998 ISAIIDLPSPRNTREVQRLIGRIAALNRFISRSTDKCLPFYQLLRANKRFEWDEKCE--- 1054
Query: 842 QDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHRKA----VAYSSRQLKIHER 897
E E +Y S + VL++ + + Y S+ L E
Sbjct: 1055 -----------------EGETLYLYIAVSTSAVSGVLVREDRGEQHPIFYVSKTLDGAEL 1097
Query: 898 NYPTHDLELAAVVFALKIWRHYLYGSTFTVFS 929
YPT + AVV + + R Y T V +
Sbjct: 1098 RYPTLEKLAFAVVISARKLRPYFKSYTVEVLT 1129
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 184 bits (466), Expect = 3e-46
Identities = 128/373 (34%), Positives = 185/373 (49%), Gaps = 54/373 (14%)
Query: 544 EDVPGIPPVRDMEFTIDIVPGTGPISIAPYRMAPAELTEL-KSQLEDLTKKGFIRP-SVS 601
+D+ GI P M + G S+ R + L ++ K ++ L G I P S S
Sbjct: 352 DDITGISPTLCMHMIH--LEGESITSVEHQRRLNSNLRDVVKKEIMKLLDAGIIYPISDS 409
Query: 602 PWGAPVLLVKKKDG------------------RSRLCVDYRQLNKVTIKNRYPLPRIDDL 643
W +PV +V KK G R+C+DYR+LN T K+ +PL ID +
Sbjct: 410 TWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGHRMCIDYRKLNSATRKDNFPLSFIDQM 469
Query: 644 MDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDY 703
+++L + +D G+ QI + +D +KT F YG + Y MPFG+ NAPA F
Sbjct: 470 LERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTFTCPYGTFAYRRMPFGLCNAPATFQHC 529
Query: 704 MNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEV 763
M IF ++ F+ VFIDD L VLQ DK L N K F + +
Sbjct: 530 MKYIFSDMIEDFMEVFIDDFL---------------VLQRCEDKHLVLNWEKSHFMVRDG 574
Query: 764 KFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTK 823
LGH IS +G+ VD +K+E I F+G AG+YRRFI+ F+KIA P T+
Sbjct: 575 IVLGHKISEKGVEVDRAKIE-------------IMRFLGHAGFYRRFIKDFSKIARPCTQ 621
Query: 824 LTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHR- 882
L K Q + + C Q+F +K+ L +A ++ P+ + P+EV CDAS +G VL Q +
Sbjct: 622 LLCKEQKSEFDDTCLQAFNIVKESLVSALIVQPPEWELPFEVICDASDYVVGAVLGQRKD 681
Query: 883 ---KAVAYSSRQL 892
A+ Y+SR L
Sbjct: 682 KKLHAIYYASRTL 694
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,287,454
Number of Sequences: 26719
Number of extensions: 882567
Number of successful extensions: 2674
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 73
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 2360
Number of HSP's gapped (non-prelim): 240
length of query: 929
length of database: 11,318,596
effective HSP length: 109
effective length of query: 820
effective length of database: 8,406,225
effective search space: 6893104500
effective search space used: 6893104500
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0004b.4


