

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
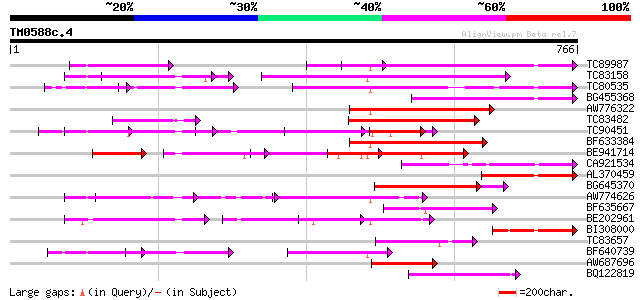
Query= TM0588c.4
(766 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat ... 251 1e-66
TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein... 232 4e-61
TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9... 194 1e-49
BG455368 weakly similar to GP|9759524|dbj selenium-binding prote... 178 8e-45
AW776322 weakly similar to PIR|H84859|H84 hypothetical protein A... 176 3e-44
TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-bin... 166 2e-41
TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13... 92 1e-37
BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30... 154 2e-37
BE941714 weakly similar to GP|11994734|dbj selenium-binding prot... 148 9e-36
CA921534 similar to PIR|D96516|D96 F16N3.14 [imported] - Arabido... 136 3e-32
AL370459 weakly similar to GP|20146256|d hypothetical protein {O... 130 2e-30
BG645370 weakly similar to PIR|T00405|T004 hypothetical protein ... 127 5e-30
AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.7... 120 3e-27
BF635667 weakly similar to PIR|C85359|C853 hypothetical protein ... 114 1e-25
BE202961 weakly similar to GP|6016735|gb|A hypothetical protein ... 106 4e-23
BI308000 similar to GP|7363286|dbj| Similar to Arabidopsis thali... 104 1e-22
TC83657 weakly similar to GP|11994382|dbj|BAB02341. gb|AAB82628.... 97 2e-20
BF640739 weakly similar to GP|7363286|dbj Similar to Arabidopsis... 97 2e-20
AW687696 weakly similar to PIR|T49965|T499 hypothetical protein ... 95 1e-19
BQ122819 similar to PIR|C84748|C84 hypothetical protein At2g3368... 93 4e-19
>TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat protein
[imported] - Arabidopsis thaliana, partial (62%)
Length = 1559
Score = 251 bits (640), Expect = 1e-66
Identities = 121/321 (37%), Positives = 193/321 (59%), Gaps = 3/321 (0%)
Frame = +2
Query: 449 QVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAG 505
+VF M + S+ M+ A+HG+ K+AL++F ++ + PD +V + SACSHAG
Sbjct: 578 RVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACSHAG 757
Query: 506 LVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSL 565
LVEEG++ F SM H I P + HY CMVDL GR G + EA +LI +M +KP+ VIW SL
Sbjct: 758 LVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSIKPNDVIWRSL 937
Query: 566 LGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVK 625
L +C+ H + +IAAE L+ NS Y+ ++N+Y+ + + IR ++ + +
Sbjct: 938 LSACKVHHNLEIGKIAAENLFMLNQNNSGDYLVLANMYAKAQKWDDVAKIRTKLAERNLV 1117
Query: 626 KQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVE 685
+ PG S +E ++V++F S P I + + Q+K GY+P+ + L D + E
Sbjct: 1118QTPGFSLIEAKRKVYKFVSQDKSIPQWNIIYEMIHQMEWQVKFEGYIPDTSQVLLDVDDE 1297
Query: 686 HKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIV 745
K+++L HS+K+A+ F +++ G ++I +N+R+C+DCH + K S ++++EI
Sbjct: 1298EKKERLKFHSQKLAIAFGLIHTSE----GXPLRITRNLRMCSDCHTYTKYISMIYEREIT 1465
Query: 746 VRDSNRFHHFKNATCSCNDYW 766
VRD RF HFKN +CSC DYW
Sbjct: 1466VRDRXRFXHFKNGSCSCKDYW 1528
Score = 70.9 bits (172), Expect = 2e-12
Identities = 41/143 (28%), Positives = 73/143 (50%), Gaps = 3/143 (2%)
Frame = +1
Query: 82 ACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNI 141
AC+ + +G+ +H ++ ++ D+ + N L+NMY KCG ++ A VF+ M +++
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMG--LEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSV 303
Query: 142 VSWTALISGYAQCGLIRECFYLFSGLLA--HYRPNEFAFASLLSACEE-HDIKCGMQVHA 198
SW+A+I +A + EC L + + R E ++LSAC G +H
Sbjct: 304 ASWSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHG 483
Query: 199 VALKISLDASVYVANALITMYSK 221
+ L+ + +V V +LI MY K
Sbjct: 484 ILLRNISELNVVVKTSLIDMYVK 552
Score = 56.6 bits (135), Expect = 3e-08
Identities = 31/113 (27%), Positives = 57/113 (50%), Gaps = 4/113 (3%)
Frame = +1
Query: 402 ACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVS 461
AC+ + + VH V K G + D ++ N+LI+ Y + G + + VF+ M + S
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 462 WNSMLKSYALHGKAKDALELFKKLD----VHPDSTTFVALLSACSHAGLVEEG 510
W++++ ++A + L L K+ + +T V +LSAC+H G + G
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLG 468
Score = 35.4 bits (80), Expect = 0.082
Identities = 19/69 (27%), Positives = 39/69 (55%)
Frame = +1
Query: 500 ACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDS 559
ACS G+V+EG+++ + G+ + ++++YG+ G+I A D+ + M K
Sbjct: 130 ACSLLGVVDEGIQVHGHVF-KMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEK-SV 303
Query: 560 VIWSSLLGS 568
WS+++G+
Sbjct: 304 ASWSAIIGA 330
Score = 29.6 bits (65), Expect = 4.5
Identities = 17/80 (21%), Positives = 35/80 (43%), Gaps = 8/80 (10%)
Frame = +2
Query: 117 VNMYSKCGHLE--------YARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLL 168
V ++S C H + F+ + + ++ + G+++E + L +
Sbjct: 725 VGVFSACSHAGLVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSM- 901
Query: 169 AHYRPNEFAFASLLSACEEH 188
+PN+ + SLLSAC+ H
Sbjct: 902 -SIKPNDVIWRSLLSACKVH 958
>TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein [imported]
- Arabidopsis thaliana, partial (42%)
Length = 1194
Score = 232 bits (592), Expect = 4e-61
Identities = 126/340 (37%), Positives = 196/340 (57%), Gaps = 4/340 (1%)
Frame = +2
Query: 341 GQISDCYRLFLDTSGKQDIVSWTAIITVLADQDP-EQAFLLFCQLHRENFVPDWHTFSIA 399
G + D +LF D +++VSWT +I ++ E+A F ++ VPD+ T
Sbjct: 2 GDVDDALKLF-DKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAI 178
Query: 400 LKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDL 459
+ ACA L VH V+K+ F+ + + N+LI YAR G + L+ QVFD M +L
Sbjct: 179 ISACANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNL 358
Query: 460 VSWNSMLKSYALHGKAKDALELF---KKLDVHPDSTTFVALLSACSHAGLVEEGVEIFNS 516
VSWNS++ +A++G A AL F KK + P+ ++ + L+ACSHAGL++EG++IF
Sbjct: 359 VSWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFAD 538
Query: 517 MSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETR 576
+ +H P+++HY C+VDLY R G++ EA D+I MPM P+ V+ SLL +CR G+
Sbjct: 539 IKRDHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKMPMMPNEVVLGSLLAACRTQGDVE 718
Query: 577 LARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVG 636
LA + EL P YV SNIY++ G + A +R+EM++ ++K S +E+
Sbjct: 719 LAEKVMKYQVELYPGGDSNYVLFSNIYAAVGKWDGASKVRREMKERGLQKNLAFSSIEID 898
Query: 637 KQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEIT 676
+H+F SG +H + I S LE+L +L GYVP+ +
Sbjct: 899 SGIHKFVSGDKYHEENDYIYSALELLSFELHLYGYVPDFS 1018
Score = 88.6 bits (218), Expect = 8e-18
Identities = 56/181 (30%), Positives = 90/181 (48%), Gaps = 2/181 (1%)
Frame = +2
Query: 124 GHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGL-LAHYRPNEFAFASLL 182
G ++ A +FD++P +N+VSWT +I G+ + E F + LA P+ +++
Sbjct: 2 GDVDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAII 181
Query: 183 SACEEHD-IKCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMF 241
SAC + G+ VH + +K +V V N+LI MY++C + A +F
Sbjct: 182 SACANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCG---------CIELARQVF 334
Query: 242 KSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFD 301
M RNL+SWNS+I GF GL DKA+ F M G+ + + S ++ + D
Sbjct: 335 DGMSQRNLVSWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLID 514
Query: 302 E 302
E
Sbjct: 515 E 517
Score = 76.6 bits (187), Expect = 3e-14
Identities = 56/212 (26%), Positives = 104/212 (48%), Gaps = 7/212 (3%)
Frame = +2
Query: 75 TYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFD 134
T ++ ACA L G+ +H ++ K+ ++++ + N L++MY++CG +E AR VFD
Sbjct: 164 TVIAIISACANLGALGLGLWVHRLVMKKE--FRDNVKVLNSLIDMYARCGCIELARQVFD 337
Query: 135 QMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAH-YRPNEFAFASLLSACEEHD-IKC 192
M +RN+VSW ++I G+A GL + F + PN ++ S L+AC I
Sbjct: 338 GMSQRNLVSWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDE 517
Query: 193 GMQVHAVALKISLDA-SVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFR-NLI 250
G+++ A + ++ + L+ +YS+ +AW + K M N +
Sbjct: 518 GLKIFADIKRDHRNSPRIEHYGCLVDLYSRAG---------RLKEAWDVIKKMPMMPNEV 670
Query: 251 SWNSMIAGFQFRG---LGDKAIRLFAHMYCSG 279
S++A + +G L +K ++ +Y G
Sbjct: 671 VLGSLLAACRTQGDVELAEKVMKYQVELYPGG 766
>TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9O13.24
{Arabidopsis thaliana}, partial (55%)
Length = 1727
Score = 194 bits (493), Expect = 1e-49
Identities = 108/387 (27%), Positives = 192/387 (48%), Gaps = 3/387 (0%)
Frame = +2
Query: 383 QLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSG 442
+L + D + F I C + + A VH ++ F+ D + N +I Y
Sbjct: 251 ELMEKGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCK 430
Query: 443 SLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKLD---VHPDSTTFVALLS 499
S+ + +VFD M ++ SW+ M++ YA + L+LF++++ + S T +A+LS
Sbjct: 431 SMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLS 610
Query: 500 ACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDS 559
AC A VE+ SM +GI P ++HY ++D+ G+ G + EAE+ I +P +P
Sbjct: 611 ACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEAEEFIEQLPFEPTV 790
Query: 560 VIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEM 619
++ +L R HG+ L E LDP + + ++
Sbjct: 791 TVFETLKNYARIHGDVDLEDHVEELIVSLDPSKA---------------------VANKI 907
Query: 620 RDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLAL 679
KK +S ++ ++ E+ + + ++ I + +K+ GYVP+ L
Sbjct: 908 PTPPPKKYTAISMLDGKNRIIEYKNPTLYKDDEKL------IAMNSMKDAGYVPDTRYVL 1069
Query: 680 YDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNL 739
+D + E KE L +HSE++A+ + +++ ++I+KN+R+C DCHN +K+ S +
Sbjct: 1070HDIDQEAKEQALLYHSERLAIAYGLISTP----PRTPLRIIKNLRVCGDCHNAIKIMSRI 1237
Query: 740 FQKEIVVRDSNRFHHFKNATCSCNDYW 766
+E++VRD+ RFHHFK+ CSC DYW
Sbjct: 1238VGRELIVRDNKRFHHFKDGKCSCGDYW 1318
Score = 66.2 bits (160), Expect = 4e-11
Identities = 39/117 (33%), Positives = 61/117 (51%)
Frame = +2
Query: 48 QGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILHKDPTIQ 107
+G ++EAL L+ +K I ++ + LF C K+K ++ +H+Y L T +
Sbjct: 227 EGKVKEALELM---EKGIKADANC----FEILFDLCGKSKSVEDAKKVHDYFLQS--TFR 379
Query: 108 NDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLF 164
+D + N ++ MY C + AR VFD MP RN+ SW +I GYA + E LF
Sbjct: 380 SDFKMHNKVIEMYGNCKSMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLF 550
Score = 51.6 bits (122), Expect = 1e-06
Identities = 39/163 (23%), Positives = 72/163 (43%), Gaps = 1/163 (0%)
Frame = +2
Query: 148 ISGYAQCGLIRECFYLFSGLLAHYRPNEFAFASLLSAC-EEHDIKCGMQVHAVALKISLD 206
++ + Q G ++E L + + + F L C + ++ +VH L+ +
Sbjct: 209 LTRFCQEGKVKEALELMEKGI---KADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFR 379
Query: 207 ASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGD 266
+ + N +I MY C + DA +F M RN+ SW+ MI G+ +GD
Sbjct: 380 SDFKMHNKVIEMYGNCK---------SMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGD 532
Query: 267 KAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRN 309
+ ++LF M G+ T+L+V S+ A ++ I L +
Sbjct: 533 EGLQLFEQMNELGLEITSETMLAVLSACGSAEAVEDAYIYLES 661
>BG455368 weakly similar to GP|9759524|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (33%)
Length = 687
Score = 178 bits (451), Expect = 8e-45
Identities = 90/225 (40%), Positives = 133/225 (59%), Gaps = 1/225 (0%)
Frame = +1
Query: 543 ISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNI 602
+ +A L+ TMPM+P+ +W +LLG+ HG +A IA+ EL+P N Y+ +S
Sbjct: 7 VEKALQLVQTMPMEPNGGVWGALLGASHIHGNPDVAEIASRSLFELEPDNLGNYLLLSKT 186
Query: 603 YSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQ-VHEFTSGGHHHPHKEAIQSRLEI 661
Y+ + + +RK MR+ +++K PG SWVE +HEF +G HP I+ L+
Sbjct: 187 YALAAKWDDVSRVRKLMREKQLRKNPGCSWVEAKNGIIHEFFAGDVKHPEINEIKKALDD 366
Query: 662 LIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMK 721
L+ +LK GY P++ YD + E K L HSEK+AL + +++ G+ IKIMK
Sbjct: 367 LLQRLKCTGYQPKLNSVPYDIDDEGKRCLLVSHSEKLALAYGLLSTD----AGSTIKIMK 534
Query: 722 NIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
N+RIC DCH M AS L ++I+VRD+ FHHF N CSCN++W
Sbjct: 535 NLRICEDCHIVMCGASXLTGRKIIVRDNMXFHHFLNGACSCNNFW 669
>AW776322 weakly similar to PIR|H84859|H84 hypothetical protein At2g42920
[imported] - Arabidopsis thaliana, partial (27%)
Length = 633
Score = 176 bits (446), Expect = 3e-44
Identities = 83/201 (41%), Positives = 130/201 (64%), Gaps = 4/201 (1%)
Frame = +1
Query: 459 LVSWNSMLKSYALHGKAKDALELFKKLD----VHPDSTTFVALLSACSHAGLVEEGVEIF 514
L WNS++ A++G ++A E F KL+ + PDS +F+ +L+AC H G + + + F
Sbjct: 22 LSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKARDYF 201
Query: 515 NSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGE 574
M + + I P + HY C+VD+ G+ G + EAE+LI MP+KPD++IW SLL SCRKH
Sbjct: 202 ELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLLSSCRKHRN 381
Query: 575 TRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVE 634
++AR AA++ EL+P ++ GYV MSN++++ F EA R M+++ +K+PG S +E
Sbjct: 382 VQIARRAAQRVYELNPSDASGYVLMSNVHAASNKFEEAIEQRLLMKENLTEKEPGCSSIE 561
Query: 635 VGKQVHEFTSGGHHHPHKEAI 655
+ +VHEF +GG HP + I
Sbjct: 562 LYGEVHEFIAGGRLHPKTQEI 624
Score = 32.7 bits (73), Expect = 0.53
Identities = 22/80 (27%), Positives = 38/80 (47%), Gaps = 8/80 (10%)
Frame = +1
Query: 117 VNMYSKCGHL---EYARYVFDQMPRR-----NIVSWTALISGYAQCGLIRECFYLFSGLL 168
+ + + C HL AR F+ M + +I +T ++ Q GL+ E L G+
Sbjct: 142 IGVLTACKHLGAINKARDYFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGM- 318
Query: 169 AHYRPNEFAFASLLSACEEH 188
+P+ + SLLS+C +H
Sbjct: 319 -PLKPDAIIWGSLLSSCRKH 375
>TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-binding
protein-like {Arabidopsis thaliana}, partial (10%)
Length = 895
Score = 166 bits (421), Expect = 2e-41
Identities = 81/179 (45%), Positives = 112/179 (62%), Gaps = 2/179 (1%)
Frame = +2
Query: 458 DLVSWNSMLKSYALHGKAKDALELFKKLDV--HPDSTTFVALLSACSHAGLVEEGVEIFN 515
DLVSWN+++ YA HG A ALE F ++ V PD TFV +LSAC HAGLVEEG + F
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRMKVVGTPDEVTFVNVLSACVHAGLVEEGEKHFT 181
Query: 516 SMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGET 575
M +GI +++HY+CMVDLYGR G+ EAE+LI MP +PD V+W +LL +C H
Sbjct: 182 DMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALLAACGLHSNL 361
Query: 576 RLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVE 634
L AAE+ + L+ + + Y +S I +G + +R M++ +KKQ +SWVE
Sbjct: 362 ELGEYAAERIRRLESSHPVSYSVLSKIQGEKGVWSSVNELRDTMKERGIKKQTAISWVE 538
Score = 44.7 bits (104), Expect = 1e-04
Identities = 31/121 (25%), Positives = 58/121 (47%), Gaps = 3/121 (2%)
Frame = +2
Query: 140 NIVSWTALISGYAQCGLIRECFYLFSGLLAHYRPNEFAFASLLSACEEHDIKCGMQVHAV 199
++VSW A+I GYA GL F + P+E F ++LSAC + + H
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRMKVVGTPDEVTFVNVLSACVHAGLVEEGEKHFT 181
Query: 200 AL--KISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFR-NLISWNSMI 256
+ K + A + + ++ +Y + +G F D+A + K+M F +++ W +++
Sbjct: 182 DMLTKYGIQAEMEHYSCMVDLYGR-AGRF--------DEAENLIKNMPFEPDVVLWGALL 334
Query: 257 A 257
A
Sbjct: 335 A 337
>TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13600
[imported] - Arabidopsis thaliana, partial (12%)
Length = 1362
Score = 92.0 bits (227), Expect = 7e-19
Identities = 60/210 (28%), Positives = 103/210 (48%), Gaps = 4/210 (1%)
Frame = +2
Query: 75 TYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFD 134
T + A +++ +Q G +H+Y + +Q++ + + +N+Y K G + A VFD
Sbjct: 491 TLPIVLKAVSQSFAIQLGQQVHSYGIKLG--LQSNEYCESGFINLYCKAGDFDSAHKVFD 664
Query: 135 QMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAH-YRPNEFAFASLLSACEE-HDIKC 192
+ + SW ALISG +Q GL + +F + H + P+ S++SAC D+
Sbjct: 665 ENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKRHGFEPDGITMVSVMSACGSIGDLYL 844
Query: 193 GMQVHAVALKISLD--ASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLI 250
+Q+H + + + ++N+LI MY KC D A+ +F +ME RN+
Sbjct: 845 ALQLHKYVFQAKTNEWTVILMSNSLIDMYGKCG---------RMDLAYEVFATMEDRNVS 997
Query: 251 SWNSMIAGFQFRGLGDKAIRLFAHMYCSGI 280
SW SMI G+ G +A+ F M G+
Sbjct: 998 SWTSMIVGYAMHGHAKEALGCFHCMKGVGV 1087
Score = 89.4 bits (220), Expect(2) = 1e-37
Identities = 39/76 (51%), Positives = 51/76 (66%)
Frame = +1
Query: 487 VHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEA 546
V P+ TF+ +LSAC H G V+EG F+ M + +GI PQL HY CMVDL GR G +A
Sbjct: 1087 VKPNYVTFIGVLSACVHGGTVQEGRFYFDMMKNIYGITPQLQHYGCMVDLLGRAGLFDDA 1266
Query: 547 EDLIHTMPMKPDSVIW 562
++ MPMKP+SV+W
Sbjct: 1267 RRMVEEMPMKPNSVVW 1314
Score = 88.2 bits (217), Expect = 1e-17
Identities = 57/214 (26%), Positives = 105/214 (48%), Gaps = 7/214 (3%)
Frame = +2
Query: 372 QDPEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLS 431
+ P+ A ++ + R +PD +T I LKA + Q VHS IK G Q +
Sbjct: 422 ESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGIKLGLQSNEYCE 601
Query: 432 NALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKLDVH--- 488
+ I+ Y ++G + +VFDE L SWN+++ + G A DA+ +F + H
Sbjct: 602 SGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKRHGFE 781
Query: 489 PDSTTFVALLSACSHAGLVEEGVE----IFNSMSDNHGIVPQLDHYACMVDLYGRVGKIS 544
PD T V+++SAC G + ++ +F + ++ ++ + ++D+YG+ G++
Sbjct: 782 PDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVILMSN---SLIDMYGKCGRMD 952
Query: 545 EAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLA 578
A ++ TM + S W+S++ HG + A
Sbjct: 953 LAYEVFATMEDRNVS-SWTSMIVGYAMHGHAKEA 1051
Score = 86.3 bits (212), Expect(2) = 1e-37
Identities = 65/235 (27%), Positives = 110/235 (46%), Gaps = 3/235 (1%)
Frame = +2
Query: 251 SWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNC 310
+WN++I + A+R++ M +G+ DR TL V ++++ A ++
Sbjct: 386 NWNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFA-------IQLG 544
Query: 311 FVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLA 370
+H K GL S + + Y G +++F D + + + SW A+I+ L+
Sbjct: 545 QQVHSYGIKLGLQSNEYCESGFINLYCK-AGDFDSAHKVF-DENHEPKLGSWNALISGLS 718
Query: 371 DQD-PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTV 429
A ++F + R F PD T + AC AL +H V + + TV
Sbjct: 719 QGGLAMDAIVVFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTV 898
Query: 430 L--SNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELF 482
+ SN+LI Y + G + L+ +VF M ++ SW SM+ YA+HG AK+AL F
Sbjct: 899 ILMSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEALGCF 1063
Score = 66.2 bits (160), Expect = 4e-11
Identities = 40/132 (30%), Positives = 62/132 (46%), Gaps = 3/132 (2%)
Frame = +2
Query: 39 DAQIRALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNY 98
+A I LS G +A+ + +H + T S+ AC L + LH Y
Sbjct: 695 NALISGLSQGGLAMDAIVVFVDMKRHGFEPDGI---TMVSVMSACGSIGDLYLALQLHKY 865
Query: 99 ILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIR 158
+ + ++N L++MY KCG ++ A VF M RN+ SWT++I GYA G +
Sbjct: 866 VFQAKTNEWTVILMSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAK 1045
Query: 159 E---CFYLFSGL 167
E CF+ G+
Sbjct: 1046EALGCFHCMKGV 1081
>BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30 -
Arabidopsis thaliana, partial (22%)
Length = 595
Score = 154 bits (388), Expect = 2e-37
Identities = 76/195 (38%), Positives = 122/195 (61%), Gaps = 9/195 (4%)
Frame = +1
Query: 460 VSWNSMLKSYALHGKAKDALELFKKL--------DVHPDSTTFVALLSACSHAGLVEEGV 511
++WN ++ +Y +HGK ++AL+LF+++ ++ P+ T++A+ ++ SH+G+V+EG+
Sbjct: 1 ITWNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFASLSHSGMVDEGL 180
Query: 512 EIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSV-IWSSLLGSCR 570
+F +M HGI P DHYAC+VDL GR G+I EA +LI TMP V WSSLLG+C+
Sbjct: 181 NLFYTMKAKHGIEPTSDHYACLVDLLGRSGQIEEAYNLIKTMPSNMKKVDAWSSLLGACK 360
Query: 571 KHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGL 630
H + IAA+ LDP + YV +SNIYSS G + +A +RK+M++ V+K +
Sbjct: 361 IHQNLEIGEIAAKNLFVLDPNVASYYVLLSNIYSSAGLWDQAIDVRKKMKEKGVRKNLDV 540
Query: 631 SWVEVGKQVHEFTSG 645
+ + + F G
Sbjct: 541 VGLNMAMRFTSFWQG 585
Score = 36.2 bits (82), Expect = 0.048
Identities = 13/22 (59%), Positives = 18/22 (81%)
Frame = +3
Query: 625 KKQPGLSWVEVGKQVHEFTSGG 646
KK+PG SW+E G +VH+F +GG
Sbjct: 522 KKEPGCSWIEHGDEVHKFLAGG 587
>BE941714 weakly similar to GP|11994734|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (10%)
Length = 619
Score = 148 bits (373), Expect = 9e-36
Identities = 76/199 (38%), Positives = 122/199 (61%), Gaps = 8/199 (4%)
Frame = +2
Query: 430 LSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELF---KKLD 486
LS +L+ YA+ G+L L++++FD M D+V WN+M+ A+HG K AL+LF +K+
Sbjct: 20 LSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEKVG 199
Query: 487 VHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEA 546
V PD TF+A+ +ACS++G+ EG+ + + M + IVP+ +HY C+VDL R G EA
Sbjct: 200 VKPDDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLVDLLSRAGLFEEA 379
Query: 547 EDLIHTMPM----KPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLG-YVQMSN 601
+I + +++ W + L +C HGET+LA +AAEK +LD G YV +SN
Sbjct: 380 MVMIRKITNSWNGSEETLAWRAFLSACCNHGETQLAELAAEKVLQLDNHIHSGVYVLLSN 559
Query: 602 IYSSEGSFIEAGLIRKEMR 620
+Y++ G +A ++ M+
Sbjct: 560 LYAASGKHNDARRVKDMMK 616
Score = 56.6 bits (135), Expect = 3e-08
Identities = 28/75 (37%), Positives = 46/75 (61%), Gaps = 1/75 (1%)
Frame = +2
Query: 112 LTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGL-LAH 170
L+ L++MY+KCG+LE A+ +FD M R++V W A+ISG A G + LF +
Sbjct: 20 LSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEKVG 199
Query: 171 YRPNEFAFASLLSAC 185
+P++ F ++ +AC
Sbjct: 200 VKPDDITFIAVFTAC 244
Score = 52.0 bits (123), Expect = 9e-07
Identities = 42/157 (26%), Positives = 75/157 (47%), Gaps = 13/157 (8%)
Frame = +2
Query: 208 SVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDK 267
SV ++ +L+ MY+KC + A +F SM R+++ WN+MI+G G G
Sbjct: 11 SVRLSTSLLDMYAKCG---------NLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKG 163
Query: 268 AIRLFAHMYCSGIGFDRATLLSVFSSLNECS-AFDELNIPLRNCFVLHC----------- 315
A++LF M G+ D T ++VF++ + A++ L + + C V +
Sbjct: 164 ALKLFYDMEKVGVKPDDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLV 343
Query: 316 -LATKTGLISEVEVVTALVKSYANLGGQISDCYRLFL 351
L ++ GL E V+ + + N G + + +R FL
Sbjct: 344 DLLSRAGLFEEAMVMIRKITNSWN-GSEETLAWRAFL 451
Score = 42.0 bits (97), Expect = 9e-04
Identities = 45/190 (23%), Positives = 85/190 (44%), Gaps = 10/190 (5%)
Frame = +2
Query: 326 VEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLA-DQDPEQAFLLFCQL 384
V + T+L+ YA G + RLF D+ +D+V W A+I+ +A D + A LF +
Sbjct: 14 VRLSTSLLDMYAKCGN-LELAKRLF-DSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDM 187
Query: 385 HRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIK-RGFQKDTVLSNALIHAYARSG- 442
+ PD TF AC+Y + L + ++ + L+ +R+G
Sbjct: 188 EKVGVKPDDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLVDLLSRAGL 367
Query: 443 ---SLALSEQVFDEM-CCHDLVSWNSMLKSYALHGK---AKDALELFKKLDVHPDSTTFV 495
++ + ++ + + ++W + L + HG+ A+ A E +LD H S +V
Sbjct: 368 FEEAMVMIRKITNSWNGSEETLAWRAFLSACCNHGETQLAELAAEKVLQLDNHIHSGVYV 547
Query: 496 ALLSACSHAG 505
L + + +G
Sbjct: 548 LLSNLYAASG 577
>CA921534 similar to PIR|D96516|D96 F16N3.14 [imported] - Arabidopsis
thaliana, partial (18%)
Length = 790
Score = 136 bits (342), Expect = 3e-32
Identities = 75/237 (31%), Positives = 123/237 (51%)
Frame = -1
Query: 530 YACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELD 589
Y +V+++G + +A + + MP++P +W +L R HG+ A E LD
Sbjct: 778 YLGVVNIFGCAVGLMKAHEFMRNMPIEPGVELWETLRNFARIHGDLEREDCADELLTVLD 599
Query: 590 PKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHH 649
P + A + R KKQ ++ +E +V E+ +
Sbjct: 598 PSKAA-----------------ADKVPLPQR----KKQSAINMLEEKNRVSEYRC---NM 491
Query: 650 PHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGN 709
P+KE +L L GQ++E GYVP+ L+D + E KE L +HSE++A+ + +++
Sbjct: 490 PYKEEGDVKLRGLTGQMREAGYVPDTRYVLHDIDEEEKEKALQYHSERLAIAYGLISTPP 311
Query: 710 LPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
++I+KN+RIC DCHN +K+ S + +E++VRD+ RFHHFK+ CSC DYW
Sbjct: 310 R----TTLRIIKNLRICGDCHNAIKIMSKIVGRELIVRDNKRFHHFKDGKCSCGDYW 152
>AL370459 weakly similar to GP|20146256|d hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (11%)
Length = 477
Score = 130 bits (327), Expect = 2e-30
Identities = 61/129 (47%), Positives = 83/129 (64%)
Frame = +1
Query: 638 QVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEK 697
QVH+F G HP + I +L+ L ++K +GYVP L+D E E KE LF HSEK
Sbjct: 7 QVHKFHVGDTLHPKAQQIYEKLDELALKIKNVGYVPNTDFVLHDVEDEQKEQYLFQHSEK 186
Query: 698 MALVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKN 757
+A+ FA+++ N I++ KN+R+C DCH +K S + +EIVVRD+NRFHH K+
Sbjct: 187 LAVAFALISTPN----PKPIRVFKNLRVCGDCHTAIKYISMVSGREIVVRDANRFHHMKD 354
Query: 758 ATCSCNDYW 766
TCSCNDYW
Sbjct: 355 GTCSCNDYW 381
>BG645370 weakly similar to PIR|T00405|T004 hypothetical protein At2g44880
[imported] - Arabidopsis thaliana, partial (11%)
Length = 784
Score = 127 bits (318), Expect(2) = 5e-30
Identities = 58/144 (40%), Positives = 97/144 (67%)
Frame = -2
Query: 494 FVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTM 553
F A+L+AC+HAG +++G E FN M N+G+ P +D YAC++DLY R G + +A DL+ M
Sbjct: 783 FTAVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEM 604
Query: 554 PMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAG 613
P P+ +IWSS L +C+ +G+ L R AA + +++P N+ Y+ +++IY+++G + EA
Sbjct: 603 PYDPNCIIWSSFLSACKIYGDVELGREAAIQLIKMEPCNAAPYLTLAHIYTTKGLWNEAS 424
Query: 614 LIRKEMRDSRVKKQPGLSWVEVGK 637
+R M+ +K PG S +E+G+
Sbjct: 423 EVRSLMQQRVKRKPPGWS-MELGR 355
Score = 23.1 bits (48), Expect(2) = 5e-30
Identities = 15/45 (33%), Positives = 19/45 (41%), Gaps = 1/45 (2%)
Frame = -3
Query: 631 SWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMG-YVPE 674
SWVEV K H F H I + E + ++ E YV E
Sbjct: 365 SWVEVDKHNHVFAVDDVTHQQSNEIYAE*ESIYFEILEFSPYVVE 231
>AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.70 -
Arabidopsis thaliana, partial (12%)
Length = 703
Score = 120 bits (300), Expect = 3e-27
Identities = 84/255 (32%), Positives = 132/255 (50%), Gaps = 6/255 (2%)
Frame = +3
Query: 116 LVNMYSKCGHLEYARYVFD--QMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAH-YR 172
LV+MY+KC + A ++F + R+N V WTA+++GYAQ G + F + A
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVE 182
Query: 173 PNEFAFASLLSACEEHDIKC-GMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYD 231
N++ F ++L+AC +C G QVH +K ++VYV +AL+ MY+KC
Sbjct: 183 CNQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKC--------- 335
Query: 232 PTGD--DAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLS 289
GD +A M ++ME +++SWNS++ GF GL ++A+RLF +M+ + D T
Sbjct: 336 --GDLKNAKNMLETMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDYT--- 500
Query: 290 VFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRL 349
F S+ C +N +H L KTG + V ALV YA G DC
Sbjct: 501 -FPSVLNCCVVGSINPK-----SVHGLIIKTGFENYKLVSNALVDMYAKTGDM--DCAYT 656
Query: 350 FLDTSGKQDIVSWTA 364
+ ++D++SWT+
Sbjct: 657 VFEKMLEKDVISWTS 701
Score = 88.6 bits (218), Expect = 8e-18
Identities = 58/215 (26%), Positives = 108/215 (49%), Gaps = 6/215 (2%)
Frame = +3
Query: 356 KQDIVSWTAIITVLADQ-DPEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALA 414
+++ V WTA++T A D +A F +H + + +TF L AC+ +
Sbjct: 75 RKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVECNQYTFPTILTACSSVLARCFGEQ 254
Query: 415 VHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGK 474
VH ++K GF + + +AL+ YA+ G L ++ + + M D+VSWNS++ + HG
Sbjct: 255 VHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNMLETMEDDDVVSWNSLMVGFVRHGL 434
Query: 475 AKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVE--IFNSMSDNHGIVPQLDH 529
++AL LFK + ++ D TF ++L+ C + + V I + +N+ +V
Sbjct: 435 EEEALRLFKNMHGRNMKIDDYTFPSVLNCCVVGSINPKSVHGLIIKTGFENYKLVSN--- 605
Query: 530 YACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSS 564
+VD+Y + G + A + M ++ D + W+S
Sbjct: 606 --ALVDMYAKTGDMDCAYTVFEKM-LEKDVISWTS 701
Score = 80.1 bits (196), Expect = 3e-15
Identities = 52/183 (28%), Positives = 97/183 (52%), Gaps = 3/183 (1%)
Frame = +3
Query: 75 TYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFD 134
T+ ++ AC+ G +H +I+ +++++ + LV+MY+KCG L+ A+ + +
Sbjct: 195 TFPTILTACSSVLARCFGEQVHGFIVKSG--FGSNVYVQSALVDMYAKCGDLKNAKNMLE 368
Query: 135 QMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLA-HYRPNEFAFASLLSACEEHDIKCG 193
M ++VSW +L+ G+ + GL E LF + + + +++ F S+L+ C I
Sbjct: 369 TMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDYTFPSVLNCCVVGSIN-P 545
Query: 194 MQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTG--DDAWTMFKSMEFRNLIS 251
VH + +K + V+NAL+ MY+K TG D A+T+F+ M +++IS
Sbjct: 546 KSVHGLIIKTGFENYKLVSNALVDMYAK-----------TGDMDCAYTVFEKMLEKDVIS 692
Query: 252 WNS 254
W S
Sbjct: 693 WTS 701
>BF635667 weakly similar to PIR|C85359|C853 hypothetical protein AT4g30700
[imported] - Arabidopsis thaliana, partial (6%)
Length = 603
Score = 114 bits (285), Expect = 1e-25
Identities = 57/158 (36%), Positives = 86/158 (54%), Gaps = 3/158 (1%)
Frame = +3
Query: 505 GLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSV---I 561
GL+++G +IF SM ++G+ P +HYACMVDL+ R G++ EA L+ M K SV +
Sbjct: 30 GLLDQGKKIFGSMISDYGLEPTAEHYACMVDLFSRCGRLEEALQLLERM--KSSSVTGSM 203
Query: 562 WSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRD 621
W +LL C H ++ +AA +L+P N+ YV + IY S G + I +MRD
Sbjct: 204 WGALLAGCVMHQNVKIGEVAAHHLFQLEPNNTSNYVALWGIYQSRGMVLGVSTIXGKMRD 383
Query: 622 SRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRL 659
+ K PG W+ + + H+F G HP I R+
Sbjct: 384 LGLVKTPGCXWINIAGRAHKFYQGDLSHPLSHIICKRV 497
>BE202961 weakly similar to GP|6016735|gb|A hypothetical protein {Arabidopsis
thaliana}, partial (14%)
Length = 613
Score = 106 bits (264), Expect = 4e-23
Identities = 60/202 (29%), Positives = 113/202 (55%), Gaps = 6/202 (2%)
Frame = +2
Query: 75 TYASLFHACAKNKCLQQGMALH----NYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYAR 130
T S+ ACA L++GM +H NY + T+ + L++MY KC E A
Sbjct: 53 TVVSVLRACACISNLEEGMKIHELAVNYGFEMETTV------STALMDMYMKCFSPEKAV 214
Query: 131 YVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAH-YRPNEFAFASLLSACEEHD 189
+F++MP++++++W L SGYA G++ E ++F +L+ RP+ A +L+ E
Sbjct: 215 DIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIALVKILTTVSELG 394
Query: 190 I-KCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRN 248
I + + HA +K + + ++ +LI +Y+KCS + +DA +FK M +++
Sbjct: 395 ILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCS---------SIEDANKVFKGMTYKD 547
Query: 249 LISWNSMIAGFQFRGLGDKAIR 270
+++W+S+IA + F G G++A++
Sbjct: 548 VVTWSSIIAAYGFHGQGEEALK 613
Score = 83.2 bits (204), Expect = 3e-16
Identities = 46/187 (24%), Positives = 93/187 (49%), Gaps = 3/187 (1%)
Frame = +2
Query: 391 PDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQV 450
P+W T L+ACA ++ + +H + GF+ +T +S AL+ Y + S + +
Sbjct: 41 PNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYMKCFSPEKAVDI 220
Query: 451 FDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAGLV 507
F+ M D+++W + YA +G +++ +F+ + PD+ V +L+ S G++
Sbjct: 221 FNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIALVKILTTVSELGIL 400
Query: 508 EEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLG 567
++ V F++ +G A ++++Y + I +A + M K D V WSS++
Sbjct: 401 QQAV-CFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYK-DVVTWSSIIA 574
Query: 568 SCRKHGE 574
+ HG+
Sbjct: 575 AYGFHGQ 595
Score = 73.9 bits (180), Expect = 2e-13
Identities = 55/198 (27%), Positives = 96/198 (47%), Gaps = 5/198 (2%)
Frame = +2
Query: 288 LSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCY 347
++V S L C+ L ++ +H LA G E V TAL+ Y
Sbjct: 50 VTVVSVLRACACISNLEEGMK----IHELAVNYGFEMETTVSTALMDMYMKCFSP-EKAV 214
Query: 348 RLFLDTSGKQDIVSWTAIITVLADQDP-EQAFLLFCQLHRENFVPDWHTFSIALKACAYF 406
+F + K+D+++W + + AD ++ +F + PD +IAL
Sbjct: 215 DIF-NRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPD----AIALVKILTT 379
Query: 407 VTE----QQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSW 462
V+E QQA+ H+ VIK GF+ + + +LI YA+ S+ + +VF M D+V+W
Sbjct: 380 VSELGILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVTW 559
Query: 463 NSMLKSYALHGKAKDALE 480
+S++ +Y HG+ ++AL+
Sbjct: 560 SSIIAAYGFHGQGEEALK 613
Score = 40.0 bits (92), Expect = 0.003
Identities = 23/87 (26%), Positives = 46/87 (52%)
Frame = +2
Query: 487 VHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEA 546
+ P+ T V++L AC+ +EEG++I + ++ N+G + ++D+Y + +A
Sbjct: 35 IKPNWVTVVSVLRACACISNLEEGMKI-HELAVNYGFEMETTVSTALMDMYMKCFSPEKA 211
Query: 547 EDLIHTMPMKPDSVIWSSLLGSCRKHG 573
D+ + MP K D + W+ L +G
Sbjct: 212 VDIFNRMP-KKDVIAWAVLFSGYADNG 289
>BI308000 similar to GP|7363286|dbj| Similar to Arabidopsis thaliana DNA
chromosome 4 BAC clone F4I10; vegetative storage
protein., partial (12%)
Length = 771
Score = 104 bits (260), Expect = 1e-22
Identities = 49/114 (42%), Positives = 73/114 (63%)
Frame = +3
Query: 653 EAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPC 712
E I + LE L ++++ GY+P+ T +++D E + KE L HSE++A+ F ++N +
Sbjct: 9 EEIYAFLEALGDKIRDAGYIPD-TNSIHDVEEKVKEQLLSSHSERLAIAFGLLNTNH--- 176
Query: 713 GGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
G I + KN+R+C DCH+ K S + +EI+VRD RFHHFKN CSC DYW
Sbjct: 177 -GTPIHVRKNLRVCGDCHDVTKYISLVTGREIIVRDLRRFHHFKNGICSCGDYW 335
>TC83657 weakly similar to GP|11994382|dbj|BAB02341.
gb|AAB82628.1~gene_id:MIL23.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1299
Score = 97.4 bits (241), Expect = 2e-20
Identities = 53/142 (37%), Positives = 84/142 (58%), Gaps = 5/142 (3%)
Frame = +3
Query: 495 VALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMP 554
+ +LSAC+H GL+ E +E+ + M + +GI + HY CMVDL GR GK+ EA +LI MP
Sbjct: 78 LXVLSACAHGGLMSEALEVISKMEE-YGIEMGIRHYGCMVDLLGRAGKLKEAYELIKRMP 254
Query: 555 MKPDSVIWSSLLGSCRKHGETRLAR-----IAAEKFKELDPKNSLGYVQMSNIYSSEGSF 609
MKP+ + +++G+C H + ++A I A+ ++ N V +SNIY++ +
Sbjct: 255 MKPNETVLGAMIGACWIHSDMKMAEQVMKMIGADSAACVNSHN----VLLSNIYAASEKW 422
Query: 610 IEAGLIRKEMRDSRVKKQPGLS 631
+A +IR M D +K PG S
Sbjct: 423 EKAEMIRSSMVDGGSEKIPGYS 488
>BF640739 weakly similar to GP|7363286|dbj Similar to Arabidopsis thaliana
DNA chromosome 4 BAC clone F4I10; vegetative storage
protein., partial (14%)
Length = 444
Score = 97.1 bits (240), Expect = 2e-20
Identities = 48/146 (32%), Positives = 80/146 (53%), Gaps = 4/146 (2%)
Frame = +1
Query: 376 QAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALI 435
+A LFC + + PD TF + A A +QA +H I+ + ++ AL+
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 436 HAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELF----KKLDVHPDS 491
YA+ G++ + ++FD M +++WN+M+ Y HG K AL+LF + + P+
Sbjct: 187 DMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPND 366
Query: 492 TTFVALLSACSHAGLVEEGVEIFNSM 517
TF++++SACSH+G VEEG+ F M
Sbjct: 367 ITFLSVISACSHSGFVEEGLYYFKIM 444
Score = 75.5 bits (184), Expect = 7e-14
Identities = 44/137 (32%), Positives = 72/137 (52%), Gaps = 2/137 (1%)
Frame = +1
Query: 51 LEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDL 110
+ EAL+L T + S T+ S+ A A +Q +H + + + ++
Sbjct: 1 VNEALNLFCTMQSQGIKPDSF---TFVSVITALADLSVTRQAKWIHGLAIRTN--MDTNV 165
Query: 111 FLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLL-- 168
F+ LV+MY+KCG +E AR +FD M R++++W A+I GY GL + LF +
Sbjct: 166 FVATALVDMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNE 345
Query: 169 AHYRPNEFAFASLLSAC 185
A +PN+ F S++SAC
Sbjct: 346 ASLKPNDITFLSVISAC 396
Score = 67.4 bits (163), Expect = 2e-11
Identities = 43/149 (28%), Positives = 79/149 (52%), Gaps = 3/149 (2%)
Frame = +1
Query: 157 IRECFYLFSGLLAH-YRPNEFAFASLLSACEEHDI-KCGMQVHAVALKISLDASVYVANA 214
+ E LF + + +P+ F F S+++A + + + +H +A++ ++D +V+VA A
Sbjct: 1 VNEALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATA 180
Query: 215 LITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAH 274
L+ MY+KC + A +F M+ R++I+WN+MI G+ GLG A+ LF
Sbjct: 181 LVDMYAKCGAI---------ETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDD 333
Query: 275 MYC-SGIGFDRATLLSVFSSLNECSAFDE 302
M + + + T LSV S+ + +E
Sbjct: 334 MQNEASLKPNDITFLSVISACSHSGFVEE 420
>AW687696 weakly similar to PIR|T49965|T499 hypothetical protein F8M21.190 -
Arabidopsis thaliana, partial (16%)
Length = 289
Score = 94.7 bits (234), Expect = 1e-19
Identities = 42/89 (47%), Positives = 59/89 (66%)
Frame = +2
Query: 489 PDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAED 548
PD TF+ +L ACSH GLV+EG F M+ ++ I P + HY CMVDL GR G EA +
Sbjct: 17 PDEITFLCVLCACSHGGLVDEGRRYFEIMNRDYNIKPTIKHYGCMVDLLGRAGLFVEAYE 196
Query: 549 LIHTMPMKPDSVIWSSLLGSCRKHGETRL 577
LI +MP++ +++IW +LL +CR +G L
Sbjct: 197 LIKSMPVECNAIIWRTLLAACRNYGNVEL 283
>BQ122819 similar to PIR|C84748|C84 hypothetical protein At2g33680 [imported]
- Arabidopsis thaliana, partial (15%)
Length = 771
Score = 92.8 bits (229), Expect = 4e-19
Identities = 51/152 (33%), Positives = 81/152 (52%)
Frame = +2
Query: 539 RVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQ 598
R GK++EA++ I + + +W LLG+C+ H L A EK EL S YV
Sbjct: 26 RAGKLNEAKEFIESATVDHGLCLWRILLGACKNHRNYELGVYAGEKLVELGSPESSAYVL 205
Query: 599 MSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSR 658
+S+IY++ G +R+ M+ V K+PG SW+E+ VH F G + HP + I+
Sbjct: 206 LSSIYTALGDRENVERVRRIMKARGVNKEPGCSWIELKGLVHVFVVGDNQHPQVDEIRLE 385
Query: 659 LEILIGQLKEMGYVPEITLALYDTEVEHKEDQ 690
LE+L + + GY P + L +T +++ DQ
Sbjct: 386 LELLTKLMIDEGYQPLLD-RLPETVIDNLTDQ 478
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.136 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,932,829
Number of Sequences: 36976
Number of extensions: 409812
Number of successful extensions: 2205
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 2019
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2140
length of query: 766
length of database: 9,014,727
effective HSP length: 104
effective length of query: 662
effective length of database: 5,169,223
effective search space: 3422025626
effective search space used: 3422025626
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0588c.4


