

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
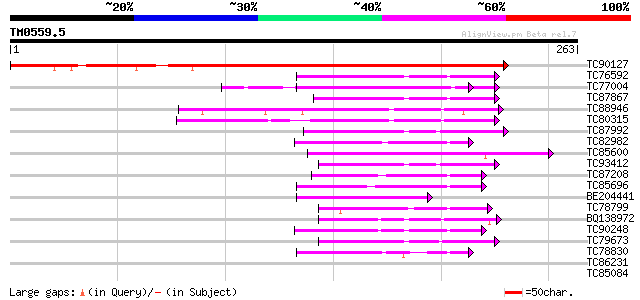
Query= TM0559.5
(263 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC90127 similar to PIR|E85439|E85439 thiol-disulfide interchange... 335 8e-93
TC76592 similar to GP|1388088|gb|AAC49358.1| thioredoxin m {Pisu... 56 2e-08
TC77004 homologue to SP|P38661|PDA6_MEDSA Probable protein disul... 54 6e-08
TC87867 similar to GP|15594012|emb|CAC69854. putative thioredoxi... 53 1e-07
TC88946 similar to GP|21592332|gb|AAM64283.1 thioredoxin putati... 52 2e-07
TC80315 similar to PIR|B96796|B96796 thioredoxin-like protein 4... 51 4e-07
TC87992 similar to SP|Q9SEU7|THM3_ARATH Thioredoxin M-type 3 ch... 51 4e-07
TC82982 weakly similar to GP|1388082|gb|AAC49355.1| thioredoxin ... 49 2e-06
TC85600 homologue to SP|P29828|PDI_MEDSA Protein disulfide isome... 49 2e-06
TC93412 similar to GP|15594012|emb|CAC69854. putative thioredoxi... 49 2e-06
TC87208 similar to SP|Q43636|THIH_RICCO Thioredoxin H-type (TRX-... 47 6e-06
TC85696 similar to SP|Q43636|THIH_RICCO Thioredoxin H-type (TRX-... 47 1e-05
BE204441 SP|P38661|PDA6 Probable protein disulfide isomerase A6 ... 45 2e-05
TC78799 weakly similar to SP|P96132|THIO_THIRO Thioredoxin (TRX)... 45 3e-05
BQ138972 similar to SP|P29429|THIO_ Thioredoxin. [Aspergillus ni... 45 4e-05
TC90248 weakly similar to GP|19548654|gb|AAL90749.1 thioredoxin ... 44 6e-05
TC79673 weakly similar to SP|Q41864|THIM_MAIZE Thioredoxin M-typ... 43 1e-04
TC78830 similar to GP|12322718|gb|AAG51342.1 thioredoxin-like pr... 42 3e-04
TC86231 similar to GP|21593313|gb|AAM65262.1 protein disulfide i... 39 0.002
TC85084 weakly similar to GP|19548654|gb|AAL90749.1 thioredoxin ... 38 0.005
>TC90127 similar to PIR|E85439|E85439 thiol-disulfide interchange like
protein [imported] - Arabidopsis thaliana, partial (53%)
Length = 754
Score = 335 bits (860), Expect = 8e-93
Identities = 177/239 (74%), Positives = 200/239 (83%), Gaps = 8/239 (3%)
Frame = +1
Query: 1 MSRLSSNPIGLHRFRPCLR---TPQFTVNP-RHLHFNTRKFHTLACQTNPNLDEKDASAT 56
M+RLS NPIG HRF+PCL + QFT+ P RH H KF TLA QT+PN +EKD + +
Sbjct: 61 MARLSLNPIGFHRFQPCLHLQPSQQFTLYPPRHRHI---KFLTLASQTDPNHNEKDNTTS 231
Query: 57 S---QEIKIVVEPSTVNGESETCKPTSSTA-DASGLPQLPTKDINRKVAIASTLAALGLF 112
+ QEIK+VVEPST + KPT+++A D++ LPQLP+KDINRK+AI STLAALGLF
Sbjct: 232 TTPPQEIKVVVEPST-----SSSKPTTTSAVDSTSLPQLPSKDINRKIAIVSTLAALGLF 396
Query: 113 LATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDR 172
+ RLDFGVSLKDL+A ALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYK++
Sbjct: 397 VFARLDFGVSLKDLSAVALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKNK 576
Query: 173 VNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALAR 231
VNFVMLNVDNT WEQELDEFGVEGIPHFAFLDKEGNEEGNVVGR+PRQ LLENV LAR
Sbjct: 577 VNFVMLNVDNTKWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRIPRQYLLENVXXLAR 753
>TC76592 similar to GP|1388088|gb|AAC49358.1| thioredoxin m {Pisum sativum},
partial (87%)
Length = 912
Score = 55.8 bits (133), Expect = 2e-08
Identities = 27/94 (28%), Positives = 51/94 (53%)
Frame = +1
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E L++ P +V+F+A WC CR +AP + ++ ++Y +++ LN D ++G
Sbjct: 349 ELVLASDTPVLVDFWAPWCGPCRMIAPIIDELAKEYAGKISCYKLNTDEN--PNIATKYG 522
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
+ IP F K G ++ +V+G +P+ L V+
Sbjct: 523 IRSIPTVLFF-KNGEKKESVIGAVPKSTLSTTVE 621
>TC77004 homologue to SP|P38661|PDA6_MEDSA Probable protein disulfide
isomerase A6 precursor (EC 5.3.4.1) (P5). [Alfalfa]
{Medicago sativa}, complete
Length = 1575
Score = 53.9 bits (128), Expect = 6e-08
Identities = 28/82 (34%), Positives = 40/82 (48%)
Frame = +2
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E L K +VEFYA WC C+ LAP K+ +K + V+ N+D + +++
Sbjct: 566 EVVLDETKDVLVEFYAPWCGHCKSLAPIYEKVAAVFKSEDDVVIANLDADKYRDLAEKYD 745
Query: 194 VEGIPHFAFLDKEGNEEGNVVG 215
V G P F K GN+ G G
Sbjct: 746 VSGFPTLKFFPK-GNKAGEDYG 808
Score = 53.1 bits (126), Expect = 1e-07
Identities = 36/129 (27%), Positives = 61/129 (46%)
Frame = +2
Query: 99 KVAIASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCREL 158
++A+AS A+ LF++ D V L + +E+ + K +VEFYA WC C++L
Sbjct: 122 RIALASFAFAI-LFVSVSADDVVVLTEEN-----FEKEVGQDKGALVEFYAPWCGHCKKL 283
Query: 159 APDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLP 218
AP+ K+ +K + ++ VD + ++GV G P + K E G
Sbjct: 284 APEYEKLGNSFKKAKSVLIAKVDCDEHKGVCSKYGVSGYPTIQWFPKGSLEPKKFEGPRT 463
Query: 219 RQLLLENVD 227
+ L E V+
Sbjct: 464 AESLAEFVN 490
>TC87867 similar to GP|15594012|emb|CAC69854. putative thioredoxin m2 {Pisum
sativum}, partial (79%)
Length = 816
Score = 53.1 bits (126), Expect = 1e-07
Identities = 25/86 (29%), Positives = 45/86 (52%)
Frame = +3
Query: 142 PTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFA 201
P +VEF+A WC CR + P + ++ ++Y ++ LN D + +G+ IP
Sbjct: 315 PVLVEFWAPWCGPCRMIHPIIDELAKEYAGKLKCYKLNTDES--PSVATRYGIRSIPTVI 488
Query: 202 FLDKEGNEEGNVVGRLPRQLLLENVD 227
F K G ++ V+G +P+ L N++
Sbjct: 489 FF-KNGEKKDTVIGAVPKATLTTNIE 563
>TC88946 similar to GP|21592332|gb|AAM64283.1 thioredoxin putative
{Arabidopsis thaliana}, partial (61%)
Length = 761
Score = 52.0 bits (123), Expect = 2e-07
Identities = 49/174 (28%), Positives = 76/174 (43%), Gaps = 23/174 (13%)
Frame = +3
Query: 79 TSSTADASGL--PQLPTKDINRKVAIASTLAALGLFLATRL------------------D 118
TSST+ + L P LP + I ++++S+L RL
Sbjct: 3 TSSTSASLTLTLPTLPVRTITASISVSSSLPLPRSHRQQRLYSQKRSFSNSSSSSVAVPK 182
Query: 119 FGVSLKDLTANALPYE-QALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVM 177
F VS N + L + +P +VEF A WC CR + P + I Q+Y DR+ V
Sbjct: 183 FTVSCGITEINESQFNVTVLKSERPVLVEFVATWCGPCRLITPAMESIAQEYADRLTVV- 359
Query: 178 LNVDNTNWEQELDEFGVEGIPHFAFLDKEGNE--EGNVVGRLPRQLLLENVDAL 229
+D+ Q ++E+ V G+P L K G E G + + L +++DAL
Sbjct: 360 -KIDHDANPQLIEEYKVYGLPTL-ILFKNGEEFPGSRKEGAITKVKLKQHLDAL 515
>TC80315 similar to PIR|B96796|B96796 thioredoxin-like protein 49720-48645
[imported] - Arabidopsis thaliana, partial (70%)
Length = 802
Score = 51.2 bits (121), Expect = 4e-07
Identities = 37/150 (24%), Positives = 68/150 (44%)
Frame = +2
Query: 78 PTSSTADASGLPQLPTKDINRKVAIASTLAALGLFLATRLDFGVSLKDLTANALPYEQAL 137
P+SS++ P R+++ +S L L A + F S DL AN+
Sbjct: 56 PSSSSSSQLQFPVRLLPIGTRRLSSSSRSRFLPLVQAKKQTFS-SFDDLLANS------- 211
Query: 138 SNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGI 197
KP V+FYA WC C+ + P + ++ + +D++ V +D + +++ +E +
Sbjct: 212 --DKPVFVDFYATWCGPCQFMVPVLEEVSARLQDQIQIV--KIDTEKYPSIANKYNIEAL 379
Query: 198 PHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
P F + K+G G L L+E ++
Sbjct: 380 PTF-IIFKDGKPFDRFEGALTADKLIERIE 466
>TC87992 similar to SP|Q9SEU7|THM3_ARATH Thioredoxin M-type 3 chloroplast
precursor (TRX-M3). [Mouse-ear cress] {Arabidopsis
thaliana}, partial (63%)
Length = 918
Score = 51.2 bits (121), Expect = 4e-07
Identities = 27/95 (28%), Positives = 51/95 (53%)
Frame = +2
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
L + P +VEFYA+WC C+ + + +I +Y R+ +LN D Q +E+G++
Sbjct: 275 LKSETPVLVEFYANWCGPCKMVHRVIDEIATEYAGRLKCFVLNTDAD--MQIAEEYGIKA 448
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALAR 231
+P L K G + VVG +P++ + ++ + +
Sbjct: 449 VP-VVVLFKNGKKCDAVVGTMPKEFYVTAIERMLK 550
>TC82982 weakly similar to GP|1388082|gb|AAC49355.1| thioredoxin h
{Arabidopsis thaliana}, partial (38%)
Length = 481
Score = 49.3 bits (116), Expect = 2e-06
Identities = 25/83 (30%), Positives = 42/83 (50%)
Frame = +1
Query: 133 YEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEF 192
+ + L+ GK +V+F A WC C+ +AP + ++YK + L VD + E+
Sbjct: 64 FNKELNKGKLVIVDFTASWCPPCKMIAPIFKNLAEEYKKVA--IFLKVDGDENPEITKEY 237
Query: 193 GVEGIPHFAFLDKEGNEEGNVVG 215
G+ P F F K+G + +VG
Sbjct: 238 GISAYPTFFFF-KDGKKIDEIVG 303
>TC85600 homologue to SP|P29828|PDI_MEDSA Protein disulfide isomerase
precursor (PDI) (EC 5.3.4.1). [Alfalfa] {Medicago
sativa}, complete
Length = 1915
Score = 49.3 bits (116), Expect = 2e-06
Identities = 30/117 (25%), Positives = 49/117 (41%), Gaps = 3/117 (2%)
Frame = +2
Query: 139 NGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIP 198
+GK ++EFYA WC C++LAP + ++ ++ + V+ +D T + D F V+G P
Sbjct: 1229 SGKNVLIEFYAPWCGHCKQLAPILDEVAVSFQSDADVVIAKLDATANDIPTDTFEVQGYP 1408
Query: 199 HFAFLDKEGNEEGNVVGRLPR---QLLLENVDALARGEASVPHARVVGKYSSAEARK 252
F G GR + + +N D V + AE K
Sbjct: 1409 TLYFRSASGKLSQYDGGRTKEDIIEFIEKNKDKTGAAHQEVEQPKAAAAQPEAEQAK 1579
Score = 35.8 bits (81), Expect = 0.017
Identities = 32/116 (27%), Positives = 49/116 (41%), Gaps = 8/116 (6%)
Frame = +2
Query: 91 LPTKDINRKVAIASTLAALGLFLATRLDFGVSLKD-----LTANALPYEQALSNGKPTVV 145
L T + + V+I L +L + +++ S D LT + + + VV
Sbjct: 35 LSTPTMAKNVSIFGLLFSLLALVPSQIFAEESSTDAKEFVLTLDNTNFHDTVKKHDFIVV 214
Query: 146 EFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELD---EFGVEGIP 198
EFYA WC C++LAP+ K V+L + N E D E V+G P
Sbjct: 215 EFYAPWCGHCKKLAPEYEKAASILSTHEPPVVLAKVDANEEHNKDLASENDVKGFP 382
>TC93412 similar to GP|15594012|emb|CAC69854. putative thioredoxin m2 {Pisum
sativum}, partial (47%)
Length = 624
Score = 48.9 bits (115), Expect = 2e-06
Identities = 23/84 (27%), Positives = 44/84 (52%)
Frame = +1
Query: 144 VVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFL 203
+VEF+A WC CR + P + ++ QY ++ LN D + +G+ IP +
Sbjct: 4 LVEFWAPWCGPCRMITPIIDELAIQYTGKLKCYKLNTDES--PSTATRYGIRSIP-TVMI 174
Query: 204 DKEGNEEGNVVGRLPRQLLLENVD 227
K+G ++ V+G +P+ L +++
Sbjct: 175 FKDGEKKDTVIGAVPKSTLTSSIE 246
>TC87208 similar to SP|Q43636|THIH_RICCO Thioredoxin H-type (TRX-H). [Castor
bean] {Ricinus communis}, partial (94%)
Length = 682
Score = 47.4 bits (111), Expect = 6e-06
Identities = 30/81 (37%), Positives = 42/81 (51%)
Frame = +3
Query: 141 KPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHF 200
K VV+F A WC CR +AP + +I ++ + + L VD + E+ VE +P F
Sbjct: 129 KLIVVDFTASWCGPCRFIAPILAEIAKKIPE---VIFLKVDIDEVKSVAKEWSVEAMPTF 299
Query: 201 AFLDKEGNEEGNVVGRLPRQL 221
FL KEG E VVG +L
Sbjct: 300 LFL-KEGKEVDKVVGARKEEL 359
>TC85696 similar to SP|Q43636|THIH_RICCO Thioredoxin H-type (TRX-H). [Castor
bean] {Ricinus communis}, partial (94%)
Length = 714
Score = 46.6 bits (109), Expect = 1e-05
Identities = 32/88 (36%), Positives = 43/88 (48%)
Frame = +2
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E+ + K VV+F A WC CR +AP + +I K N L VD + +E+
Sbjct: 164 EKGNGSKKLIVVDFTASWCGPCRFIAPILAEIA---KKLPNVTFLKVDVDELKTVAEEWA 334
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQL 221
V+ +P F FL KEG VVG QL
Sbjct: 335 VDAMPTFLFL-KEGKLVDKVVGAQKEQL 415
>BE204441 SP|P38661|PDA6 Probable protein disulfide isomerase A6 precursor
(EC 5.3.4.1) (P5). [Alfalfa] {Medicago sativa}, partial
(38%)
Length = 416
Score = 45.4 bits (106), Expect = 2e-05
Identities = 21/63 (33%), Positives = 32/63 (50%)
Frame = +3
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E L K +VEFYA WC C+ LAP K+ +K + V+ N+D + +++
Sbjct: 228 EVVLDETKDVLVEFYAPWCGHCKSLAPIYEKVAAVFKSEDDVVIANLDADKYRDLAEKYD 407
Query: 194 VEG 196
V G
Sbjct: 408 VSG 416
>TC78799 weakly similar to SP|P96132|THIO_THIRO Thioredoxin (TRX)
(Fragment). {Thiocapsa roseopersicina}, partial (37%)
Length = 734
Score = 45.1 bits (105), Expect = 3e-05
Identities = 23/82 (28%), Positives = 44/82 (53%), Gaps = 1/82 (1%)
Frame = +2
Query: 144 VVEFYADWC-EVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAF 202
+VEFYA WC + C + + ++ Y +V+F LN+D + + +G++ +P F
Sbjct: 260 LVEFYAPWCGQECINIHSIMVELANDYAGKVDFYKLNIDENPY--ITNRYGIQNLPTVVF 433
Query: 203 LDKEGNEEGNVVGRLPRQLLLE 224
+ K G + +VG +P+ +E
Sbjct: 434 I-KYGMQRDRLVGYVPKATFIE 496
>BQ138972 similar to SP|P29429|THIO_ Thioredoxin. [Aspergillus nidulans]
{Emericella nidulans}, partial (73%)
Length = 653
Score = 44.7 bits (104), Expect = 4e-05
Identities = 30/88 (34%), Positives = 43/88 (48%), Gaps = 3/88 (3%)
Frame = +1
Query: 144 VVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFL 203
V++ +A WC C+ +AP V K Y + F L+VD E G+ +P F L
Sbjct: 223 VLDCFATWCGPCKVIAPQVVKFSSTYPN-ARFYKLDVDEV--PDVAQELGIRAMPTF-LL 390
Query: 204 DKEGNEEGNVVGRLPRQL---LLENVDA 228
K G++ VVG P+ L + NVDA
Sbjct: 391 FKGGDKVAEVVGANPKALEAAIKANVDA 474
>TC90248 weakly similar to GP|19548654|gb|AAL90749.1 thioredoxin H {Populus
tremula x Populus tremuloides}, partial (63%)
Length = 660
Score = 43.9 bits (102), Expect = 6e-05
Identities = 28/89 (31%), Positives = 44/89 (48%)
Frame = +1
Query: 133 YEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEF 192
+E + K V++F A WC C+ + P + ++ +YKD V F+ ++VD F
Sbjct: 172 FEASKVTNKLMVIDFTATWCGPCKYMDPIIKELAAKYKD-VEFIKIDVDEL--MDVASAF 342
Query: 193 GVEGIPHFAFLDKEGNEEGNVVGRLPRQL 221
V+ +P F L K+G VVG QL
Sbjct: 343 QVQAMPTFILL-KKGKVVEKVVGAKKEQL 426
>TC79673 weakly similar to SP|Q41864|THIM_MAIZE Thioredoxin M-type
chloroplast precursor (TRX-M). [Maize] {Zea mays},
partial (37%)
Length = 743
Score = 43.1 bits (100), Expect = 1e-04
Identities = 21/84 (25%), Positives = 43/84 (51%)
Frame = +1
Query: 144 VVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFL 203
+VEF+A C C+ + + ++ +Y V F NVD+ + ++G++GIP+ +
Sbjct: 214 LVEFFAPLCSPCKNVDFKMVELANEYAGEVEFFKFNVDDN--QLIPSKYGIKGIPN-VLI 384
Query: 204 DKEGNEEGNVVGRLPRQLLLENVD 227
K G + + G LP+ + ++
Sbjct: 385 FKNGEQRDTLFGNLPKATFIRRME 456
>TC78830 similar to GP|12322718|gb|AAG51342.1 thioredoxin-like protein;
56513-57227 {Arabidopsis thaliana}, partial (88%)
Length = 975
Score = 41.6 bits (96), Expect = 3e-04
Identities = 25/87 (28%), Positives = 47/87 (53%), Gaps = 5/87 (5%)
Frame = +3
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVD-----NTNWEQE 188
E+A +GK + F A WC C+ +AP ++ ++Y + F++++VD +T+W+
Sbjct: 327 EEAKKDGKIVIANFSAVWCGPCKVIAPYYCEMSEKYTSMM-FLLVDVDELTDFSTSWD-- 497
Query: 189 LDEFGVEGIPHFAFLDKEGNEEGNVVG 215
++ P F FL K+G + +VG
Sbjct: 498 -----IKATPTFFFL-KDGQQLDKLVG 560
>TC86231 similar to GP|21593313|gb|AAM65262.1 protein disulfide isomerase
precursor-like {Arabidopsis thaliana}, partial (79%)
Length = 2185
Score = 38.9 bits (89), Expect = 0.002
Identities = 21/70 (30%), Positives = 34/70 (48%)
Frame = +1
Query: 133 YEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEF 192
+ + N + +VEFYA WC C+ LAP+ + K + V+ VD + + E+
Sbjct: 388 FTTVIENNQFVMVEFYAPWCGHCQALAPEYAAAATELK-KDGVVLAKVDASVENELAYEY 564
Query: 193 GVEGIPHFAF 202
V+G P F
Sbjct: 565 NVQGFPTVYF 594
>TC85084 weakly similar to GP|19548654|gb|AAL90749.1 thioredoxin H {Populus
tremula x Populus tremuloides}, partial (41%)
Length = 601
Score = 37.7 bits (86), Expect = 0.005
Identities = 24/87 (27%), Positives = 43/87 (48%)
Frame = +3
Query: 133 YEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEF 192
++ + K V+EF A WC C+ + P + +Y +V+F+ ++VD + EF
Sbjct: 147 FDASKETNKLMVIEFTAAWCGPCKYMDPIIQDFAAKY-IKVDFIKIDVDELMSISQ--EF 317
Query: 193 GVEGIPHFAFLDKEGNEEGNVVGRLPR 219
V+ +P F + K G VV ++ R
Sbjct: 318 QVQAMPTFILMKK-----GKVVDKVVR 383
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,437,535
Number of Sequences: 36976
Number of extensions: 95073
Number of successful extensions: 526
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 517
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 522
length of query: 263
length of database: 9,014,727
effective HSP length: 94
effective length of query: 169
effective length of database: 5,538,983
effective search space: 936088127
effective search space used: 936088127
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0559.5


