

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
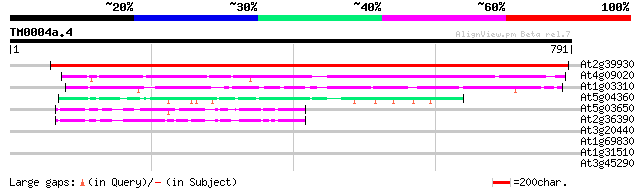
Query= TM0004a.4
(791 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g39930 putative isoamylase 1238 0.0
At4g09020 isoamylase-like protein 607 e-174
At1g03310 putative isoamylase 311 9e-85
At5g04360 pullulanase-like protein (starch debranching enzyme) 88 2e-17
At5g03650 1,4-alpha-glucan branching enzyme protein soform SBE2.... 74 2e-13
At2g36390 starch branching enzyme class II (sbe2-1) 70 6e-12
At3g20440 1,4-alpha-glucan branching enzyme- like protein 42 0.001
At1g69830 alpha-amylase like protein 42 0.002
At1g31510 putative protein 30 4.0
At3g45290 membrane protein Mlo3 (Mlo3) 29 9.0
>At2g39930 putative isoamylase
Length = 783
Score = 1238 bits (3204), Expect = 0.0
Identities = 565/730 (77%), Positives = 639/730 (87%), Gaps = 1/730 (0%)
Query: 58 ETETTLVVDKPQLGGRFQVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQD 117
E E VV+KP RF +S G P+PFG TVRD GVNF++YS N+VSAT+CL +LSD +
Sbjct: 51 EAENIAVVEKPLKSDRFFISDGLPSPFGPTVRDDGVNFSVYSTNSVSATICLISLSDLRQ 110
Query: 118 NQVTEYITLDPLMNKTGSVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYA 177
N+VTE I LDP N+TG VWHVFL+GDF DMLYGY+FDGKFSP EGHYYDSS ILLDPYA
Sbjct: 111 NKVTEEIQLDPSRNRTGHVWHVFLRGDFKDMLYGYRFDGKFSPEEGHYYDSSNILLDPYA 170
Query: 178 KAVISRGEFGSLGPDGNCWPQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIYEMHVRGFTK 237
KA+ISR EFG LGPD NCWPQMA MVP+ ++EFDWEGD+ LK PQKDL+IYEMHVRGFT+
Sbjct: 171 KAIISRDEFGVLGPDDNCWPQMACMVPTREEEFDWEGDMHLKLPQKDLVIYEMHVRGFTR 230
Query: 238 HESSKTKFPGTYLGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGY 297
HESSK +FPGTY GV EKLDHLKELG+NCIEL+PCHEFNELEY+SYN++ GD+RVNFWGY
Sbjct: 231 HESSKIEFPGTYQGVAEKLDHLKELGINCIELMPCHEFNELEYYSYNTILGDHRVNFWGY 290
Query: 298 STVNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPI 357
ST+ +FSPMIRY+SA N INE K L+KEAHKRGIEVIMDVV NHT EGNE GPI
Sbjct: 291 STIGFFSPMIRYASASSNNFAGRAINEFKILVKEAHKRGIEVIMDVVLNHTAEGNEKGPI 350
Query: 358 ISFRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFD 417
SFRGVDNS+YYM+APKGEFYNYSGCGNT NCNHPVVRQFI+DCLRYWVTEMHVDGFRFD
Sbjct: 351 FSFRGVDNSVYYMLAPKGEFYNYSGCGNTFNCNHPVVRQFILDCLRYWVTEMHVDGFRFD 410
Query: 418 LASIMTRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWDA 477
L SIM+RSSSLW+ NV+G +EGDLL TGTP+ PP+ID+ISNDPIL GVKLIAEAWDA
Sbjct: 411 LGSIMSRSSSLWDAANVYGADVEGDLLTTGTPISCPPVIDMISNDPILRGVKLIAEAWDA 470
Query: 478 GGLYQVGTFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWN 537
GGLYQVG FPHWGIWSEWNGK+RD VRQF+KGTDGF+GAFAECLCGSPN+YQ GGRKPW+
Sbjct: 471 GGLYQVGMFPHWGIWSEWNGKFRDVVRQFIKGTDGFSGAFAECLCGSPNLYQ-GGRKPWH 529
Query: 538 SINFVCTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKR 597
SINF+C HDGFTLADLVTYNNK+NL NGE+NNDGENHN SWNCG+EG+FAS SVK+LRKR
Sbjct: 530 SINFICAHDGFTLADLVTYNNKNNLANGEENNDGENHNYSWNCGEEGDFASISVKRLRKR 589
Query: 598 QMRNFFLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFC 657
QMRNFF+SLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNY+NYF+WD KEE+ SDFFRFC
Sbjct: 590 QMRNFFVSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYMNYFRWDKKEEAHSDFFRFC 649
Query: 658 CLMTKFRYECESLGLDDFPTSDRLQWHGHFPGKPDWSETSRFVAFTMVDSVKREIYIAFN 717
++ KFR ECESLGL+DFPT+ RLQWHG P P+WSETSRFVAF++VDSVK+EIY+AFN
Sbjct: 650 RILIKFRDECESLGLNDFPTAKRLQWHGLAPEIPNWSETSRFVAFSLVDSVKKEIYVAFN 709
Query: 718 TSHLPVTITLPERPGYRWEPLVDTGKSAPYDFLTPDLPGRDIAIQQYAQFLDANLYPMLS 777
TSHL ++LP RPGYRWEP VDT K +PYD +TPDLP R+ A++QY FLDAN+YPMLS
Sbjct: 710 TSHLATLVSLPNRPGYRWEPFVDTSKPSPYDCITPDLPERETAMKQYRHFLDANVYPMLS 769
Query: 778 YSSIILLRTP 787
YSSIILL +P
Sbjct: 770 YSSIILLLSP 779
>At4g09020 isoamylase-like protein
Length = 702
Score = 607 bits (1566), Expect = e-174
Identities = 332/737 (45%), Positives = 441/737 (59%), Gaps = 71/737 (9%)
Query: 74 FQVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLS----------DFQDNQVTEY 123
F+VS G +P G + D G+NFA++S NA S TLCL +LS D D+ + E
Sbjct: 7 FKVSSGEVSPLGVSQVDKGINFALFSQNATSVTLCL-SLSQRYITSSGKDDTDDDGMIEL 65
Query: 124 ITLDPLMNKTGSVWHVFLKG-DFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVIS 182
+ LDP +NKTG WH+ ++ ++LYGY+ DG +GH +D S++LLDPYAK V
Sbjct: 66 V-LDPSVNKTGDTWHICVEDLPLNNVLYGYRVDGPGEWQQGHRFDRSILLLDPYAKLVKG 124
Query: 183 RGEFGSLGPDGNCWPQMAGMVPSNDDEFDWEGDLPL-KYPQKDLIIYEMHVRGFTKHESS 241
FG + Q G FDW D P+KDL+IYEM+VR FT ESS
Sbjct: 125 HSSFGD---SSQKFAQFYGTYDFESSPFDWGDDYKFPNIPEKDLVIYEMNVRAFTADESS 181
Query: 242 --KTKFPGTYLGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYST 299
G+YLG +EK+ HL++LG+N +ELLP EF+ELE S D+ VN WGYST
Sbjct: 182 GMDPAIGGSYLGFIEKIPHLQDLGINAVELLPVFEFDELE-LQRRSNPRDHMVNTWGYST 240
Query: 300 VNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIE---------VIMDVVFNHTVE 350
VN+F+PM RY+S + E K ++K H GIE VI+DVV+NHT E
Sbjct: 241 VNFFAPMSRYASG--EGDPIKASKEFKEMVKALHSAGIEKYSYKFSLQVILDVVYNHTNE 298
Query: 351 GNENGPII-SFRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEM 409
++ P SFRG+DN +YYM+ P + N+SGCGNTLNCNHPVV + I+D LR+WVTE
Sbjct: 299 ADDKYPYTTSFRGIDNKVYYMLDPNNQLLNFSGCGNTLNCNHPVVMELILDSLRHWVTEY 358
Query: 410 HVDGFRFDLASIMTRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVK 469
HVDGFRFDLAS++ R++ G+PL +PPLI I+ D +L K
Sbjct: 359 HVDGFRFDLASVLCRATD-------------------GSPLSAPPLIRAIAKDSVLSRCK 399
Query: 470 LIAEAWDAGGLYQVGTFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQ 529
+IAE WD GGLY VG FP+W W+EWNG YRD VR+F+KG G G+FA + GS ++YQ
Sbjct: 400 IIAEPWDCGGLYLVGKFPNWDRWAEWNGMYRDDVRRFIKGDSGMKGSFATRVSGSSDLYQ 459
Query: 530 GGGRKPWNSINFVCTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASS 589
RKP++ +NFV HDGFTL DLV+YN KHN NGE NDG N N+SWNCG EGE +
Sbjct: 460 VNQRKPYHGVNFVIAHDGFTLRDLVSYNFKHNEANGEGGNDGCNDNHSWNCGFEGETGDA 519
Query: 590 SVKKLRKRQMRNFFLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEES 649
+K LR RQM+NF L+LM+SQG PM+ MGDEYGHT+ GNNN+Y HD LN FQW +
Sbjct: 520 HIKSLRTRQMKNFHLALMISQGTPMMLMGDEYGHTRYGNNNSYGHDTSLNNFQWKELDAK 579
Query: 650 SSDFFRFCCLMTKFRYECESLGLDDFPTSDRLQWHGHFPGKPDWSET-SRFVAFTMVDSV 708
+ FRF + KFR+ L ++F T + WH + +W + S+F+AFT+ D +
Sbjct: 580 KQNHFRFFSEVIKFRHSHHVLKHENFLTQGEITWH-----EDNWDNSESKFLAFTLHDGI 634
Query: 709 -KREIYIAFNTSHLPVTITLPE-RPGYRWEPLVDTGKSAPYDFLTPDLPGRDIAIQQYAQ 766
R+IY+AFN V +P+ PG +W + DT +P DF+ + G
Sbjct: 635 GGRDIYVAFNAHDYFVKALIPQPPPGKQWFRVADTNLESPDDFVREGVAG---------- 684
Query: 767 FLDANLYPMLSYSSIIL 783
A+ Y + +SSI+L
Sbjct: 685 --VADTYNVAPFSSILL 699
>At1g03310 putative isoamylase
Length = 882
Score = 311 bits (797), Expect = 9e-85
Identities = 227/730 (31%), Positives = 343/730 (46%), Gaps = 117/730 (16%)
Query: 79 GYPAPFGATVR--DGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTGSV 136
G+P P G + D NF+ +S ++ + LCL+ D ++ + LDP +N+TG V
Sbjct: 230 GHPLPLGLSSGPDDDSWNFSFFSRSSTNVVLCLY--DDSTTDKPALELDLDPYVNRTGDV 287
Query: 137 WHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAV---ISRGEFGSLGPDG 193
WH + + + YGY+ E + I+LDPYA V +S+ GSL
Sbjct: 288 WHASVDNTWDFVRYGYRCKETAHSKEDVDVEGEPIVLDPYATVVGKSVSQKYLGSL---- 343
Query: 194 NCWPQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIYEMHVRGFTKHESSK--TKFPGTYLG 251
S FDW D+ P + L++Y ++V+GFT+H SSK + GT+ G
Sbjct: 344 -----------SKSPSFDWGEDVSPNIPLEKLLVYRLNVKGFTQHRSSKLPSNVAGTFSG 392
Query: 252 VVEKLDHLKELGVNCIELLPCHEFNELE--YFSYNSVQGDYRVNFWGYSTVNYFSPMIRY 309
V EK+ HLK LG N + L P F+E + YF ++ +FSPM Y
Sbjct: 393 VAEKVSHLKTLGTNAVLLEPIFSFSEQKGPYFPFH-----------------FFSPMDIY 435
Query: 310 SSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRGVDNSIYY 369
G N +N +K ++K+ H GIEV+++VVF HT + + RG+D+S YY
Sbjct: 436 ---GPSNSLESAVNSMKVMVKKLHSEGIEVLLEVVFTHTADSG------ALRGIDDSSYY 486
Query: 370 MIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMTRSSSLW 429
+ + S LNCN+PVV+Q +++ LRYWVTE HVDGF F +SSL
Sbjct: 487 YKGRANDLDSKS----YLNCNYPVVQQLVLESLRYWVTEFHVDGFCF------INASSLL 536
Query: 430 NGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWDAGGLY-QVGTFPH 488
GV+ G L PPL++ I+ DP+L KLIA+ WD + + FPH
Sbjct: 537 RGVH-------------GEQLSRPPLVEAIAFDPLLAETKLIADCWDPLEMMPKEVRFPH 583
Query: 489 WGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNSINFVCTHDGF 548
W W+E N +Y VR F++G G A +CGS +V+ GR P S N++ + G
Sbjct: 584 WKRWAELNTRYCRNVRNFLRGR-GVLSDLATRICGSGDVFT-DGRGPAFSFNYISRNSGL 641
Query: 549 TLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNFFLSLMV 608
+L D+V+++ SWNCG+EG S+V + R +Q+RNF +
Sbjct: 642 SLVDIVSFSGPELA-----------SELSWNCGEEGATNKSAVLQRRLKQIRNFLFIQYI 690
Query: 609 SQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEES-SSDFFRFCCLMTKFR-YE 666
S GVP++ MGDE G + G+ F W++ + + +F MT R
Sbjct: 691 SLGVPVLNMGDECGISTRGSPLLESR----KPFDWNLLASAFGTQITQFISFMTSVRARR 746
Query: 667 CESLGLDDFPTSDRLQWHGHFPGKPDWSE-TSRFVAFTMVDSVKRE-------------- 711
+ DF + + W+ + P W + S+F+A + + E
Sbjct: 747 SDVFQRRDFLKPENIVWYANDQTTPKWEDPASKFLALEIKSESEEEETASLAEPNEPKSN 806
Query: 712 -IYIAFNTSHLPVTITLPERP-GYRWEPLVDTGKSAPYDFLTPDLPGRDIAIQQYAQFLD 769
++I FN S P ++ LP P G +W LVDT P F + G + ++ Q L
Sbjct: 807 DLFIGFNASDHPESVVLPSLPDGSKWRRLVDTALPFPGFF---SVEGETVVAEEPLQQL- 862
Query: 770 ANLYPMLSYS 779
+Y M YS
Sbjct: 863 -VVYEMKPYS 871
>At5g04360 pullulanase-like protein (starch debranching enzyme)
Length = 965
Score = 88.2 bits (217), Expect = 2e-17
Identities = 149/642 (23%), Positives = 243/642 (37%), Gaps = 119/642 (18%)
Query: 69 QLGGRFQVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDP 128
QL G Y P GA GV+ +++ A + ++C++ + D E + P
Sbjct: 193 QLPGVLDELFSYDGPLGAHFTPEGVSLHLWAPTAQAVSVCIY--KNPLDKSPME---ICP 247
Query: 129 LMNKTGSVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEFGS 188
L G VW +G Y YK Y+ S++ L YA +RG
Sbjct: 248 LKEANG-VWSTEGACSWGGCYYVYKVSV--------YHPSTMKLETCYANDPYARG---- 294
Query: 189 LGPDGNCWPQMAGMVPSNDDEFDWEG--DLPLKYPQ----KDLIIYEMHVRGFTKH-ESS 241
L DG + +V + D+ EG +L K P D+ IYE+HVR F+ + E+
Sbjct: 295 LSADG----RKTFLVNLDSDDLKPEGWDNLADKKPCLRSFSDISIYELHVRDFSANDETV 350
Query: 242 KTKFPGTYLGVVEK----LDHLKEL---GVNCIELLPCHEFNELEYFSYN---------- 284
+ + G YL K + HL++L G+ + LLP +F +++ N
Sbjct: 351 EPENRGGYLAFTSKDSAGVKHLQKLVDAGLTHLHLLPTFQFGDVDDEKENWKSVDTSLLE 410
Query: 285 --------------SVQGDYRVNFWGYSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLIK 330
+Q D N WGY+ V + P Y+S C I E + +++
Sbjct: 411 GLRPDSTEAQARITEIQNDDGYN-WGYNPVLWGVPKGSYASDPTGPCR---IIEFRKMVQ 466
Query: 331 EAHKRGIEVIMDVVFNHTVEGNENGPIISFRGVDNSI--YYMIAPKGEFYNYSGCGNTLN 388
+ G+ V++DVV+NH + +GP +D + YY+ F S C N
Sbjct: 467 ALNCTGLNVVLDVVYNHL---HASGPHDKESVLDKIVPGYYLRRNSDGFIENSTCVNNTA 523
Query: 389 CNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMTRSSSLWNGVNVFGTSIEGDLLATGT 448
H +V + I D L WV VDGFRFDL + + +++ N + G+ + G+
Sbjct: 524 SEHYMVDRLIRDDLLNWVVNYKVDGFRFDLMGHIMK-ATIVNAKSAIGSLRKETDGVDGS 582
Query: 449 PLVSPPLIDLISNDPILHGVKLIAEAWDAGGLYQVG---TFPHWGIWSEWNGKYRDTVRQ 505
+ L E W+ G + + G + + G + D +R
Sbjct: 583 ------------------RIYLYGEGWNFGEVAENGRGINASQFNLGGTGIGSFNDRIRD 624
Query: 506 FVKGTDGFA----GAFAECLCGSPNVYQGGGRKPWNSI-----NFVCTHDGFTLADLVTY 556
G F F L PN + G + N + T L D +
Sbjct: 625 ATLGGSPFGHPLQQGFITGLLLQPNAHDHGSEATQELMLSTAKNHIQTGMAANLKDYMLT 684
Query: 557 NNKHNLPNGED------------NNDGENHNNSWNCGQEGEFASSSVK-----KLRKRQM 599
N++ G + + E N E F S+K + +R
Sbjct: 685 NHEGKEVKGSEVLMHDATPVAYASLPTETINYVSAHDNETLFDIISLKTPMEISVDERCR 744
Query: 600 RNFFLSLMV--SQGVPMIYMGDEYGHTKGGNNNTYCHDNYLN 639
N S M+ SQG+P + GDE +K + ++Y ++ N
Sbjct: 745 INHLASSMIALSQGIPFFHAGDEILRSKSLDRDSYNSGDWFN 786
>At5g03650 1,4-alpha-glucan branching enzyme protein soform SBE2.2
precursor
Length = 805
Score = 74.3 bits (181), Expect = 2e-13
Identities = 99/361 (27%), Positives = 152/361 (41%), Gaps = 70/361 (19%)
Query: 65 VDKPQLGGRFQVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQD-NQVTEY 123
+DK + GG SRGY G + D G+ + ++ A +A+L + DF + N +
Sbjct: 153 IDKYE-GGLEAFSRGYEK-LGFSRSDAGITYREWAPGAKAASL----IGDFNNWNSNADI 206
Query: 124 ITLDPLMNKTGSVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISR 183
+T N+ G VW +FL + DG SP H + + P
Sbjct: 207 MT----RNEFG-VWEIFLPNNT---------DG--SPAIPHGSRVKIRMDTP-------S 243
Query: 184 GEFGSLGPDGNCWPQMAGMVPSNDDEFD--WEGDLPLKYPQ----KDLIIYEMHVRGFTK 237
G S+ Q G +P N +D E K+PQ K L IYE HV
Sbjct: 244 GIKDSIPAWIKFSVQAPGEIPFNGIYYDPPEEEKYVFKHPQPKRPKSLRIYEAHVG---- 299
Query: 238 HESSKTKFPGTYLGVVEK-LDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWG 296
SS TY + L +K+LG N ++++ E + Y +F G
Sbjct: 300 -MSSTEPMVNTYANFRDDVLPRIKKLGYNAVQIMAIQEHS-------------YYASF-G 344
Query: 297 YSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGP 356
Y N+F+P R CG E+K LI AH+ G+ V+MD+V +H + +G
Sbjct: 345 YHVTNFFAPSSR--------CGTP--EELKSLIDRAHELGLVVLMDIVHSHASKNTLDG- 393
Query: 357 IISFRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRF 416
+ F G D + Y+ P+G Y++ N V ++++ R+W+ E DGFRF
Sbjct: 394 LNMFDGTD-AHYFHSGPRG--YHWMWDSRLFNYGSWEVLRYLLSNARWWLEEYKFDGFRF 450
Query: 417 D 417
D
Sbjct: 451 D 451
>At2g36390 starch branching enzyme class II (sbe2-1)
Length = 858
Score = 69.7 bits (169), Expect = 6e-12
Identities = 89/354 (25%), Positives = 145/354 (40%), Gaps = 56/354 (15%)
Query: 65 VDKPQLGGRFQVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYI 124
+DK + GG SRGY FG T G+ + ++ A +A+L + DF +
Sbjct: 188 IDKNE-GGLEAFSRGYEI-FGFTRSATGITYREWAPGAKAASL----IGDFNNWNAKS-- 239
Query: 125 TLDPLMNKTGSVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYA-KAVISR 183
D + VW +FL + DG SP H + + P K I
Sbjct: 240 --DVMARNDFGVWEIFLPNNA---------DG--SPAIPHGSRVKIRMDTPSGIKDSIPA 286
Query: 184 GEFGSLGPDGNCWPQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIYEMHVRGFTKHESSKT 243
S+ P G P +D++ ++ P K L IYE HV G + E
Sbjct: 287 WIKYSVQPPGEIPYNGVYYDPPEEDKYAFKHPRPKK--PTSLRIYESHV-GMSSTEPKIN 343
Query: 244 KFPGTYLGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYF 303
+ V L +K+LG N ++++ E Y +F GY N+F
Sbjct: 344 TYANFRDDV---LPRIKKLGYNAVQIMAIQEHA-------------YYASF-GYHVTNFF 386
Query: 304 SPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRGV 363
+P R+ + +++K LI +AH+ G+ V+MD+V +H + +G + F G
Sbjct: 387 APSSRFGTP----------DDLKSLIDKAHELGLVVLMDIVHSHASKNTLDG-LDMFDGT 435
Query: 364 DNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFD 417
D Y+ +G Y++ N V ++++ R+W+ E DGFRFD
Sbjct: 436 DGQ-YFHSGSRG--YHWMWDSRLFNYGSWEVLRYLLSNARWWLEEYKFDGFRFD 486
>At3g20440 1,4-alpha-glucan branching enzyme- like protein
Length = 869
Score = 42.4 bits (98), Expect = 0.001
Identities = 31/123 (25%), Positives = 55/123 (44%), Gaps = 15/123 (12%)
Query: 301 NYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISF 360
N+F+ RY + ++ K L+ EAH G+ V +D+V ++ G +S
Sbjct: 426 NFFAASSRYGTP----------DDFKRLVDEAHGLGLLVFLDIVHSYAAADQMVG--LSL 473
Query: 361 RGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRF-DLA 419
N Y+ +G ++ V F++ L +W+TE VDG++F LA
Sbjct: 474 FDGSNDCYFHYGKRGHHKHWGT--RMFKYGDLDVLHFLISNLNWWITEYQVDGYQFHSLA 531
Query: 420 SIM 422
S++
Sbjct: 532 SMI 534
>At1g69830 alpha-amylase like protein
Length = 887
Score = 41.6 bits (96), Expect = 0.002
Identities = 57/232 (24%), Positives = 91/232 (38%), Gaps = 49/232 (21%)
Query: 205 SNDDEFDWEGDLPLKYPQKDLII-------YEMHVRGFTKHESSKTKFPGTYLGVVEKLD 257
S F EG L + + D+ I +E+ +GF + ++ YL + EK D
Sbjct: 465 STTPAFSEEGVLEAEADKPDIKISSGTGSGFEILCQGFNWESNKSGRW---YLELQEKAD 521
Query: 258 HLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYFSPMIRYSSAGIQNC 317
L LG + L P E E GY + ++ RY +
Sbjct: 522 ELASLGFTVLWLPPPTESVSPE----------------GYMPKDLYNLNSRYGT------ 559
Query: 318 GRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVE--GNENGPIISFRG----------VDN 365
I+E+K +K+ HK GI+V+ D V NH N+NG F G D+
Sbjct: 560 ----IDELKDTVKKFHKVGIKVLGDAVLNHRCAHFKNQNGVWNLFGGRLNWDDRAVVADD 615
Query: 366 SIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFD 417
+ K N+ N ++ + VR+ I + L + + E+ DG+R D
Sbjct: 616 PHFQGRGNKSSGDNFHAAPN-IDHSQDFVRKDIKEWLCWMMEEVGYDGWRLD 666
>At1g31510 putative protein
Length = 309
Score = 30.4 bits (67), Expect = 4.0
Identities = 29/111 (26%), Positives = 48/111 (43%), Gaps = 14/111 (12%)
Query: 328 LIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRGVDNSIYYMIAPKGEFYNYSGCGNTL 387
L+KEA R I+ F+H+++ G I + + Y M+ + C N L
Sbjct: 80 LLKEA--RWIKCGYSFEFDHSIDAYGLGYISNESPSGCNDYKMVRFR--------CPNNL 129
Query: 388 NCNHPVVRQFIVDCLRYWVTEMHVDGF-RFDLASIMTRSSSLWNGVNVFGT 437
+ + F DC + V + GF R L+S+ + W G N++GT
Sbjct: 130 SVE---IYDFKSDCWKVVVVDKTFHGFFRLPLSSVCIGGTPYWIGYNIYGT 177
>At3g45290 membrane protein Mlo3 (Mlo3)
Length = 508
Score = 29.3 bits (64), Expect = 9.0
Identities = 15/55 (27%), Positives = 31/55 (56%), Gaps = 2/55 (3%)
Query: 555 TYNNKHNLPNGEDNNDGENHNNSW--NCGQEGEFASSSVKKLRKRQMRNFFLSLM 607
T + +++ +D++DG+NH+NS+ C +G+ + S + L + F L+ M
Sbjct: 107 TIKSHNDVSEDDDDDDGDNHDNSFFHQCSSKGKTSLISEEGLTQLSYFFFVLACM 161
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.139 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,431,204
Number of Sequences: 26719
Number of extensions: 989703
Number of successful extensions: 2112
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 2071
Number of HSP's gapped (non-prelim): 14
length of query: 791
length of database: 11,318,596
effective HSP length: 107
effective length of query: 684
effective length of database: 8,459,663
effective search space: 5786409492
effective search space used: 5786409492
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0004a.4


