

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0004a.8
(1041 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
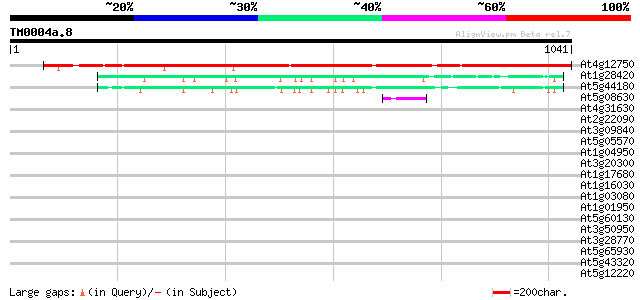
Score E
Sequences producing significant alignments: (bits) Value
At4g12750 putative protein 930 0.0
At1g28420 hypothetical protein 209 7e-54
At5g44180 unknown protein 186 8e-47
At5g08630 unknown protein 45 2e-04
At4g31630 putative protein 35 0.17
At2g22090 putative RNA-binding protein 35 0.17
At3g09840 putative transitional endoplasmic reticulum ATPase 35 0.29
At5g05570 putative protein 33 0.65
At1g04950 putative TATA binding protein-associated factor (F13M7.6) 33 0.65
At3g20300 unknown protein 33 0.84
At1g17680 unknown protein 33 1.1
At1g16030 unknown protein 33 1.1
At1g03080 unknown protein 33 1.1
At1g01950 unknown protein 33 1.1
At5g60130 putative protein 32 1.4
At3g50950 putative disease resistance protein 32 1.4
At3g28770 hypothetical protein 32 1.4
At5g65930 kinesin-like calmodulin-binding protein 32 1.9
At5g43320 casein kinase I 32 1.9
At5g12220 unknown protein 32 1.9
>At4g12750 putative protein
Length = 1108
Score = 930 bits (2404), Expect = 0.0
Identities = 523/1037 (50%), Positives = 675/1037 (64%), Gaps = 87/1037 (8%)
Query: 63 LGTVLRNDGPSIGREFDYLPSGHKSYN-----STCREDQQPAKRSKVSKGTGKGLMNASV 117
L V R DGPS+G EFD+LPSG + + S ++ Q+ A++ K+S+ +
Sbjct: 86 LAKVFRKDGPSLGSEFDHLPSGARKASWLGTSSVGQQKQKVARKRKISELMDHTSQDCIQ 145
Query: 118 KKHGIGKGLMNASVKKHGIGKGLMTIWRATNPDSGELPVGFGFADREVHLISNSKKSVRE 177
+ NA+V KHGIGKGLMT+WR NP+ ++ D L +S ++
Sbjct: 146 E---------NATVMKHGIGKGLMTVWRVMNPNRRDVSPCVDLLDERATLPQSSARN--P 194
Query: 178 NNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVTRNQSLKEKCELSLDS 237
++ K + +LK KL KR+ + +R + + E N+ T+ + KE CEL+ D
Sbjct: 195 PHQKKKQRQLASILKQKLLQKRSTEKKRRSI---HREAELNKDETQRE-FKENCELAADG 250
Query: 238 EISEEGVDRISMLIDDEELELRELQERTNLLICSDHLAASGMLGCSLGK----------- 286
E+ +E IS L+DDEELE+RE ER N L CS H +SG GC L K
Sbjct: 251 EVFKETCQTISTLVDDEELEMRERHERGNPLTCSCHHPSSGSHGCFLCKGIAMRSSDSSL 310
Query: 287 ---DVLVKYPPDTVKMKKPIHLQPWDSSPELVKKLFKDSMLLGKIHVALLTLLLSDIEVE 343
D+L K+PP++V+M+ P L PW+SSPE VKKLFKDS+LLGKIH++LL LLL D+E E
Sbjct: 311 LFPDLLPKFPPNSVQMRMPFGLHPWNSSPESVKKLFKDSLLLGKIHLSLLKLLLLDVETE 370
Query: 344 LSNGFAPHLDKSCNFLALLHSVESQEYSPDFWRRSLNSLTWIEILRQVLVASGFGSKQGA 403
L G +L SC FLALL SVESQ D WR SLNSLTW E+LRQ+LVA+G+GS + A
Sbjct: 371 LERGSFSNLSISCKFLALLQSVESQILILDMWRNSLNSLTWTELLRQILVAAGYGSLKCA 430
Query: 404 LRREALSKEL--------------------------------NLLINYGISPGTLKGELF 431
++ E LSK+L L+ YG+ GTLKGELF
Sbjct: 431 VQSEELSKQLASTCFVLGDRSVICGELKALARLYFVIDDIHMKLMKKYGLRLGTLKGELF 490
Query: 432 KILSERGNNGCKVSDLAKATQIAELNLASTTEELESLICSTLSSDITLFEKISSSAYRLR 491
++L+ +GNNG K+S+LA A ++A LNLA+ EE E+ ICSTL+SDITLFEKIS S YR+R
Sbjct: 491 RMLNGQGNNGLKISELADAPEVAVLNLATVPEERENSICSTLASDITLFEKISESTYRVR 550
Query: 492 MSTVTKDDDECQSDMEDYGSVDDELDDSDTCSCGDDFESGSVDSNIRKLKRVKSHKTKSA 551
++ ++D D+ QSD +D GSVDDE DD + S GD+ E S + +RK+K K K KS
Sbjct: 551 VNCFSEDPDKSQSDSDDSGSVDDESDDC-SISSGDEIEHVSENPALRKVKCRKRRKHKSK 609
Query: 552 KLTVFNEIDESHPGEVWLLGLMESEYSDLNIEEKLNALAALTDLLSSGSSIRMKDPVKVT 611
V +EIDESHPGE WLLGLME EYSDL++EEKL+ AL DLLSSGS+IRM+D +
Sbjct: 610 MREVCSEIDESHPGEPWLLGLMEGEYSDLSVEEKLDVFVALIDLLSSGSTIRMEDLPRAV 669
Query: 612 ADYDSSIQLRGSGAKIKRSVVKK---PVPFWNQFGQMQRVKEGHLNSHPCPVDSSSLMSK 668
AD SI GSG KIKRS + P W G++ +K +S PVDSSS++
Sbjct: 670 ADCAPSIYSHGSGGKIKRSSSNQYSYPRGSWVHGGELYGIKALSKSSDSHPVDSSSIVGA 729
Query: 669 FRSHEPSFEKGKGSTDSHPIQSVYLGSDRRYNRYWLFLGPCNADDPGHKRVYFESSEDGH 728
F G + + HP+QSVYLGSDRR+NRYWLFLG CNA+DPGH+ V+FESSEDGH
Sbjct: 730 FAKLA-----GNRANNVHPMQSVYLGSDRRFNRYWLFLGTCNANDPGHRCVFFESSEDGH 784
Query: 729 WEVIDTEEALCTLLSVLDDRGKREALLIESLERRQTSLCRFMSRVKVNGIGTGCMSHSDQ 788
WEVI+ +EAL LLSVLDDRG+REA LIESLE+R++ LC+ M +V T +
Sbjct: 785 WEVINNKEALRALLSVLDDRGRREARLIESLEKRESFLCQAMLSRQVTQSETAHFT---- 840
Query: 789 SELDMVTEDSNSPVSDVDN-LNLSDSAKSLPSA--GAVVIEAGKGVEEQVQKWIRVQEYD 845
D+V EDS+SPVSD+DN L L++ A S+ A+V E G E+ + W +QE+D
Sbjct: 841 ---DIVREDSSSPVSDIDNNLCLNEIANDQFSSQHAAIVFEIGSKREKSLL-WSLIQEFD 896
Query: 846 SWIWNSFYLDLNVVKYGRRSYLDSLARCKSCHDLYWRDERHCKICHMTFELDFDLEERYA 905
WIW +F +LN VK+ RRSYLDSL RCKSCHDLYWRDE+HCKICH TFE+D DLEERYA
Sbjct: 897 DWIWANFNFNLNSVKHRRRSYLDSLTRCKSCHDLYWRDEKHCKICHATFEVDIDLEERYA 956
Query: 906 IHIATCREKENSKTHPNHKVLSSQIQSLKAAIYAIESVMPEDSLVGAWRKSAHKLWVKRL 965
IH ATC KE T P+HKVLSSQ+QSLKAA+YAIES MPED+L+GAWRKSAH+LW KRL
Sbjct: 957 IHAATCMRKEECDTFPDHKVLSSQLQSLKAAVYAIESAMPEDALIGAWRKSAHRLWAKRL 1016
Query: 966 RRTSTLVELVQVLTDFVGAINESWLFQCKFP-DGMVEEIIASFASMPHTSSALALWLVKL 1024
RR+S++ E+ QV+ DFVGAINE WL+ C ++ EII F SMP T+SA+ALWLVKL
Sbjct: 1017 RRSSSVSEITQVIGDFVGAINEEWLWHCSDQGQTLMGEIINCFPSMPQTTSAIALWLVKL 1076
Query: 1025 DTIIAPYLDRVYLQKKQ 1041
DT+IAPY+++ ++ Q
Sbjct: 1077 DTLIAPYVEKAPPERDQ 1093
>At1g28420 hypothetical protein
Length = 1703
Score = 209 bits (532), Expect = 7e-54
Identities = 248/1063 (23%), Positives = 418/1063 (38%), Gaps = 227/1063 (21%)
Query: 164 EVHLISNSKKSVRENNRSSKTATMNRMLKN-KLQNKRNNLQDKRKLLMQRKAGESNQHVT 222
E + +++ RE + + R+ + +LQ ++ ++R+ +QR+ + +
Sbjct: 394 EERMRKEMERNERERRKEEERLMRERIKEEERLQREQRREVERREKFLQRENERAEKKKQ 453
Query: 223 RNQSLKEKCELSLDSEISEEGVDRISM----LIDDEELELRELQERTNLLICSDHLAASG 278
+++ +EK + I + RI+ LI+DE+LEL EL + L L
Sbjct: 454 KDEIRREKDAIRRKLAIEKATARRIAKESMDLIEDEQLELMELAAISKGLPSVLQLDHDT 513
Query: 279 MLGCSLGKDVLVKYPPDTVKMKKPIHLQPWDSSPELVKKLFK------------------ 320
+ + +D L +PP ++++K P + PW S E V L
Sbjct: 514 LQNLEVYRDSLSTFPPKSLQLKMPFAISPWKDSDETVGNLLMVWRFLISFSDVLDLWPFT 573
Query: 321 -----------DSMLLGKIHVALLTLLLSDIE-------VELSNGFAPHLDKSCNFLALL 362
DS LLG+IHV LL ++ D+E + N + ++
Sbjct: 574 LDEFIQAFHDYDSRLLGEIHVTLLRSIIRDVEDVARTPFSGIGNNQYTTANPEGGHPQIV 633
Query: 363 HSVESQEYSPDFWRRSLNSLTWIEILRQVLVASGFGSK---------------------- 400
+ + W++ LN LTW EILRQ+ +++GFG K
Sbjct: 634 EGAYAWGFDIRSWKKHLNPLTWPEILRQLALSAGFGPKLKKKHSRLTNTGDKDEAKGCED 693
Query: 401 ------QGALRREALS--KELNLLI----NYGISPGTLKGELFKILSERGNNGCKVSDLA 448
G A + +E LL + ++PGT+K F +LS G+ G V +LA
Sbjct: 694 VISTIRNGTAAESAFASMREKGLLAPRKSRHRLTPGTVKFAAFHVLSLEGSKGLTVLELA 753
Query: 449 KATQIAELNLASTTEELESLICSTLSSDITLFEKISSSAYRLRMSTVTKDDD-------- 500
Q + L +T++ E+ I L+ D+ LFE+I+ S Y +R V D
Sbjct: 754 DKIQKSGLRDLTTSKTPEASISVALTRDVKLFERIAPSTYCVRAPYVKDPKDGEAILADA 813
Query: 501 --------------------ECQSDMEDYGSVDDELDDSDTCSCGDDFE----------- 529
E D E D E+DD T +
Sbjct: 814 RKKIRAFENGFTGPEDVNDLERDEDFEIDIDEDPEVDDLATLASASKSAVLGEANVLSGK 873
Query: 530 ---------SGSVDSNIRK---------LKRVKSHKTKSAKLTVFNE----IDESHPGEV 567
V S + K +K + ++ K TV IDES+ G+
Sbjct: 874 GVDTMFCDVKADVKSELEKEFSSPPPSTMKSIVPQHSERHKNTVVGGVDAVIDESNQGQS 933
Query: 568 WLLGLMESEYSDLNIEEKLNALAALTDLLSSGSSIR------------MKDPVKVTADYD 615
W+ GL E +Y L++EE+LNAL AL + + G+SIR +K + A D
Sbjct: 934 WIQGLTEGDYCHLSVEERLNALVALVGIANEGNSIRTGLEDRMEAANALKKQMWAEAQLD 993
Query: 616 SS-------IQLRGSGAKIKRSVVKKPV-----------------------PFWNQFGQM 645
+S + L+ + S + P+ P + +
Sbjct: 994 NSCMRDVLKLDLQNLASSKTESTIGLPIIQSSTRERDSFDRDPSQLLDETKPLEDLSNDL 1053
Query: 646 QRVK-EGHLNSHPCPVDSSSLMSKFRSHEPSFEKGKGSTDSHPIQSVYLGSDRRYNRYWL 704
+ E L + + + SK + G + + +P +S+ LG DRR+NRYW
Sbjct: 1054 HKSSAERALINQDANISQENYASKRSRSQLKSYIGHKAEEVYPYRSLPLGQDRRHNRYWH 1113
Query: 705 FLGPCNADDPGHKRVYFESSEDGHWEVIDTEEALCTLLSVLDDRGKREALLIESLERRQT 764
F + DP R+ F DG W +ID+EEA L++ LD RG RE+ L L++ +
Sbjct: 1114 FAVSVSKSDPC-SRLLFVELHDGKWLLIDSEEAFDILVASLDMRGIRESHLRIMLQKIEG 1172
Query: 765 SL----CRFMSRVKVNGIGTGCMSHSDQSELDMVTEDSNSPVSDVDNLNLSDSAKSLPSA 820
S C+ + + + +++S ++ DS SP S + + SDS ++ S
Sbjct: 1173 SFKENACKDIKLARNPFL-------TEKSVVNHSPTDSVSPSSSAISGSNSDSMETSTS- 1224
Query: 821 GAVVIEAGKGVEEQVQKWIRVQEYDSWIWNSFYLDLNVV--KYGRRSYLDSLARCKSCHD 878
+ ++ G+ E R ++ W+W Y L KYG++ + LA C +C
Sbjct: 1225 --IRVDLGRNDTENKNLSKRFHDFQRWMWTETYSSLPSCARKYGKKRS-ELLATCDACVA 1281
Query: 879 LYWRDERHCKICHMTFELDFDLEERYAIHIATCREKENSKTHPNHKVLSSQIQSLKAAIY 938
Y + C CH ++ D E +A L ++ LK +
Sbjct: 1282 SYLSEYTFCSSCHQRLDV-VDSSEILDSGLAV-------------SPLPFGVRLLKPLLV 1327
Query: 939 AIESVMPEDSLVGAWRKSAHKLWVKRLRRTSTLVELVQVLTDFVGAINESWLFQCKFPDG 998
+E+ +P+++L W + K W RL +S+ EL+QVLT AI + L F
Sbjct: 1328 FLEASVPDEALESFWTEDQRKKWGFRLNTSSSPGELLQVLTSLESAIKKESL-SSNFMS- 1385
Query: 999 MVEEIIASFAS-------------MPHTSSALALWLVKLDTII 1028
+E++ + + +P T SA+AL L +LD I
Sbjct: 1386 -AKELLGAANAEADDQGSVDVLPWIPKTVSAVALRLSELDASI 1427
Score = 32.3 bits (72), Expect = 1.4
Identities = 15/27 (55%), Positives = 19/27 (69%), Gaps = 1/27 (3%)
Query: 59 CLQ-DLGTVLRNDGPSIGREFDYLPSG 84
C++ LG LR+DGP +G EFD LP G
Sbjct: 185 CIEAQLGEPLRDDGPILGMEFDPLPPG 211
>At5g44180 unknown protein
Length = 1694
Score = 186 bits (471), Expect = 8e-47
Identities = 250/1085 (23%), Positives = 420/1085 (38%), Gaps = 259/1085 (23%)
Query: 163 REVHLISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVT 222
RE + ++ RE + + R+L+ K + + L+++ + L QR+ +
Sbjct: 360 REEQIRKEMERQDRERRKEEE-----RLLREKQREEERYLKEQMREL-QRREKFLKKETI 413
Query: 223 RNQSLKEKCELSLDSEISE----------EGVDRISM-LIDDEELELRELQERTNLLICS 271
R + +++K E+ + E++ + + SM LI+DE LEL E+ T L
Sbjct: 414 RAEKMRQKEEMRKEKEVARLKAANERAIARKIAKESMELIEDERLELMEVAALTKGLPSM 473
Query: 272 DHLAASGMLGCSLGKDVLVKYPPDTVKMKKPIHLQPWDSSPELVKKLFK----------- 320
L + +D +PP +VK+KKP ++PW+ S E V L
Sbjct: 474 LALDFETLQNLDEYRDKQAIFPPTSVKLKKPFAVKPWNGSDENVANLLMVWRFLITFADV 533
Query: 321 ------------------DSMLLGKIHVALLTLLLSDIEV---ELSNGFAPHLDKSCNFL 359
D L+G+IH+ LL ++ DIE LS G + + + N
Sbjct: 534 LGLWPFTLDEFAQAFHDYDPRLMGEIHIVLLKTIIKDIEGVVRTLSTGVGANQNVAANPG 593
Query: 360 ALLHSVESQEYSPDF----WRRSLNSLTWIEILRQVLVASGFGSKQGALRREALS----- 410
V Y+ F WR++LN TW EILRQ+ +++G G + + +S
Sbjct: 594 GGHPHVVEGAYAWGFDIRSWRKNLNVFTWPEILRQLALSAGLGPQLKKMNIRTVSVHDDN 653
Query: 411 ---KELNLLIN--------------------------YGISPGTLKGELFKILSERGNNG 441
N++ N + ++PGT+K F +LS G G
Sbjct: 654 EANNSENVIFNLRKGVAAENAFAKMQERGLSNPRRSRHRLTPGTVKFAAFHVLSLEGEKG 713
Query: 442 CKVSDLAKATQIAELNLASTTEELESLICSTLSSDITLFEKISSSAYRLRMSTVTKDDDE 501
+ ++A+ Q + L +T+ E+ + + LS D LFE+++ S Y +R S KD +
Sbjct: 714 LNILEVAEKIQKSGLRDLTTSRTPEASVAAALSRDTKLFERVAPSTYCVRAS-YRKDAGD 772
Query: 502 CQ--------------SDMEDYGSVDD-ELDDSDTCSCGDD------------------- 527
+ S + D VDD E D+ G+D
Sbjct: 773 AETIFAEARERIRAFKSGITDVEDVDDAERDEDSESDVGEDPEVDVNLKKEDPNPLKVEN 832
Query: 528 -------FESGSVDS-----------------NIRKLKRVKSHKTKSAKLTVFNE----- 558
E+G +D+ ++ KR + +S + V N
Sbjct: 833 LIGVEPLLENGKLDTVPMKTELGLPLTPSLPEEMKDEKRDDTLADQSLEDAVANGEDSAC 892
Query: 559 IDESHPGEVWLLGLMESEYSDLNIEEKLNALA--------------ALTDLLSSGSSI-- 602
DES GE W+ GL+E +YS+L+ EE+LNAL AL + L S++
Sbjct: 893 FDESKLGEQWVQGLVEGDYSNLSSEERLNALVALIGIATEGNTIRIALEERLEVASALKK 952
Query: 603 ----------RMKDPVKVTADYDS--------SIQLRGSGAKIKRSVVKKPVPFWNQFG- 643
R K+ + A+Y S +I SG + S P+ +
Sbjct: 953 QMWGEVQLDKRWKEESLIRANYLSYPTAKPGLNIATPASGNQESSSADVTPISSQDPVSL 1012
Query: 644 --------------QMQRVKEGHLN-----SHPCPVDSSSLMSKFRSHEPSFEKGKGSTD 684
Q+Q G N D L ++ +++ G + +
Sbjct: 1013 PQIDVNNVIAGPSLQLQENVPGVENLQYQQQQGYTADRERLRAQLKAYV-----GYKAEE 1067
Query: 685 SHPIQSVYLGSDRRYNRYWLFLGPCNADDPGHKRVYFESSEDGHWEVIDTEEALCTLLSV 744
+ +S+ LG DRR NRYW F + +DPG R++ E +DG W +ID+EEA L+
Sbjct: 1068 LYVYRSLPLGQDRRRNRYWRFSASASRNDPGCGRIFVE-LQDGRWRLIDSEEAFDYLVKS 1126
Query: 745 LDDRGKREALLIESLERRQTSLCRFMSRVKVNGIGTGCMSHSDQSELDMVTEDSNSPVSD 804
LD RG RE+ L L + + S + R G +S S S+ + +S
Sbjct: 1127 LDVRGVRESHLHFMLLKIEASFKEALRRNVAANPGVCSISSSLDSD--------TAEIST 1178
Query: 805 VDNLNLSDSAKSLPSAGAVVIEAGKGVEEQVQKWIRVQEYDSWIWNSFY--LDLNVVKYG 862
+ L DS E+ R ++ W+W++ L+ KYG
Sbjct: 1179 TFKIELGDS----------------NAVERCSVLQRFHSFEKWMWDNMLHPSALSAFKYG 1222
Query: 863 RRSYLDSLARCKSCHDLYWRDERHCKICHMTFELDFDLEERYAIHIATCREKENSKTHPN 922
+ C+ C +L++ + C C E +A +A + +N +
Sbjct: 1223 AKQSSPLFRICRICAELHFVGDICCPSCGQMHAGPDVGELCFAEQVA--QLGDNLRRGDT 1280
Query: 923 HKVLSSQIQS------LKAAIYAIESVMPEDSLVGAWRKSAHKLWVKRLRRTSTLVELVQ 976
+L S I S LK + +E+ +P + L W ++ K W +L +S+ +L Q
Sbjct: 1281 GFILRSSILSPLRIRLLKVQLALVEASLPPEGLEAFWTENLRKSWGMKLLSSSSHEDLYQ 1340
Query: 977 VLTDFVGAINESWLFQCKFPD-----GMVEEIIASFAS--------MPHTSSALALWLVK 1023
VLT A+ +L F G+ E +AS + +P T+ +AL L
Sbjct: 1341 VLTTLEAALKRDFL-SSNFETTSELLGLQEGALASDLTCGVNVLPWIPKTAGGVALRLFD 1399
Query: 1024 LDTII 1028
D+ I
Sbjct: 1400 FDSSI 1404
Score = 34.7 bits (78), Expect = 0.29
Identities = 22/55 (40%), Positives = 29/55 (52%), Gaps = 2/55 (3%)
Query: 30 KGKGPLGRIIQQREKANVPSSAKKNQKRKCLQDLGTVLRNDGPSIGREFDYLPSG 84
+G G G + +R N PSSA+ LG LR++GP +G EFD LP G
Sbjct: 117 RGSGGSGVTVVRR--FNEPSSAEVRAIGYVEAQLGERLRDNGPVLGMEFDPLPPG 169
>At5g08630 unknown protein
Length = 723
Score = 45.4 bits (106), Expect = 2e-04
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 8/80 (10%)
Query: 693 LGSDRRYNRYWLFLGPCNADDPGHKRVYFESSEDGHWEVIDTEEALCTLLSVLDDRGKRE 752
LG DR YNRYW F + R++ E+S+ W +E L L+ L+ +G+RE
Sbjct: 608 LGKDRDYNRYWWFRS--------NGRIFVENSDSEEWGYYTAKEELDALMGSLNRKGERE 659
Query: 753 ALLIESLERRQTSLCRFMSR 772
L LE +C + +
Sbjct: 660 LSLYTQLEIFYDRICSTLQK 679
Score = 30.4 bits (67), Expect = 5.5
Identities = 37/119 (31%), Positives = 53/119 (44%), Gaps = 12/119 (10%)
Query: 402 GALRREALSKELNLLINYGISPGTLKGELFKILSERGNNGC--KVSDLAKATQIAELNLA 459
G L K L L+N+ G KGE+ +++ +R G + LA+A QI E
Sbjct: 406 GLLDPNVKLKILRELVNHITETGMFKGEIDELVEQRHALGAARREEALAEARQIREEKER 465
Query: 460 STT-EELESLICSTLSSDITLFEKISSSAYRLRMSTVTKDDDECQ--SDMEDYGSVDDE 515
S T EE ++ D EK +SA L S +K ++ S ME+ GSV E
Sbjct: 466 SKTGEEANGVL------DNNRLEKKKNSAQVLESSEDSKKNESTAGGSKMEN-GSVSSE 517
>At4g31630 putative protein
Length = 512
Score = 35.4 bits (80), Expect = 0.17
Identities = 21/59 (35%), Positives = 28/59 (46%)
Query: 492 MSTVTKDDDECQSDMEDYGSVDDELDDSDTCSCGDDFESGSVDSNIRKLKRVKSHKTKS 550
+S+ T DDD+ + + D DD DD D DDF S + S R K +S KS
Sbjct: 110 ISSSTSDDDDDERTVFDDDEDDDVGDDDDNSISEDDFCSKKISSKKRARKETESSSDKS 168
>At2g22090 putative RNA-binding protein
Length = 343
Score = 35.4 bits (80), Expect = 0.17
Identities = 30/136 (22%), Positives = 61/136 (44%), Gaps = 15/136 (11%)
Query: 186 TMNRMLKNKL-QNKRNNLQDKRKLLMQRKAGESNQHVTRNQSLKEKCELSLDSEISEEGV 244
T+++ K KL ++K N K+K+ Q++ ES+ + + S + + D+E E
Sbjct: 4 TLDKSKKRKLVKSKSTNFDKKQKINKQQQQPESSTPYSSSSSSSDSSDSESDNEFDPE-- 61
Query: 245 DRISMLIDDEELELRELQERTNLLICSDHLAASGMLGCSLGKDVLVKYPPDTVKMKKPIH 304
ELREL + + D + ++ +G S+ V+ D K ++
Sbjct: 62 ------------ELRELLQPYSKDQLVDLVCSASRIGSSIYSAVVEAADRDVTHRKIFVY 109
Query: 305 LQPWDSSPELVKKLFK 320
PW+++ E + +F+
Sbjct: 110 GLPWETTRETLVGVFE 125
>At3g09840 putative transitional endoplasmic reticulum ATPase
Length = 809
Score = 34.7 bits (78), Expect = 0.29
Identities = 55/250 (22%), Positives = 97/250 (38%), Gaps = 27/250 (10%)
Query: 424 GTLKGELFKILSERGNNGCKVSDLAKATQIAELNLASTTEELESLICSTLSSDITLFEKI 483
G + L K E N + + + IA T E+E I S L +TL + +
Sbjct: 283 GESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKR-EKTNGEVERRIVSQL---LTLMDGL 338
Query: 484 SSSAYRLRMSTVTKDDDECQSDMEDYGSVDDELDDSDTCSCGDDFESGSVDSNIRKLKRV 543
S A+ + M T + + +G D E+D V I +L+ +
Sbjct: 339 KSRAHVIVMGA-TNRPNSIDPALRRFGRFDREIDIG-------------VPDEIGRLEVL 384
Query: 544 KSHKTKSAKLTV---FNEIDESHPGEVWL-LGLMESEYSDLNIEEKLNALAALTDLLSSG 599
+ H TK+ KL I + G V L + +E + I EK++ + DL
Sbjct: 385 RIH-TKNMKLAEDVDLERISKDTHGYVGADLAALCTEAALQCIREKMDVI----DLEDDS 439
Query: 600 SSIRMKDPVKVTADYDSSIQLRGSGAKIKRSVVKKPVPFWNQFGQMQRVKEGHLNSHPCP 659
+ + + VT ++ + + + ++ +VV+ P WN G ++ VK + P
Sbjct: 440 IDAEILNSMAVTNEHFHTALGNSNPSALRETVVEVPNVSWNDIGGLENVKRELQETVQYP 499
Query: 660 VDSSSLMSKF 669
V+ KF
Sbjct: 500 VEHPEKFEKF 509
>At5g05570 putative protein
Length = 1124
Score = 33.5 bits (75), Expect = 0.65
Identities = 25/111 (22%), Positives = 46/111 (40%), Gaps = 11/111 (9%)
Query: 708 PCNADDPGHKRVYFESSEDGHWEVIDTEEALCTLLSVLDDRGKREALLIESLERRQTSLC 767
P + DD +R+Y +DG + D +L+ +L+ K + I ++ T+ C
Sbjct: 482 PSHVDDYKLERLYMAGYQDGSMRIWDATYPCLSLIYILEP--KASVIDITGVDASVTAFC 539
Query: 768 RFMSRVKVNGIGTGC--------MSHSDQSELDMVTEDSNSPVSDVDNLNL 810
F S+ +G C + H+ L++VT ++ V L L
Sbjct: 540 -FCSKTSCLAVGNECGMVRLYKLVGHTSGGTLEVVTNTEKKGLAIVTTLTL 589
>At1g04950 putative TATA binding protein-associated factor (F13M7.6)
Length = 549
Score = 33.5 bits (75), Expect = 0.65
Identities = 40/164 (24%), Positives = 66/164 (39%), Gaps = 25/164 (15%)
Query: 20 PSCSFASHGGKGKGPLGRIIQQREKANVPSSAKKNQKRKCLQDLGTVLRNDGPSIGREFD 79
PS SF H GKGKG + + K +V SS ++ +++ ++ DGP D
Sbjct: 401 PSPSFL-HKGKGKGKIISTDPHKRKLSVDSSENQSPQKR-------LITMDGPDGVHSQD 452
Query: 80 YLPSGHKSYNSTCREDQ------QPAKRSKVSKGTGKGLMNASVKKHGIGKGLMNASVKK 133
S ++ D QP+ + S N VK+ G + + ++
Sbjct: 453 QSGSAPMQVDNPVENDNPPQNSVQPSSSEQASDANESESRNGKVKESGRSRAITMKAI-- 510
Query: 134 HGIGKGLMTIWRATNPDSGELPVGFG--FADREVHLISNSKKSV 175
L IW+ + DSG L V + DR + I +++ SV
Sbjct: 511 ------LDQIWK-DDLDSGRLLVKLHELYGDRILPFIPSTEMSV 547
>At3g20300 unknown protein
Length = 452
Score = 33.1 bits (74), Expect = 0.84
Identities = 25/79 (31%), Positives = 38/79 (47%), Gaps = 9/79 (11%)
Query: 447 LAKATQIAELNLASTTEELESLICSTLSSDITLFEKISSSAYRL--RMST---VTKDDDE 501
L A++I A T + +C+T+ S FE + RL R S DDD
Sbjct: 315 LRSASKITHKAQAVTCLAAKWHVCATIES----FETVDGETPRLVDRASGHGYYPTDDDN 370
Query: 502 CQSDMEDYGSVDDELDDSD 520
+SD EDYG +D+ D+++
Sbjct: 371 GESDSEDYGDEEDDFDNNN 389
>At1g17680 unknown protein
Length = 896
Score = 32.7 bits (73), Expect = 1.1
Identities = 21/48 (43%), Positives = 26/48 (53%), Gaps = 8/48 (16%)
Query: 498 DDDECQSDMEDYGSVDDELDDSDTCSCG------DDFESGSVDSNIRK 539
DD + SD E G VDD+ DDSD G DDFE+GSV + +
Sbjct: 41 DDKDLNSDEE--GLVDDDDDDSDDDDEGDESEEEDDFEAGSVPNTFER 86
>At1g16030 unknown protein
Length = 646
Score = 32.7 bits (73), Expect = 1.1
Identities = 40/160 (25%), Positives = 74/160 (46%), Gaps = 28/160 (17%)
Query: 125 GLMNASVKKHGIG-KGLMTIWRATNPDSGELPVGFGFADREVH-LISNSKKSVRENNRSS 182
G++N S + G K +TI TN D G L + E+ ++ +++K E+ +
Sbjct: 489 GILNVSAEDKTAGVKNQITI---TN-DKGRL------SKEEIEKMVQDAEKYKAEDEQVK 538
Query: 183 KTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVTRNQSLKEKCELSLDSEISEE 242
K L+N N RN ++D++ L Q+ E Q + +++ E E ++++E
Sbjct: 539 KKVEAKNSLENYAYNMRNTIKDEK--LAQKLTQEDKQKI--EKAIDETIEWIEGNQLAE- 593
Query: 243 GVDRISMLIDDEELELRELQERTNLLICSDH---LAASGM 279
+D+ E +L+EL+ N +I + AA GM
Sbjct: 594 --------VDEFEYKLKELEGICNPIISKMYQGGAAAGGM 625
>At1g03080 unknown protein
Length = 1744
Score = 32.7 bits (73), Expect = 1.1
Identities = 31/140 (22%), Positives = 64/140 (45%), Gaps = 26/140 (18%)
Query: 155 PVGFGFADREVHLISNSKKSVRENNRSSKTATM------------NRMLKNK-------- 194
P FG + +E+ ++ K +RE KTA + N +L+N
Sbjct: 628 PESFGSSVKELQEENSKLKEIRERESIEKTALIEKLEMMEKLVQKNLLLENSISDLNAEL 687
Query: 195 --LQNKRNNLQDKRKLLMQRKAG---ESNQHVTRNQSLKEKC-ELSLDSEISEEGVDRIS 248
++ K L++ L + K+G E + ++R QS E +LS ++ + E + +
Sbjct: 688 ETIRGKLKTLEEASMSLAEEKSGLHSEKDMLISRLQSATENSKKLSEENMVLENSLFNAN 747
Query: 249 MLIDDEELELRELQERTNLL 268
+ +++ + +L+ L+E +LL
Sbjct: 748 VELEELKSKLKSLEESCHLL 767
>At1g01950 unknown protein
Length = 893
Score = 32.7 bits (73), Expect = 1.1
Identities = 25/106 (23%), Positives = 52/106 (48%), Gaps = 12/106 (11%)
Query: 164 EVHLISNSKKSVRENNRSSKTATMNRMLK-NKLQNKRNNLQDKRKLLMQRKAGESNQHVT 222
E LISN R + + +N ++ ++ + +L+++ KL RK+ E
Sbjct: 480 EEKLISNQ----RNHENGKRNGEVNGVVTASEFTRLKESLENEMKL---RKSAEEEVSKV 532
Query: 223 RNQSLKEKCELSLDSEISEEGVDRISMLIDDEELELRELQERTNLL 268
++QS + + E + G+ R+ L++DE L+ ++L+E +L
Sbjct: 533 KSQSTLK----TRSGEGEDAGITRLQKLLEDEALQKKKLEEEVTIL 574
>At5g60130 putative protein
Length = 300
Score = 32.3 bits (72), Expect = 1.4
Identities = 19/53 (35%), Positives = 30/53 (55%), Gaps = 1/53 (1%)
Query: 478 TLFEKISSSAYRLRMSTVTKDDDECQSDMEDYGSVDDELDDSDTCSCGDDFES 530
T+ +KI S+ + V D DE +D S+D++ DDSD +CG+D +S
Sbjct: 85 TMCKKIRRSSDQSEEIKVESDSDEQNQASDDVLSLDEDDDDSD-YNCGEDNDS 136
>At3g50950 putative disease resistance protein
Length = 852
Score = 32.3 bits (72), Expect = 1.4
Identities = 17/39 (43%), Positives = 24/39 (60%), Gaps = 6/39 (15%)
Query: 437 RGNNGCKVSDLAKATQIAELNLAST------TEELESLI 469
R NNGCK+S++ T + +L L+ T EEL+SLI
Sbjct: 670 RSNNGCKLSEVKNLTNLRKLGLSLTRGDQIEEEELDSLI 708
>At3g28770 hypothetical protein
Length = 2081
Score = 32.3 bits (72), Expect = 1.4
Identities = 43/226 (19%), Positives = 84/226 (37%), Gaps = 17/226 (7%)
Query: 31 GKGPLGRIIQQREKANVPSSAKKNQKRKCLQDLGTVLRNDGPS-IGREFDYLPSGHKSYN 89
G + + ++ +E A KN ++LG N+G S I E + G +
Sbjct: 285 GDSAIEKNLESKEDVKSEVEAAKNDGSSMTENLGEAQGNNGVSTIDNEKEVEGQGESIED 344
Query: 90 STCREDQQPAKRSKVSKGTGKGLMNASVKKHGIGKGLMNASVKKHGIGKGLMTIWRATNP 149
S ++ + SK K + A+ G + + + +G+ + N
Sbjct: 345 SDIEKNLE-------SKEDVKSEVEAAKNAGSSMTGKLEEAQRNNGVSTN--ETMNSENK 395
Query: 150 DSGELPVGFGFADREVHLISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLL 209
SGE D+ V+ +N + +EN + + L+NK N + +
Sbjct: 396 GSGE-----STNDKMVNATTNDEDHKKENKEETHENNGESVKGENLENKAGNEESMKGEN 450
Query: 210 MQRKAGESNQHVTRNQSLKEKCELSLDSEISEEGVDRISMLIDDEE 255
++ K G N+ + N S++ K E E R + + ++E
Sbjct: 451 LENKVG--NEELKGNASVEAKTNNESSKEEKREESQRSNEVYMNKE 494
Score = 30.8 bits (68), Expect = 4.2
Identities = 17/82 (20%), Positives = 39/82 (46%), Gaps = 4/82 (4%)
Query: 168 ISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVT----R 223
+ K +E N+ T+N K K ++K+ ++ + M++K + ++V +
Sbjct: 914 VKYKKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKK 973
Query: 224 NQSLKEKCELSLDSEISEEGVD 245
+ K++ S +S++ EE D
Sbjct: 974 QEDNKKETTKSENSKLKEENKD 995
Score = 30.0 bits (66), Expect = 7.1
Identities = 36/162 (22%), Positives = 69/162 (42%), Gaps = 14/162 (8%)
Query: 104 VSKGTGKGLM-NASVKKHGI---GKGLMNASVKKHGIGKGLMTIWRATNPDSGELPVGFG 159
V KG+G+ + KK G K +N S K+ G K + +S +
Sbjct: 906 VQKGSGESVKYKKDEKKEGNKEENKDTINTSSKQKGKDK------KKKKKESKNSNMKKK 959
Query: 160 FADREVHLISNSKKSVRENNRSSKTATMNRMLK--NKLQNKRNNLQDKRKLLMQRKAGES 217
D++ ++ + KK +E+N+ T + N LK NK ++ +D ++K E
Sbjct: 960 EEDKKEYVNNELKK--QEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEE 1017
Query: 218 NQHVTRNQSLKEKCELSLDSEISEEGVDRISMLIDDEELELR 259
+ T+ ++ KEK + ++ +R S +E +L+
Sbjct: 1018 KKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLK 1059
>At5g65930 kinesin-like calmodulin-binding protein
Length = 1260
Score = 32.0 bits (71), Expect = 1.9
Identities = 25/104 (24%), Positives = 53/104 (50%), Gaps = 7/104 (6%)
Query: 162 DREVHL-ISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQH 220
++E+H+ + NSK+ + EN + + + K +++ + + ++K+L R + N+
Sbjct: 743 NKELHVAVDNSKRLLSENKILEQNLNIEKKKKEEVEIHQKRYEQEKKVLKLRVSELENKL 802
Query: 221 VTRNQSLKEKCELSLDSEISEEGVDRISMLIDDEEL-ELRELQE 263
Q L S +S I + D + + + +EL ELRE++E
Sbjct: 803 EVLAQDLD-----SAESTIESKNSDMLLLQNNLKELEELREMKE 841
>At5g43320 casein kinase I
Length = 480
Score = 32.0 bits (71), Expect = 1.9
Identities = 31/150 (20%), Positives = 65/150 (42%), Gaps = 11/150 (7%)
Query: 93 REDQQPAKRSKVSKGTGKGLMNASVKKHGIGKGLMNASVKKHGIGKGLMTIWRATNP--- 149
R D+ A + S+ G + A +++G G G++ A + + ++ + T P
Sbjct: 325 RPDKPSAGAGQDSRDRFSGALEAYARRNGSGSGVVQADRSRPRTSENVLASSKDTTPQNY 384
Query: 150 DSGELPVGFGFADREVHLISNSKKSVRENNRSSKTA--TMNRMLKNKLQNKRNNLQDKRK 207
+ E P+ S+S+K+V + R++ +A T NR + N R++ + +
Sbjct: 385 ERVERPIS------STRHASSSRKAVVSSVRATSSADFTENRSSRVVPSNGRSSTAQRTQ 438
Query: 208 LLMQRKAGESNQHVTRNQSLKEKCELSLDS 237
L+ S+ TR + +++L S
Sbjct: 439 LVPDPTTRPSSSSFTRAAPSRTARDITLQS 468
>At5g12220 unknown protein
Length = 611
Score = 32.0 bits (71), Expect = 1.9
Identities = 28/105 (26%), Positives = 48/105 (45%), Gaps = 7/105 (6%)
Query: 152 GELPVGFGFADREVHLISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQ 211
G LP G + R L + + KS+ SK A N K + RN L+ +
Sbjct: 482 GMLPRIIGSSGRLPLLDNQNAKSI------SKQAQGNNNAKRGAECNRNQLEKSPCKRAR 535
Query: 212 RKAGESNQHVTRNQSLKEKCELSLDSEISE-EGVDRISMLIDDEE 255
+ AG+S + +S +E+ E+ ++ E E V +++ +DEE
Sbjct: 536 KSAGDSESNDVTLESYEEEAEMEIEHAYEETETVAEENLMWNDEE 580
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,951,737
Number of Sequences: 26719
Number of extensions: 1078695
Number of successful extensions: 4030
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 48
Number of HSP's that attempted gapping in prelim test: 3927
Number of HSP's gapped (non-prelim): 108
length of query: 1041
length of database: 11,318,596
effective HSP length: 109
effective length of query: 932
effective length of database: 8,406,225
effective search space: 7834601700
effective search space used: 7834601700
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0004a.8


