

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
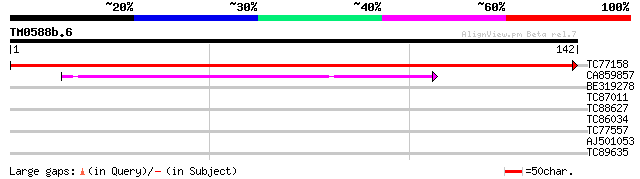
Query= TM0588b.6
(142 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77158 weakly similar to GP|9828633|gb|AAG00256.1| F1N21.7 {Ara... 221 1e-58
CA859857 52 6e-08
BE319278 weakly similar to GP|14334802|gb| putative polygalactur... 30 0.42
TC87011 weakly similar to GP|22136814|gb|AAM91751.1 unknown prot... 30 0.42
TC88627 apyrase-like protein [Medicago truncatula] 30 0.42
TC86034 similar to GP|6815067|dbj|BAA90354.1 ESTs AU082210(C5365... 27 2.7
TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine prot... 26 4.7
AJ501053 similar to GP|17064832|gb putative VP1/ABI3 family regu... 26 6.1
TC89635 weakly similar to GP|20473411|gb|AAM22488.1 36I5.7 {Oryz... 25 8.0
>TC77158 weakly similar to GP|9828633|gb|AAG00256.1| F1N21.7 {Arabidopsis
thaliana}, partial (45%)
Length = 677
Score = 221 bits (562), Expect = 1e-58
Identities = 107/142 (75%), Positives = 119/142 (83%)
Frame = +2
Query: 1 MEEAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLY 60
MEE +K+I HQIGG+ D LRFGL GVKSDIVG+HPL+S+ +S + E MKRQC VNLY
Sbjct: 65 MEEESKTIPHQIGGVHTDVLRFGLPGVKSDIVGAHPLESSLQSVRGVEEAMKRQCKVNLY 244
Query: 61 GAAFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPL 120
GAAFPLK +LDRQILSRFQRP G IPSSMLGLE TG+LD FGFEDYL+D RESET RPL
Sbjct: 245 GAAFPLKEELDRQILSRFQRPPGVIPSSMLGLETVTGTLDHFGFEDYLNDSRESETFRPL 424
Query: 121 DMHHGMEVRLGLSKGPVCPSFM 142
DMHHGMEVRLGLSKGPV PS +
Sbjct: 425 DMHHGMEVRLGLSKGPVYPSII 490
>CA859857
Length = 334
Score = 52.4 bits (124), Expect = 6e-08
Identities = 32/94 (34%), Positives = 49/94 (52%)
Frame = +2
Query: 14 GIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQ 73
G+Q D +G + + S+ SHPL+S + +K LYG P++ ++R
Sbjct: 59 GVQ-DTFHYGPKTLHSEFSPSHPLESILNKWEETQTNLKFTMQKRLYGVHAPIRHLMERS 235
Query: 74 ILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDY 107
I+S+ QR A+PSS LGLE G + FED+
Sbjct: 236 IVSKMQR-LPALPSSNLGLEILMGKDETIDFEDF 334
>BE319278 weakly similar to GP|14334802|gb| putative polygalacturonase
{Arabidopsis thaliana}, partial (20%)
Length = 359
Score = 29.6 bits (65), Expect = 0.42
Identities = 20/58 (34%), Positives = 28/58 (47%)
Frame = +2
Query: 48 NEVMKRQCMVNLYGAAFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFE 105
N V C V++ A LK D+ ++ + RPS + + L AFTGS FG E
Sbjct: 62 NNVCIEDCFVSIGFDAISLKSGWDQYGIN-YGRPSEKVHIRRVHLRAFTGSAISFGTE 232
>TC87011 weakly similar to GP|22136814|gb|AAM91751.1 unknown protein
{Arabidopsis thaliana}, partial (79%)
Length = 1405
Score = 29.6 bits (65), Expect = 0.42
Identities = 20/58 (34%), Positives = 28/58 (47%)
Frame = +3
Query: 48 NEVMKRQCMVNLYGAAFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFE 105
N V C V++ A LK D+ ++ + RPS + + L AFTGS FG E
Sbjct: 897 NNVCIEDCFVSIGFDAISLKSGWDQYGIN-YGRPSEKVHIRRVHLRAFTGSAISFGTE 1067
>TC88627 apyrase-like protein [Medicago truncatula]
Length = 1614
Score = 29.6 bits (65), Expect = 0.42
Identities = 13/44 (29%), Positives = 22/44 (49%), Gaps = 11/44 (25%)
Frame = +1
Query: 103 GFEDYLSDP-----------RESETIRPLDMHHGMEVRLGLSKG 135
G Y +DP +++E + P+D+HH +RLG + G
Sbjct: 325 GLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGATAG 456
>TC86034 similar to GP|6815067|dbj|BAA90354.1 ESTs AU082210(C53655)
AU056546(S20671) AU056545(S20671) correspond to a
region of the predicted, partial (42%)
Length = 2412
Score = 26.9 bits (58), Expect = 2.7
Identities = 19/85 (22%), Positives = 35/85 (40%)
Frame = +3
Query: 23 GLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILSRFQRPS 82
G + + + P+ + +A ++E +R+ V+ + D+D Q L F +
Sbjct: 789 GNRTTSCQLASAGPVPAPPPNAPPVSEYTQRKIFVS------NVSSDIDPQKLLEFFKQF 950
Query: 83 GAIPSSMLGLEAFTGSLDDFGFEDY 107
G + LGL+ TG F Y
Sbjct: 951 GEVDDGPLGLDKATGKPKGFALFVY 1025
>TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (88%)
Length = 2682
Score = 26.2 bits (56), Expect = 4.7
Identities = 13/40 (32%), Positives = 22/40 (54%)
Frame = +3
Query: 15 IQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQ 54
+QN AL++ S I+ +P + +S SKI V K++
Sbjct: 105 LQNSALKYCFH*SSSVILSINPFSTLTQSHSKIKRVRKKK 224
>AJ501053 similar to GP|17064832|gb putative VP1/ABI3 family regulatory
protein {Arabidopsis thaliana}, partial (13%)
Length = 682
Score = 25.8 bits (55), Expect = 6.1
Identities = 8/21 (38%), Positives = 16/21 (76%)
Frame = +1
Query: 108 LSDPRESETIRPLDMHHGMEV 128
+ +PR SE ++P+ +H G++V
Sbjct: 103 MMNPRFSERLKPMPLHQGLQV 165
>TC89635 weakly similar to GP|20473411|gb|AAM22488.1 36I5.7 {Oryza sativa
(japonica cultivar-group)}, partial (54%)
Length = 1133
Score = 25.4 bits (54), Expect = 8.0
Identities = 11/30 (36%), Positives = 17/30 (56%)
Frame = +2
Query: 24 LQGVKSDIVGSHPLQSAQESASKINEVMKR 53
L G+ +DIV SHP + ++ N + KR
Sbjct: 197 LNGLLADIVSSHPKEVTPDATIARNSLFKR 286
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,298,644
Number of Sequences: 36976
Number of extensions: 30803
Number of successful extensions: 146
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 145
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 145
length of query: 142
length of database: 9,014,727
effective HSP length: 87
effective length of query: 55
effective length of database: 5,797,815
effective search space: 318879825
effective search space used: 318879825
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0588b.6


