

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0002.14
(340 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
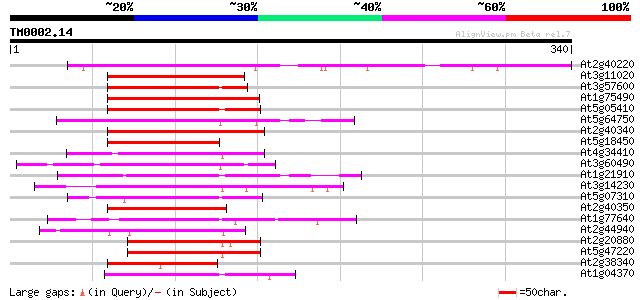
At2g40220 AP2 domain transcription factor (ABI4:abscisic acid-in... 223 9e-59
At3g11020 DREB2B transcription factor 107 9e-24
At3g57600 AP2 transcription factor - like protein 106 2e-23
At1g75490 transcription factor DREB2A like protein 106 2e-23
At5g05410 DREB2A (dbj|BAA33794.1) 105 4e-23
At5g64750 unknown protein 101 5e-22
At2g40340 AP2 domain transcription factor 100 1e-21
At5g18450 AP2-domain DNA-binding protein -like 99 3e-21
At4g34410 unknown protein 95 5e-20
At3g60490 transcription factor - like protein 95 6e-20
At1g21910 TINY like protein 94 8e-20
At3g14230 transcription factor EREBP like protein 94 1e-19
At5g07310 putative transcription factor 93 2e-19
At2g40350 AP2 domain transcription factor 93 2e-19
At1g77640 hypothetical protein 93 2e-19
At2g44940 putative AP2 domain transcription factor 92 3e-19
At2g20880 AP2 domain transcription factor 92 3e-19
At5g47220 ethylene responsive element binding factor 2 (ATERF2) ... 92 4e-19
At2g38340 DREB-like AP2 domain transcription factor 92 4e-19
At1g04370 hypothetical protein 92 4e-19
>At2g40220 AP2 domain transcription factor (ABI4:abscisic
acid-insensitive 4 )
Length = 328
Score = 223 bits (569), Expect = 9e-59
Identities = 148/329 (44%), Positives = 180/329 (53%), Gaps = 42/329 (12%)
Query: 36 NSNSSHNN--NITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKR 93
N +HNN + +T ++ST+ RK KGKGGPDN+KFRYRGVRQRSWGKWVAEIREPRKR
Sbjct: 17 NQTLTHNNPQSDSTTDSSTSSAQRKRKGKGGPDNSKFRYRGVRQRSWGKWVAEIREPRKR 76
Query: 94 TRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQSSSSSSRASS---SSS 150
TRKWLGTF+TAEDAARAYDRAA+ LYGSRAQLNL PS SS S SSSS S ASS SSS
Sbjct: 77 TRKWLGTFATAEDAARAYDRAAVYLYGSRAQLNLTPSSPSSVSSSSSSVSAASSPSTSSS 136
Query: 151 STQTLRPLLPRPSGYGFSFSGSHLPVVFSAAASGPFG--FYN---ANNGYMQLQPQQHHF 205
STQTLRPLLPRP+ V A GP+G F N N G L P F
Sbjct: 137 STQTLRPLLPRPAA----------ATVGGGANFGPYGIPFNNNIFLNGGTSMLCPSYGFF 186
Query: 206 HHQEVVQSQL----QPQLQQCRQPEPDVKGGGVVD-HVRSTSYQNQHSHHHQDQVVMQNY 260
Q+ Q+Q+ Q Q QQ + + + D + N S HH+
Sbjct: 187 PQQQQQQNQMVQMGQFQHQQYQNLHSNTNNNKISDIELTDVPVTNSTSFHHE-------- 238
Query: 261 PVLNHQQHNQNCMGEGVSD--NTLVGPSLASQNFVH-------IDAAPMDPDPGSGIGSP 311
L +Q C + N+L G +S + H + + +DP G GS
Sbjct: 239 VALGQEQGGSGCNNNSSMEDLNSLAGSVGSSLSITHPPPLVDPVCSMGLDPGYMVGDGSS 298
Query: 312 SIWPLTSTELEDYNPVCLWDYNDPFFLDF 340
+IWP E +N +WD+ DP +F
Sbjct: 299 TIWPFGGEEEYSHNWGSIWDFIDPILGEF 327
>At3g11020 DREB2B transcription factor
Length = 330
Score = 107 bits (267), Expect = 9e-24
Identities = 53/83 (63%), Positives = 60/83 (71%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGPDN+ +RGVRQR WGKWVAEIREP+ TR WLGTF TAE AA AYD AA +Y
Sbjct: 66 KGKGGPDNSHCSFRGVRQRIWGKWVAEIREPKIGTRLWLGTFPTAEKAASAYDEAATAMY 125
Query: 120 GSRAQLNLQPSPTSSSSQSSSSS 142
GS A+LN S S + +SS S
Sbjct: 126 GSLARLNFPQSVGSEFTSTSSQS 148
>At3g57600 AP2 transcription factor - like protein
Length = 277
Score = 106 bits (265), Expect = 2e-23
Identities = 53/85 (62%), Positives = 64/85 (74%), Gaps = 1/85 (1%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
+GKGGP N +YRGVRQR+WGKWVAEIREP+KR R WLG+F+TAE+AA AYD AA+ LY
Sbjct: 16 RGKGGPQNALCQYRGVRQRTWGKWVAEIREPKKRARLWLGSFATAEEAAMAYDEAALKLY 75
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSR 144
G A LNL P ++ S S+S R
Sbjct: 76 GHDAYLNL-PHLQRNTRPSLSNSQR 99
>At1g75490 transcription factor DREB2A like protein
Length = 206
Score = 106 bits (264), Expect = 2e-23
Identities = 52/92 (56%), Positives = 64/92 (69%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
+GKGGPDN Y+GVRQR+WGKWVAEIREP + R WLGTF T+ +AA AYD AA LY
Sbjct: 30 RGKGGPDNASCTYKGVRQRTWGKWVAEIREPNRGARLWLGTFDTSREAALAYDSAARKLY 89
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSSS 151
G A LNL S S +SS +S+ + SS++
Sbjct: 90 GPEAHLNLPESLRSYPKTASSPASQTTPSSNT 121
>At5g05410 DREB2A (dbj|BAA33794.1)
Length = 335
Score = 105 bits (262), Expect = 4e-23
Identities = 51/93 (54%), Positives = 68/93 (72%), Gaps = 3/93 (3%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N++ +RGVRQR WGKWVAEIREP + +R WLGTF TA++AA AYD AA +Y
Sbjct: 67 KGKGGPENSRCSFRGVRQRIWGKWVAEIREPNRGSRLWLGTFPTAQEAASAYDEAAKAMY 126
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSSST 152
G A+LN P S +S+ +S+SS++ + T
Sbjct: 127 GPLARLNF---PRSDASEVTSTSSQSEVCTVET 156
>At5g64750 unknown protein
Length = 391
Score = 101 bits (252), Expect = 5e-22
Identities = 69/190 (36%), Positives = 94/190 (49%), Gaps = 23/190 (12%)
Query: 29 SETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKWVAEIR 88
S TS ++S+ + T +TT D + RYRGVRQR WGKW AEIR
Sbjct: 142 SSTSKVREASSNMSGPGPTYEYTTTATASSETSSFSGDQPRRRYRGVRQRPWGKWAAEIR 201
Query: 89 EPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLN-------LQPSPTSSSSQSSSS 141
+P K R WLGTF AE AARAYD AA+ G++A+LN ++P+ T + ++
Sbjct: 202 DPFKAARVWLGTFDNAESAARAYDEAALRFRGNKAKLNFPENVKLVRPASTEAQPVHQTA 261
Query: 142 SSRASSS--SSSTQTLRPLLPRPSGYGFSFSGSHLPVVFSAAASGPFGFYNANNGYMQLQ 199
+ R + S S ST TL P+ P + S P++ S YN + M Q
Sbjct: 262 AQRPTQSRNSGSTTTLLPIRPASNQ-----SVHSQPLMQS---------YNLSYSEMARQ 307
Query: 200 PQQHHFHHQE 209
QQ HHQ+
Sbjct: 308 QQQFQQHHQQ 317
>At2g40340 AP2 domain transcription factor
Length = 341
Score = 100 bits (249), Expect = 1e-21
Identities = 50/95 (52%), Positives = 65/95 (67%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N YRGVRQR WGKWVAEIREP R WLGTFS++ +AA AYD AA +Y
Sbjct: 60 KGKGGPENGICDYRGVRQRRWGKWVAEIREPDGGARLWLGTFSSSYEAALAYDEAAKAIY 119
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSSSTQT 154
G A+LNL SSS +++++ S ++ S ++
Sbjct: 120 GQSARLNLPEITNRSSSTAATATVSGSVTAFSDES 154
>At5g18450 AP2-domain DNA-binding protein -like
Length = 307
Score = 99.4 bits (246), Expect = 3e-21
Identities = 46/68 (67%), Positives = 54/68 (78%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N +RGVRQR+WGKWVAEIREP + TR WLGTF+T+ +AA AYD AA LY
Sbjct: 22 KGKGGPENATCTFRGVRQRTWGKWVAEIREPNRGTRLWLGTFNTSVEAAMAYDEAAKKLY 81
Query: 120 GSRAQLNL 127
G A+LNL
Sbjct: 82 GHEAKLNL 89
>At4g34410 unknown protein
Length = 268
Score = 95.1 bits (235), Expect = 5e-20
Identities = 56/124 (45%), Positives = 74/124 (59%), Gaps = 7/124 (5%)
Query: 35 ENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRT 94
E ++SS+ ++P K + + N K YRGVRQR WGK+ AEIR+P++ T
Sbjct: 102 EITSSSNRRRESSPVAKKAEGGGKIRKR---KNKKNGYRGVRQRPWGKFAAEIRDPKRAT 158
Query: 95 RKWLGTFSTAEDAARAYDRAAIILYGSRAQLNL----QPSPTSSSSQSSSSSSRASSSSS 150
R WLGTF TAEDAARAYDRAAI G RA+LN S SS + ++AS+S+S
Sbjct: 159 RVWLGTFETAEDAARAYDRAAIGFRGPRAKLNFPFVDYTSSVSSPVAADDIGAKASASAS 218
Query: 151 STQT 154
+ T
Sbjct: 219 VSAT 222
>At3g60490 transcription factor - like protein
Length = 256
Score = 94.7 bits (234), Expect = 6e-20
Identities = 60/164 (36%), Positives = 93/164 (56%), Gaps = 15/164 (9%)
Query: 5 TTTPHHPPQEITKPTATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGG 64
++ HH ++ TAT + + S ++S +S S+ ++ N S ++ K
Sbjct: 9 SSATHHQDNIVSVITATIS---SSSVVTSSSDSWSTSKRSLVQDNDS---GGKRRKSNVS 62
Query: 65 PDN-NKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRA 123
DN N YRGVR RSWGKWV+EIREPRK++R WLGT+ TAE AARA+D AA+ + G+
Sbjct: 63 DDNKNPTSYRGVRMRSWGKWVSEIREPRKKSRIWLGTYPTAEMAARAHDVAALAIKGNSG 122
Query: 124 QLN------LQPSPTSSSSQSSSSSSRASSSSSSTQTLRPLLPR 161
LN L P P S S + ++ A+ ++ +T +P++ +
Sbjct: 123 FLNFPELSGLLPRPVSCSPKDIQAA--ATKAAEATTWHKPVIDK 164
>At1g21910 TINY like protein
Length = 230
Score = 94.4 bits (233), Expect = 8e-20
Identities = 60/184 (32%), Positives = 97/184 (52%), Gaps = 25/184 (13%)
Query: 30 ETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKWVAEIRE 89
+TS+++ ++ ++ ++S++ ++ CK K K +Y+GVR RSWG WV+EIR
Sbjct: 9 QTSSTKKEMPLSSSPSSSSSSSSSSSSSSCKNKNKKSKIK-KYKGVRMRSWGSWVSEIRA 67
Query: 90 PRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQSSSSSSRASSSS 149
P ++TR WLG++STAE AARAYD A + L G +A LN PTSSSS + ++
Sbjct: 68 PNQKTRIWLGSYSTAEAAARAYDVALLCLKGPQANLNF---PTSSSSHHLLDNLLDENTL 124
Query: 150 SSTQTLRPLLPRPSGYGFSFSGSHLPVVFSAAASGPFGFYNANNGYMQLQPQQHHFHHQE 209
S ++++ + + + S +H SA +S P H HH +
Sbjct: 125 LSPKSIQRVAAQAAN-----SFNHFAPTSSAVSS----------------PSDHDHHHDD 163
Query: 210 VVQS 213
+QS
Sbjct: 164 GMQS 167
>At3g14230 transcription factor EREBP like protein
Length = 375
Score = 94.0 bits (232), Expect = 1e-19
Identities = 68/204 (33%), Positives = 94/204 (45%), Gaps = 35/204 (17%)
Query: 16 TKPTATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRYRGV 75
TKP A+A ++T S + T+ + + K K +YRG+
Sbjct: 86 TKPVASAFVSTVGSAYAKK------------------TVESAEQAEKSSKRKRKNQYRGI 127
Query: 76 RQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNL----QPSP 131
RQR WGKW AEIR+PRK +R+WLGTF TAE+AARAYD AA + G++A++N PS
Sbjct: 128 RQRPWGKWAAEIRDPRKGSREWLGTFDTAEEAARAYDAAARRIRGTKAKVNFPEEKNPSV 187
Query: 132 TSSSSQSSSSS----SRASSSSSSTQTLRPLLPRPSGYGFSFSGSHLPVVFSAAA----S 183
S S+ ++ S A + S T +P SF S + F +
Sbjct: 188 VSQKRPSAKTNNLQKSVAKPNKSVTLVQQPTHLSQQYCNNSFDNSFGDMSFMEEKPQMYN 247
Query: 184 GPFGFYNA-----NNGYMQLQPQQ 202
FG N+ NNGY Q
Sbjct: 248 NQFGLTNSFDAGGNNGYQYFSSDQ 271
>At5g07310 putative transcription factor
Length = 263
Score = 93.2 bits (230), Expect = 2e-19
Identities = 54/124 (43%), Positives = 71/124 (56%), Gaps = 11/124 (8%)
Query: 36 NSNSSHNNNITTPNTSTTINNRKCKGKGGPDNN------KFRYRGVRQRSWGKWVAEIRE 89
N +SSH+NN P + N++ P K YRGVRQR WGKW AEIR+
Sbjct: 54 NQSSSHDNNQHQP----VVYNQQDPNPPAPPTQDQGLLRKRHYRGVRQRPWGKWAAEIRD 109
Query: 90 PRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQSSSSSSRASSSS 149
P+K R WLGTF TAE AA AYD AA+ GS+A+LN P +S +S+++ + S
Sbjct: 110 PQKAARVWLGTFETAEAAALAYDNAALKFKGSKAKLNF-PERAQLASNTSTTTGPPNYYS 168
Query: 150 SSTQ 153
S+ Q
Sbjct: 169 SNNQ 172
>At2g40350 AP2 domain transcription factor
Length = 177
Score = 92.8 bits (229), Expect = 2e-19
Identities = 43/72 (59%), Positives = 53/72 (72%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N Y GVRQR+WGKWVAEIREP + + WLGTFS++ +AA AYD A+ +Y
Sbjct: 55 KGKGGPENGICDYTGVRQRTWGKWVAEIREPGRGAKLWLGTFSSSYEAALAYDEASKAIY 114
Query: 120 GSRAQLNLQPSP 131
G A+LNL P
Sbjct: 115 GQSARLNLPLLP 126
>At1g77640 hypothetical protein
Length = 244
Score = 92.8 bits (229), Expect = 2e-19
Identities = 68/216 (31%), Positives = 101/216 (46%), Gaps = 46/216 (21%)
Query: 24 ITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKW 83
+TT+ S S+S +S+SS +++ + ++ CK N +Y+GVR RSWG W
Sbjct: 10 VTTSSSSLSHSSSSSSS---------STSALRHQSCK------NKIKKYKGVRMRSWGSW 54
Query: 84 VAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSS-------- 135
V EIR P ++TR WLG++STAE AARAYD A + L G +A LN P+ T++S
Sbjct: 55 VTEIRAPNQKTRIWLGSYSTAEAAARAYDAALLCLKGPKANLNF-PNITTTSPFLMNIDE 113
Query: 136 ----SQSSSSSSRASSSSSSTQTLRPLLPRPSGYGFSFSGSHLPVVFSAAASGP------ 185
S S A +++SS+ P + + H P S+AAS P
Sbjct: 114 KTLLSPKSIQKVAAQAANSSSDHFTPPSDE-NDHDHDDGLDHHPSASSSAASSPPDDDHH 172
Query: 186 -----------FGFYNANNGYMQLQPQQHHFHHQEV 210
F + N + P + F H E+
Sbjct: 173 NDDDGDLVSLMESFVDYNEHVSLMDPSLYEFGHNEI 208
>At2g44940 putative AP2 domain transcription factor
Length = 295
Score = 92.4 bits (228), Expect = 3e-19
Identities = 56/149 (37%), Positives = 86/149 (57%), Gaps = 26/149 (17%)
Query: 19 TATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKC-----------KGKGGPDN 67
TA+A+++++P T++S +S+S+++N I N+ + R + GG
Sbjct: 31 TASASLSSSP--TTSSSSSSSTNSNFIEEDNSKRKASRRSLSSLVSVEDDDDQNGGGGKR 88
Query: 68 NKFR-------YRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYG 120
K YRGVR RSWGKWV+EIREPRK++R WLGT+ TAE AARA+D AA+ + G
Sbjct: 89 RKTNGGDKHPTYRGVRMRSWGKWVSEIREPRKKSRIWLGTYPTAEMAARAHDVAALAIKG 148
Query: 121 SRAQLNLQ------PSPTSSSSQSSSSSS 143
+ A LN P P ++S + +++
Sbjct: 149 TTAYLNFPKLAGELPRPVTNSPKDIQAAA 177
>At2g20880 AP2 domain transcription factor
Length = 336
Score = 92.4 bits (228), Expect = 3e-19
Identities = 50/93 (53%), Positives = 57/93 (60%), Gaps = 12/93 (12%)
Query: 72 YRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNL---- 127
YRGVRQR WGKWVAEIR+PR RTR WLGTF TAE+AA AYDR A L G A+LN
Sbjct: 187 YRGVRQRHWGKWVAEIRKPRNRTRLWLGTFDTAEEAAMAYDREAFKLRGETARLNFPELF 246
Query: 128 ----QPSPT----SSSSQSSSSSSRASSSSSST 152
+P+P + +S SSR SST
Sbjct: 247 LNKQEPTPVHQKQCETGTTSEDSSRRGEDDSST 279
>At5g47220 ethylene responsive element binding factor 2 (ATERF2)
(sp|O80338)
Length = 243
Score = 92.0 bits (227), Expect = 4e-19
Identities = 52/89 (58%), Positives = 61/89 (68%), Gaps = 8/89 (8%)
Query: 72 YRGVRQRSWGKWVAEIREPRKR-TRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNL--- 127
YRGVRQR WGK+ AEIR+P K R WLGTF TAEDAA AYD AA + GSRA LN
Sbjct: 117 YRGVRQRPWGKFAAEIRDPAKNGARVWLGTFETAEDAALAYDIAAFRMRGSRALLNFPLR 176
Query: 128 ----QPSPTSSSSQSSSSSSRASSSSSST 152
+P P +S+ SSSSS +SSSS+S+
Sbjct: 177 VNSGEPDPVRITSKRSSSSSSSSSSSTSS 205
>At2g38340 DREB-like AP2 domain transcription factor
Length = 244
Score = 92.0 bits (227), Expect = 4e-19
Identities = 45/75 (60%), Positives = 54/75 (72%), Gaps = 8/75 (10%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREP--------RKRTRKWLGTFSTAEDAARAY 111
+GKGGP+N R+RGVRQR WGKWVAEIREP + R WLGTF+TA +AA AY
Sbjct: 58 RGKGGPENPVCRFRGVRQRVWGKWVAEIREPVSHRGANSSRSKRLWLGTFATAAEAALAY 117
Query: 112 DRAAIILYGSRAQLN 126
DRAA ++YG A+LN
Sbjct: 118 DRAASVMYGPYARLN 132
>At1g04370 hypothetical protein
Length = 133
Score = 92.0 bits (227), Expect = 4e-19
Identities = 56/123 (45%), Positives = 72/123 (58%), Gaps = 10/123 (8%)
Query: 58 KCKGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKR-TRKWLGTFSTAEDAARAYDRAAI 116
+ G GG + +YRGVR+R WGK+ AEIR+ RK R WLGTF TAEDAARAYDRAA
Sbjct: 6 RSSGSGGGGAEQGKYRGVRRRPWGKYAAEIRDSRKHGERVWLGTFDTAEDAARAYDRAAY 65
Query: 117 ILYGSRAQLNLQPSPTSSSSQSSSSSSRASSSSSSTQTLR------PLLPRPSGYGFSFS 170
+ G A LN P + + SSS+ A+SSSSS Q +L YG +++
Sbjct: 66 SMRGKAAILNF---PHEYNMGTGSSSTAANSSSSSQQVFEFEYLDDSVLDELLEYGENYN 122
Query: 171 GSH 173
+H
Sbjct: 123 KTH 125
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.127 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,604,705
Number of Sequences: 26719
Number of extensions: 398157
Number of successful extensions: 4540
Number of sequences better than 10.0: 326
Number of HSP's better than 10.0 without gapping: 220
Number of HSP's successfully gapped in prelim test: 110
Number of HSP's that attempted gapping in prelim test: 3418
Number of HSP's gapped (non-prelim): 731
length of query: 340
length of database: 11,318,596
effective HSP length: 100
effective length of query: 240
effective length of database: 8,646,696
effective search space: 2075207040
effective search space used: 2075207040
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0002.14


