

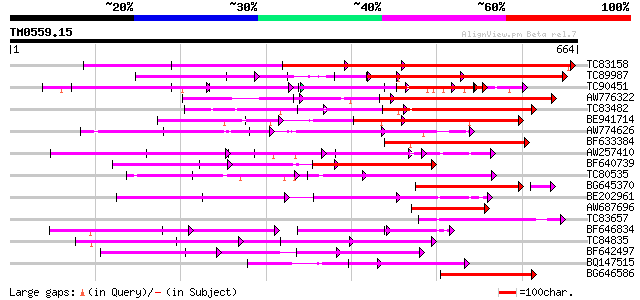
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0559.15
(664 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein... 274 7e-74
TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat ... 191 2e-67
TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13... 163 2e-40
AW776322 weakly similar to PIR|H84859|H84 hypothetical protein A... 161 9e-40
TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-bin... 157 2e-38
BE941714 weakly similar to GP|11994734|dbj selenium-binding prot... 155 4e-38
AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.7... 143 2e-34
BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30... 142 4e-34
AW257410 weakly similar to GP|10177533|db selenium-binding prote... 140 1e-33
BF640739 weakly similar to GP|7363286|dbj Similar to Arabidopsis... 124 1e-28
TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9... 118 8e-27
BG645370 weakly similar to PIR|T00405|T004 hypothetical protein ... 112 1e-26
BE202961 weakly similar to GP|6016735|gb|A hypothetical protein ... 114 1e-25
AW687696 weakly similar to PIR|T49965|T499 hypothetical protein ... 109 4e-24
TC83657 weakly similar to GP|11994382|dbj|BAB02341. gb|AAB82628.... 106 3e-23
BF646834 weakly similar to PIR|A84784|A847 hypothetical protein ... 103 3e-22
TC84835 weakly similar to PIR|T02328|T02328 hypothetical protein... 102 6e-22
BF642497 weakly similar to PIR|H96713|H96 hypothetical protein T... 101 1e-21
BQ147515 weakly similar to PIR|T05355|T05 hypothetical protein F... 100 2e-21
BG646586 weakly similar to GP|10177683|dbj selenium-binding prot... 100 2e-21
>TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein [imported]
- Arabidopsis thaliana, partial (42%)
Length = 1194
Score = 274 bits (701), Expect = 7e-74
Identities = 140/347 (40%), Positives = 217/347 (62%), Gaps = 4/347 (1%)
Frame = +2
Query: 320 GLIDHALEIFDQMPVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSIL 379
G +D AL++FD++PVKN VSW +I V++ + EA+E F M ++GV+PD T+++I+
Sbjct: 2 GDVDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAII 181
Query: 380 SSCSHMGDLALGKKAHDYICDNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQKNVV 439
S+C+++G L LG H + +V + NSL++MYA+CG ++ A +F GM Q+N+V
Sbjct: 182 SACANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLV 361
Query: 440 SWNVIIGALALHGSGKEAIEMFEKMRASGLFPDEITFTGLLSACSHSGLVEMGRYYFGLM 499
SWN II A++G +A+ F M+ GL P+ +++T L+ACSH+GL++ G F +
Sbjct: 362 SWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADI 541
Query: 500 SSTFGISPDVAHYACMVDLLGRGGFLGEAMSLIQKLPMKPDAVVWGALLGACKTFGNLEM 559
SP + HY C+VDL R G L EA +I+K+PM P+ VV G+LL AC+T G++E+
Sbjct: 542 KRDHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKMPMMPNEVVLGSLLAACRTQGDVEL 721
Query: 560 GKQILKQLLELGRYDSGLYVLLSNMYSESQRWDDMKKIRKIMNDSGIKKCRAISSIEIDG 619
++++K +EL YVL SN+Y+ +WD K+R+ M + G++K A SSIEID
Sbjct: 722 AEKVMKYQVELYPGGDSNYVLFSNIYAAVGKWDGASKVRREMKERGLQKNLAFSSIEIDS 901
Query: 620 CCYQFMVDDKNHEVSTSIYSMVDQLMDHLKSVGY----PCKHSDVED 662
++F+ DK HE + IYS ++ L L GY K SDV+D
Sbjct: 902 GIHKFVSGDKYHEENDYIYSALELLSFELHLYGYVPDFSGKESDVDD 1042
Score = 120 bits (301), Expect = 2e-27
Identities = 81/276 (29%), Positives = 134/276 (48%), Gaps = 1/276 (0%)
Frame = +2
Query: 190 IRSARQVFDDISDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEADMFTLVGLLSV 249
+ A ++FD + + +VSW +I G+ K +EA+ F+EMQ GV D T++ ++S
Sbjct: 8 VDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAIISA 187
Query: 250 ALRHGNLDLGRFVHLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRMLDRNVVSW 309
G L LG +VH ++ + V N+LIDMYA+CG ++ A VFD M RN+VSW
Sbjct: 188 CANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLVSW 367
Query: 310 TCMINAYANHGLIDHALEIFDQMPVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVM 369
+I +A +GL D AL F M + G+
Sbjct: 368 NSIIVGFAVNGLADKALSFFRSMKKE-------------------------------GLE 454
Query: 370 PDNATLVSILSSCSHMGDLALGKKAH-DYICDNNITVSVTLCNSLMNMYAKCGALQTAMD 428
P+ + S L++CSH G + G K D D+ + + L+++Y++ G L+ A D
Sbjct: 455 PNGVSYTSALTACSHAGLIDEGLKIFADIKRDHRNSPRIEHYGCLVDLYSRAGRLKEAWD 634
Query: 429 IFFGMPQKNVVSWNVIIGALALHGSGKEAIEMFEKM 464
+ MP ++ V++G+L + +E+ EK+
Sbjct: 635 VIKKMP---MMPNEVVLGSLLAACRTQGDVELAEKV 733
Score = 102 bits (253), Expect = 6e-22
Identities = 74/311 (23%), Positives = 131/311 (41%)
Frame = +2
Query: 87 GDLRYAHLLFDQTPQPNKFMYNHLIRGYSNSDDPNKSLLLYRKMVCAGIMPNQFTIPFVL 146
GD+ A LFD+ P N + +I G+ + ++L +R+M AG++P+ T+ ++
Sbjct: 2 GDVDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAII 181
Query: 147 KACSAKPSCWDVAVVHAQAIKLGMGSHVCVQNAFLSLYVACRLIRSARQVFDDISDRTLV 206
AC+ + VH +K +V V N+ + +Y C I ARQVFD +S R LV
Sbjct: 182 SACANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLV 361
Query: 207 SWNSMIVGYSKMGFSKEAILLFKEMQYLGVEADMFTLVGLLSVALRHGNLDLGRFVHLYI 266
SWNS+IVG++ G + +A+ F+ M+ G+E + + L+ G +D G +
Sbjct: 362 SWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKI---- 529
Query: 267 VVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRMLDRNVVSWTCMINAYANHGLIDHAL 326
+A+ D + + C+++ Y+ G + A
Sbjct: 530 --------------------------FADIKRDHRNSPRIEHYGCLVDLYSRAGRLKEAW 631
Query: 327 EIFDQMPVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMG 386
++ +MP +MP+ L S+L++C G
Sbjct: 632 DVIKKMP----------------------------------MMPNEVVLGSLLAACRTQG 709
Query: 387 DLALGKKAHDY 397
D+ L +K Y
Sbjct: 710 DVELAEKVMKY 742
>TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat protein
[imported] - Arabidopsis thaliana, partial (62%)
Length = 1559
Score = 191 bits (485), Expect(2) = 2e-67
Identities = 98/236 (41%), Positives = 144/236 (60%)
Frame = +2
Query: 418 AKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKEAIEMFEKMRASGLFPDEITFT 477
++ L+ + +F M +KN S+ V+I LA+HG GKEA+++F +M GL PD++ +
Sbjct: 548 SRVDVLRKGLRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYV 727
Query: 478 GLLSACSHSGLVEMGRYYFGLMSSTFGISPDVAHYACMVDLLGRGGFLGEAMSLIQKLPM 537
G+ SACSH+GLVE G F M I P V HY CMVDLLGR G L EA LI+ + +
Sbjct: 728 GVFSACSHAGLVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSI 907
Query: 538 KPDAVVWGALLGACKTFGNLEMGKQILKQLLELGRYDSGLYVLLSNMYSESQRWDDMKKI 597
KP+ V+W +LL ACK NLE+GK + L L + +SG Y++L+NMY+++Q+WDD+ KI
Sbjct: 908 KPNDVIWRSLLSACKVHHNLEIGKIAAENLFMLNQNNSGDYLVLANMYAKAQKWDDVAKI 1087
Query: 598 RKIMNDSGIKKCRAISSIEIDGCCYQFMVDDKNHEVSTSIYSMVDQLMDHLKSVGY 653
R + + + + S IE Y+F+ DK+ IY M+ Q+ +K GY
Sbjct: 1088RTKLAERNLVQTPGFSLIEAKRKVYKFVSQDKSIPQWNIIYEMIHQMEWQVKFEGY 1255
Score = 86.7 bits (213), Expect = 3e-17
Identities = 50/155 (32%), Positives = 83/155 (53%), Gaps = 1/155 (0%)
Frame = +1
Query: 381 SCSHMGDLALGKKAHDYICDNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQKNVVS 440
+CS +G + G + H ++ + V + NSL+NMY KCG ++ A D+F GM +K+V S
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 441 WNVIIGALALHGSGKEAIEMFEKMRASG-LFPDEITFTGLLSACSHSGLVEMGRYYFGLM 499
W+ IIGA A E + + KM + G +E T +LSAC+H G ++G+ G++
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGIL 489
Query: 500 SSTFGISPDVAHYACMVDLLGRGGFLGEAMSLIQK 534
+V ++D+ + G L + + IQK
Sbjct: 490 LRNIS-ELNVVVKTSLIDMYVKSGCLEKRFTRIQK 591
Score = 84.0 bits (206), Expect(2) = 2e-67
Identities = 55/171 (32%), Positives = 90/171 (52%)
Frame = +1
Query: 254 GNLDLGRFVHLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRMLDRNVVSWTCMI 313
G +D G VH ++ G+E D IV N+LI+MY KCG ++ A VF+ M +++V SW+ +I
Sbjct: 145 GVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVASWSAII 324
Query: 314 NAYANHGLIDHALEIFDQMPVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNA 373
A+A + + L + +M EG+ CR+ + +
Sbjct: 325 GAHACVEMWNECLMLLGKMS---------------SEGR--------CRV-------EES 414
Query: 374 TLVSILSSCSHMGDLALGKKAHDYICDNNITVSVTLCNSLMNMYAKCGALQ 424
TLV++LS+C+H+G LGK H + N ++V + SL++MY K G L+
Sbjct: 415 TLVNVLSACTHLGSPDLGKCIHGILLRNISELNVVVKTSLIDMYVKSGCLE 567
Score = 76.6 bits (187), Expect(2) = 8e-25
Identities = 44/146 (30%), Positives = 80/146 (54%), Gaps = 1/146 (0%)
Frame = +1
Query: 148 ACSAKPSCWDVAVVHAQAIKLGMGSHVCVQNAFLSLYVACRLIRSARQVFDDISDRTLVS 207
ACS + VH K+G+ V VQN+ +++Y C I++A VF+ + ++++ S
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 208 WNSMIVGYSKMGFSKEAILLFKEMQYLG-VEADMFTLVGLLSVALRHGNLDLGRFVHLYI 266
W+++I ++ + E ++L +M G + TLV +LS G+ DLG+ +H +
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGIL 489
Query: 267 VVAGIEIDSIVTNALIDMYAKCGHLQ 292
+ E++ +V +LIDMY K G L+
Sbjct: 490 LRNISELNVVVKTSLIDMYVKSGCLE 567
Score = 55.8 bits (133), Expect(2) = 8e-25
Identities = 39/139 (28%), Positives = 68/139 (48%), Gaps = 5/139 (3%)
Frame = +2
Query: 326 LEIFDQMPVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHM 385
L +F M KN S+ +IS G+ EA+++F M G+ PD+ V + S+CSH
Sbjct: 575 LRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACSHA 754
Query: 386 GDLALGKKAHDYI-CDNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQK-NVVSWNV 443
G + G + + ++ I +V ++++ + G L+ A ++ M K N V W
Sbjct: 755 GLVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSIKPNDVIWRS 934
Query: 444 IIGALALHGS---GKEAIE 459
++ A +H + GK A E
Sbjct: 935 LLSACKVHHNLEIGKIAAE 991
>TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13600
[imported] - Arabidopsis thaliana, partial (12%)
Length = 1362
Score = 163 bits (413), Expect = 2e-40
Identities = 98/266 (36%), Positives = 149/266 (55%), Gaps = 6/266 (2%)
Frame = +2
Query: 73 ILTLGKLLSFCVHLGDLR--YAHLLFDQ--TPQPNKFMYNHLIRGYSNSDDPNKSLLLYR 128
+ + LLS + DL YAH+L + P F +N++IR Y+ + P +L +Y
Sbjct: 275 VTVIATLLSNTTRIRDLNQIYAHILLTRFLESNPASFNWNNIIRSYTRLESPQNALRIYV 454
Query: 129 KMVCAGIMPNQFTIPFVLKACSAKPSCWDVAVVHAQAIKLGMGSHVCVQNAFLSLYVACR 188
M+ AG++P+++T+P VLKA S + VH+ IKLG+ S+ ++ F++LY
Sbjct: 455 SMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGIKLGLQSNEYCESGFINLYCKAG 634
Query: 189 LIRSARQVFDDISDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEADMFTLVGLLS 248
SA +VFD+ + L SWN++I G S+ G + +AI++F +M+ G E D T+V ++S
Sbjct: 635 DFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKRHGFEPDGITMVSVMS 814
Query: 249 VALRHGNLDLGRFVHLYIVVAGIEIDSIV--TNALIDMYAKCGHLQYAETVFDRMLDRNV 306
G+L L +H Y+ A +++ +N+LIDMY KCG + A VF M DRNV
Sbjct: 815 ACGSIGDLYLALQLHKYVFQAKTNEWTVILMSNSLIDMYGKCGRMDLAYEVFATMEDRNV 994
Query: 307 VSWTCMINAYANHGLIDHALEIFDQM 332
SWT MI YA HG AL F M
Sbjct: 995 SSWTSMIVGYAMHGHAKEALGCFHCM 1072
Score = 126 bits (317), Expect = 2e-29
Identities = 84/289 (29%), Positives = 136/289 (46%), Gaps = 9/289 (3%)
Frame = +2
Query: 190 IRSARQVFDDI-------SDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEADMFT 242
IR Q++ I S+ +WN++I Y+++ + A+ ++ M GV D +T
Sbjct: 314 IRDLNQIYAHILLTRFLESNPASFNWNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYT 493
Query: 243 LVGLLSVALRHGNLDLGRFVHLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRML 302
L +L + + LG+ VH Y + G++ + +Y E+ F
Sbjct: 494 LPIVLKAVSQSFAIQLGQQVHSYGIKLGLQSN-----------------EYCESGF---- 610
Query: 303 DRNVVSWTCMINAYANHGLIDHALEIFDQMPVKNAVSWNSIISCHVQEGQHAEAMELFCR 362
IN Y G D A ++FD+ SWN++IS Q G +A+ +F
Sbjct: 611 ----------INLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIVVFVD 760
Query: 363 MCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDYI--CDNNITVSVTLCNSLMNMYAKC 420
M G PD T+VS++S+C +GDL L + H Y+ N + + NSL++MY KC
Sbjct: 761 MKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVILMSNSLIDMYGKC 940
Query: 421 GALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKEAIEMFEKMRASGL 469
G + A ++F M +NV SW +I A+HG KEA+ F M+ G+
Sbjct: 941 GRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEALGCFHCMKGVGV 1087
Score = 101 bits (252), Expect(2) = 9e-23
Identities = 44/96 (45%), Positives = 66/96 (67%)
Frame = +1
Query: 454 GKEAIEMFEKMRASGLFPDEITFTGLLSACSHSGLVEMGRYYFGLMSSTFGISPDVAHYA 513
G + ++E R G+ P+ +TF G+LSAC H G V+ GR+YF +M + +GI+P + HY
Sbjct: 1048 GTWLLSLYEGSR--GVKPNYVTFIGVLSACVHGGTVQEGRFYFDMMKNIYGITPQLQHYG 1221
Query: 514 CMVDLLGRGGFLGEAMSLIQKLPMKPDAVVWGALLG 549
CMVDLLGR G +A +++++PMKP++VVWG G
Sbjct: 1222 CMVDLLGRAGLFDDARRMVEEMPMKPNSVVWGVFDG 1329
Score = 81.3 bits (199), Expect = 1e-15
Identities = 48/189 (25%), Positives = 91/189 (47%), Gaps = 4/189 (2%)
Frame = +2
Query: 339 SWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDYI 398
+WN+II + + A+ ++ M +GV+PD TL +L + S + LG++ H Y
Sbjct: 386 NWNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYG 565
Query: 399 CDNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKEAI 458
+ + + +N+Y K G +A +F + + SWN +I L+ G +AI
Sbjct: 566 IKLGLQSNEYCESGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAI 745
Query: 459 EMFEKMRASGLFPDEITFTGLLSACSHSG----LVEMGRYYFGLMSSTFGISPDVAHYAC 514
+F M+ G PD IT ++SAC G +++ +Y F ++ + + +
Sbjct: 746 VVFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTV---ILMSNS 916
Query: 515 MVDLLGRGG 523
++D+ G+ G
Sbjct: 917 LIDMYGKCG 943
Score = 75.1 bits (183), Expect(2) = 7e-18
Identities = 55/203 (27%), Positives = 92/203 (45%), Gaps = 5/203 (2%)
Frame = +2
Query: 39 PTHQTLHCLLDQCSSLRQLKL---LHAQIILHGLTGQILTLGKLLSFCVHLGDLRYAHLL 95
P TL +L S ++L +H+ I GL ++ GD AH +
Sbjct: 479 PDRYTLPIVLKAVSQSFAIQLGQQVHSYGIKLGLQSNEYCESGFINLYCKAGDFDSAHKV 658
Query: 96 FDQTPQPNKFMYNHLIRGYSNSDDPNKSLLLYRKMVCAGIMPNQFTIPFVLKACSAKPSC 155
FD+ +P +N LI G S +++++ M G P+ T+ V+ AC +
Sbjct: 659 FDENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKRHGFEPDGITMVSVMSACGSIGDL 838
Query: 156 WDVAVVHAQAI--KLGMGSHVCVQNAFLSLYVACRLIRSARQVFDDISDRTLVSWNSMIV 213
+ +H K + + + N+ + +Y C + A +VF + DR + SW SMIV
Sbjct: 839 YLALQLHKYVFQAKTNEWTVILMSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMIV 1018
Query: 214 GYSKMGFSKEAILLFKEMQYLGV 236
GY+ G +KEA+ F M+ +GV
Sbjct: 1019GYAMHGHAKEALGCFHCMKGVGV 1087
Score = 46.2 bits (108), Expect = 4e-05
Identities = 42/204 (20%), Positives = 87/204 (42%), Gaps = 37/204 (18%)
Frame = +2
Query: 440 SWNVIIGALALHGSGKEAIEMFEKMRASGLFPDEITFTGLLSACSHSGLVEMGRYY---- 495
+WN II + S + A+ ++ M +G+ PD T +L A S S +++G+
Sbjct: 386 NWNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYG 565
Query: 496 --FGLMSSTFGIS------------------------PDVAHYACMVDLLGRGGFLGEAM 529
GL S+ + S P + + ++ L +GG +A+
Sbjct: 566 IKLGLQSNEYCESGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAI 745
Query: 530 SL---IQKLPMKPDAVVWGALLGACKTFGNLEMGKQILKQLLELGRYDSGLYVLLSN--- 583
+ +++ +PD + +++ AC + G+L + Q+ K + + + + +L+SN
Sbjct: 746 VVFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQ-AKTNEWTVILMSNSLI 922
Query: 584 -MYSESQRWDDMKKIRKIMNDSGI 606
MY + R D ++ M D +
Sbjct: 923 DMYGKCGRMDLAYEVFATMEDRNV 994
Score = 33.9 bits (76), Expect(2) = 7e-18
Identities = 26/113 (23%), Positives = 45/113 (39%), Gaps = 1/113 (0%)
Frame = +1
Query: 235 GVEADMFTLVGLLSVALRHGNLDLGRFVHLYIVVAGIEIDSIVTNALIDMYAKCGHLQYA 294
GV+ + T +G+LS + G + GRF + + ++Y LQ+
Sbjct: 1084 GVKPNYVTFIGVLSACVHGGTVQEGRFYF---------------DMMKNIYGITPQLQH- 1215
Query: 295 ETVFDRMLDRNVVSWTCMINAYANHGLIDHALEIFDQMPVK-NAVSWNSIISC 346
+ CM++ GL D A + ++MP+K N+V W C
Sbjct: 1216 --------------YGCMVDLLGRAGLFDDARRMVEEMPMKPNSVVWGVFDGC 1332
Score = 23.9 bits (50), Expect(2) = 9e-23
Identities = 8/15 (53%), Positives = 12/15 (79%)
Frame = +2
Query: 545 GALLGACKTFGNLEM 559
G L+GAC+ GN++M
Sbjct: 1316 GCLMGACEKHGNVDM 1360
>AW776322 weakly similar to PIR|H84859|H84 hypothetical protein At2g42920
[imported] - Arabidopsis thaliana, partial (27%)
Length = 633
Score = 161 bits (407), Expect = 9e-40
Identities = 76/208 (36%), Positives = 132/208 (62%), Gaps = 1/208 (0%)
Frame = +1
Query: 434 PQKNVVSWNVIIGALALHGSGKEAIEMFEKMRASGLF-PDEITFTGLLSACSHSGLVEMG 492
P++ + WN II LA++G +EA E F K+ +S L PD ++F G+L+AC H G +
Sbjct: 10 PRRGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKA 189
Query: 493 RYYFGLMSSTFGISPDVAHYACMVDLLGRGGFLGEAMSLIQKLPMKPDAVVWGALLGACK 552
R YF LM + + I P + HY C+VD+LG+ G L EA LI+ +P+KPDA++WG+LL +C+
Sbjct: 190 RDYFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLLSSCR 369
Query: 553 TFGNLEMGKQILKQLLELGRYDSGLYVLLSNMYSESQRWDDMKKIRKIMNDSGIKKCRAI 612
N+++ ++ +++ EL D+ YVL+SN+++ S ++++ + R +M ++ +K
Sbjct: 370 KHRNVQIARRAAQRVYELNPSDASGYVLMSNVHAASNKFEEAIEQRLLMKENLTEKEPGC 549
Query: 613 SSIEIDGCCYQFMVDDKNHEVSTSIYSM 640
SSIE+ G ++F+ + H + IY +
Sbjct: 550 SSIELYGEVHEFIAGGRLHPKTQEIYHL 633
Score = 53.1 bits (126), Expect = 3e-07
Identities = 33/125 (26%), Positives = 61/125 (48%), Gaps = 6/125 (4%)
Frame = +1
Query: 333 PVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVM-PDNATLVSILSSCSHMGDLALG 391
P + WNSII G EA E F ++ S ++ PD+ + + +L++C H+G +
Sbjct: 10 PRRGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAI--- 180
Query: 392 KKAHDY----ICDNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQK-NVVSWNVIIG 446
KA DY + I S+ ++++ + G L+ A ++ GMP K + + W ++
Sbjct: 181 NKARDYFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLLS 360
Query: 447 ALALH 451
+ H
Sbjct: 361 SCRKH 375
Score = 43.5 bits (101), Expect = 3e-04
Identities = 31/146 (21%), Positives = 65/146 (44%), Gaps = 3/146 (2%)
Frame = +1
Query: 203 RTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLG-VEADMFTLVGLLSVALRHGNLDLGRF 261
R L WNS+I+G + G +EA F +++ ++ D + +G+L+ G ++ R
Sbjct: 16 RGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKAR- 192
Query: 262 VHLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRM-LDRNVVSWTCMINAYANHG 320
Y E + ++ ++ ++ +TC+++ G
Sbjct: 193 ------------------------------DYFELMMNKYEIEPSIKHYTCIVDVLGQAG 282
Query: 321 LIDHALEIFDQMPVK-NAVSWNSIIS 345
L++ A E+ MP+K +A+ W S++S
Sbjct: 283 LLEEAEELIKGMPLKPDAIIWGSLLS 360
>TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-binding
protein-like {Arabidopsis thaliana}, partial (10%)
Length = 895
Score = 157 bits (396), Expect = 2e-38
Identities = 79/180 (43%), Positives = 111/180 (60%)
Frame = +2
Query: 437 NVVSWNVIIGALALHGSGKEAIEMFEKMRASGLFPDEITFTGLLSACSHSGLVEMGRYYF 496
++VSWN IIG A HG A+E F++M+ G PDE+TF +LSAC H+GLVE G +F
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRMKVVGT-PDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 497 GLMSSTFGISPDVAHYACMVDLLGRGGFLGEAMSLIQKLPMKPDAVVWGALLGACKTFGN 556
M + +GI ++ HY+CMVDL GR G EA +LI+ +P +PD V+WGALL AC N
Sbjct: 179 TDMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALLAACGLHSN 358
Query: 557 LEMGKQILKQLLELGRYDSGLYVLLSNMYSESQRWDDMKKIRKIMNDSGIKKCRAISSIE 616
LE+G+ +++ L Y +LS + E W + ++R M + GIKK AIS +E
Sbjct: 359 LELGEYAAERIRRLESSHPVSYSVLSKIQGEKGVWSSVNELRDTMKERGIKKQTAISWVE 538
Score = 67.0 bits (162), Expect = 2e-11
Identities = 42/135 (31%), Positives = 69/135 (51%), Gaps = 5/135 (3%)
Frame = +2
Query: 338 VSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAH-D 396
VSWN+II + G A+E F RM V G PD T V++LS+C H G + G+K D
Sbjct: 8 VSWNAIIGGYASHGLATRALEEFDRMKVVGT-PDEVTFVNVLSACVHAGLVEEGEKHFTD 184
Query: 397 YICDNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMP-QKNVVSWNVIIGALALHGS-- 453
+ I + + ++++Y + G A ++ MP + +VV W ++ A LH +
Sbjct: 185 MLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALLAACGLHSNLE 364
Query: 454 -GKEAIEMFEKMRAS 467
G+ A E ++ +S
Sbjct: 365 LGEYAAERIRRLESS 409
Score = 57.0 bits (136), Expect = 2e-08
Identities = 45/175 (25%), Positives = 84/175 (47%), Gaps = 6/175 (3%)
Frame = +2
Query: 205 LVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEADMFTLVGLLSVALRHGNLDLG--RFV 262
LVSWN++I GY+ G + A+ F M+ +G D T V +LS + G ++ G F
Sbjct: 5 LVSWNAIIGGYASHGLATRALEEFDRMKVVGT-PDEVTFVNVLSACVHAGLVEEGEKHFT 181
Query: 263 HLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRM-LDRNVVSWTCMINA---YAN 318
+ + GI+ + + ++D+Y + G AE + M + +VV W ++ A ++N
Sbjct: 182 DM-LTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALLAACGLHSN 358
Query: 319 HGLIDHALEIFDQMPVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNA 373
L ++A E ++ + VS++ + ++G + EL M G+ A
Sbjct: 359 LELGEYAAERIRRLESSHPVSYSVLSKIQGEKGVWSSVNELRDTMKERGIKKQTA 523
>BE941714 weakly similar to GP|11994734|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (10%)
Length = 619
Score = 155 bits (393), Expect = 4e-38
Identities = 78/204 (38%), Positives = 123/204 (60%), Gaps = 5/204 (2%)
Frame = +2
Query: 403 ITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKEAIEMFE 462
+ +SV L SL++MYAKCG L+ A +F M ++VV WN +I +A+HG GK A+++F
Sbjct: 2 VPLSVRLSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFY 181
Query: 463 KMRASGLFPDEITFTGLLSACSHSGLVEMGRYYFGLMSSTFGISPDVAHYACMVDLLGRG 522
M G+ PD+ITF + +ACS+SG+ G M S + I P HY C+VDLL R
Sbjct: 182 DMEKVGVKPDDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLVDLLSRA 361
Query: 523 GFLGEAMSLIQKLPM----KPDAVVWGALLGACKTFGNLEMGKQILKQLLELGRY-DSGL 577
G EAM +I+K+ + + W A L AC G ++ + +++L+L + SG+
Sbjct: 362 GLFEEAMVMIRKITNSWNGSEETLAWRAFLSACCNHGETQLAELAAEKVLQLDNHIHSGV 541
Query: 578 YVLLSNMYSESQRWDDMKKIRKIM 601
YVLLSN+Y+ S + +D ++++ +M
Sbjct: 542 YVLLSNLYAASGKHNDARRVKDMM 613
Score = 67.8 bits (164), Expect = 1e-11
Identities = 51/194 (26%), Positives = 87/194 (44%), Gaps = 6/194 (3%)
Frame = +2
Query: 277 VTNALIDMYAKCGHLQYAETVFDRMLDRNVVSWTCMINAYANHGLIDHALEIFDQMPVKN 336
++ +L+DMYAKCG+L+ A+ +FD M R+VV W MI+ A HG
Sbjct: 20 LSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHG---------------- 151
Query: 337 AVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHD 396
+G+ A++LF M GV PD+ T +++ ++CS+ G G D
Sbjct: 152 -------------DGK--GALKLFYDMEKVGVKPDDITFIAVFTACSYSGMAYEGLMLLD 286
Query: 397 YICD-NNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMP-----QKNVVSWNVIIGALAL 450
+C NI L+++ ++ G + AM + + + ++W + A
Sbjct: 287 KMCSVYNIVPKSEHYGCLVDLLSRAGLFEEAMVMIRKITNSWNGSEETLAWRAFLSACCN 466
Query: 451 HGSGKEAIEMFEKM 464
HG + A EK+
Sbjct: 467 HGETQLAELAAEKV 508
Score = 60.5 bits (145), Expect = 2e-09
Identities = 38/157 (24%), Positives = 75/157 (47%), Gaps = 10/157 (6%)
Frame = +2
Query: 174 VCVQNAFLSLYVACRLIRSARQVFDDISDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQY 233
V + + L +Y C + A+++FD ++ R +V WN+MI G + G K A+ LF +M+
Sbjct: 14 VRLSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEK 193
Query: 234 LGVEADMFTLVGLLSVA-----LRHGNLDLGRFVHLYIVVAGIEIDSIVTNALIDMYAKC 288
+GV+ D T + + + G + L + +Y +V E L+D+ ++
Sbjct: 194 VGVKPDDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKSE----HYGCLVDLLSRA 361
Query: 289 GHLQYAETVFDRMLD-----RNVVSWTCMINAYANHG 320
G + A + ++ + ++W ++A NHG
Sbjct: 362 GLFEEAMVMIRKITNSWNGSEETLAWRAFLSACCNHG 472
>AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.70 -
Arabidopsis thaliana, partial (12%)
Length = 703
Score = 143 bits (361), Expect = 2e-34
Identities = 78/228 (34%), Positives = 135/228 (59%)
Frame = +3
Query: 83 CVHLGDLRYAHLLFDQTPQPNKFMYNHLIRGYSNSDDPNKSLLLYRKMVCAGIMPNQFTI 142
CV + + L FD+ N ++ ++ GY+ + D K++ +R M G+ NQ+T
Sbjct: 30 CVSEAEFLFKGLEFDRK---NHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVECNQYTF 200
Query: 143 PFVLKACSAKPSCWDVAVVHAQAIKLGMGSHVCVQNAFLSLYVACRLIRSARQVFDDISD 202
P +L ACS+ + VH +K G GS+V VQ+A + +Y C +++A+ + + + D
Sbjct: 201 PTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNMLETMED 380
Query: 203 RTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEADMFTLVGLLSVALRHGNLDLGRFV 262
+VSWNS++VG+ + G +EA+ LFK M ++ D +T +L+ + G+++ + V
Sbjct: 381 DDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDYTFPSVLNCCV-VGSIN-PKSV 554
Query: 263 HLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRMLDRNVVSWT 310
H I+ G E +V+NAL+DMYAK G + A TVF++ML+++V+SWT
Sbjct: 555 HGLIIKTGFENYKLVSNALVDMYAKTGDMDCAYTVFEKMLEKDVISWT 698
Score = 110 bits (276), Expect = 1e-24
Identities = 74/263 (28%), Positives = 123/263 (46%), Gaps = 2/263 (0%)
Frame = +3
Query: 181 LSLYVACRLIRSARQVFD--DISDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEA 238
+ +Y C+ + A +F + + V W +M+ GY++ G +A+ F+ M GVE
Sbjct: 6 VDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVEC 185
Query: 239 DMFTLVGLLSVALRHGNLDLGRFVHLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVF 298
+ +T +L+ G VH +IV +G + V +AL+DMYAKCG L+ A+ +
Sbjct: 186 NQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNML 365
Query: 299 DRMLDRNVVSWTCMINAYANHGLIDHALEIFDQMPVKNAVSWNSIISCHVQEGQHAEAME 358
+ M D +VVSW ++ + HGL + AL +F M H M+
Sbjct: 366 ETMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNM--------------------HGRNMK 485
Query: 359 LFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDYICDNNITVSVTLCNSLMNMYA 418
+ D+ T S+L+ C +G + K H I + N+L++MYA
Sbjct: 486 I-----------DDYTFPSVLNCCV-VGSIN-PKSVHGLIIKTGFENYKLVSNALVDMYA 626
Query: 419 KCGALQTAMDIFFGMPQKNVVSW 441
K G + A +F M +K+V+SW
Sbjct: 627 KTGDMDCAYTVFEKMLEKDVISW 695
Score = 99.8 bits (247), Expect = 3e-21
Identities = 72/273 (26%), Positives = 129/273 (46%), Gaps = 9/273 (3%)
Frame = +3
Query: 281 LIDMYAKCGHLQYAETVFDRMLDRNVVSWTCMINAYANHGLIDHALEIFDQMPVKNAVSW 340
L+DMYAKC + AE +F LE FD+ KN V W
Sbjct: 3 LVDMYAKCKCVSEAEFLFK-------------------------GLE-FDR---KNHVLW 95
Query: 341 NSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDYICD 400
++++ + Q G +A+E F M GV + T +IL++CS + G++ H +I
Sbjct: 96 TAMVTGYAQNGDGYKAVEFFRYMHAQGVECNQYTFPTILTACSSVLARCFGEQVHGFIVK 275
Query: 401 NNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKEAIEM 460
+ +V + ++L++MYAKCG L+ A ++ M +VVSWN ++ HG +EA+ +
Sbjct: 276 SGFGSNVYVQSALVDMYAKCGDLKNAKNMLETMEDDDVVSWNSLMVGFVRHGLEEEALRL 455
Query: 461 FEKMRASGLFPDEITFTGLLSAC---------SHSGLVEMGRYYFGLMSSTFGISPDVAH 511
F+ M + D+ TF +L+ C H +++ G + L+S+
Sbjct: 456 FKNMHGRNMKIDDYTFPSVLNCCVVGSINPKSVHGLIIKTGFENYKLVSN---------- 605
Query: 512 YACMVDLLGRGGFLGEAMSLIQKLPMKPDAVVW 544
+VD+ + G + A ++ +K+ ++ D + W
Sbjct: 606 --ALVDMYAKTGDMDCAYTVFEKM-LEKDVISW 695
>BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30 -
Arabidopsis thaliana, partial (22%)
Length = 595
Score = 142 bits (358), Expect = 4e-34
Identities = 73/176 (41%), Positives = 107/176 (60%), Gaps = 6/176 (3%)
Frame = +1
Query: 439 VSWNVIIGALALHGSGKEAIEMFEKMRASG-----LFPDEITFTGLLSACSHSGLVEMGR 493
++WNV+I A +HG G+EA+++F +M G + P+E+T+ + ++ SHSG+V+ G
Sbjct: 1 ITWNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFASLSHSGMVDEGL 180
Query: 494 YYFGLMSSTFGISPDVAHYACMVDLLGRGGFLGEAMSLIQKLPMKPDAV-VWGALLGACK 552
F M + GI P HYAC+VDLLGR G + EA +LI+ +P V W +LLGACK
Sbjct: 181 NLFYTMKAKHGIEPTSDHYACLVDLLGRSGQIEEAYNLIKTMPSNMKKVDAWSSLLGACK 360
Query: 553 TFGNLEMGKQILKQLLELGRYDSGLYVLLSNMYSESQRWDDMKKIRKIMNDSGIKK 608
NLE+G+ K L L + YVLLSN+YS + WD +RK M + G++K
Sbjct: 361 IHQNLEIGEIAAKNLFVLDPNVASYYVLLSNIYSSAGLWDQAIDVRKKMKEKGVRK 528
>AW257410 weakly similar to GP|10177533|db selenium-binding protein-like
{Arabidopsis thaliana}, partial (21%)
Length = 724
Score = 140 bits (354), Expect = 1e-33
Identities = 78/212 (36%), Positives = 120/212 (55%), Gaps = 2/212 (0%)
Frame = +1
Query: 49 DQCSSLRQLKLLHAQIILHGLTGQILTLGKLLSF--CVHLGDLRYAHLLFDQTPQPNKFM 106
++CS++ +LK ++ Q + G LT+ +LL+ + +L YA ++FD+ PN M
Sbjct: 1 ERCSNIGELKQIYVQFLKKGTIRHKLTVSRLLTTYASMEFSNLTYARMVFDRISSPNTVM 180
Query: 107 YNHLIRGYSNSDDPNKSLLLYRKMVCAGIMPNQFTIPFVLKACSAKPSCWDVAVVHAQAI 166
+N +IR YSNS+DP ++LLLY +M+ I N +T PF+LKACSA + + +H Q I
Sbjct: 181 WNTMIRAYSNSNDPEEALLLYHQMLHHSIPHNAYTFPFLLKACSALSALAETHQIHVQII 360
Query: 167 KLGMGSHVCVQNAFLSLYVACRLIRSARQVFDDISDRTLVSWNSMIVGYSKMGFSKEAIL 226
K G GS V N+ L +Y I+SA +FD + R +VSWN+MI GY K G + A
Sbjct: 361 KRGFGSEVYATNSLLRVYAISGSIKSAHVLFDLLPSRDIVSWNTMIDGYIKCGNVEMAYK 540
Query: 227 LFKEMQYLGVEADMFTLVGLLSVALRHGNLDL 258
+F+ M V + +VG + + L L
Sbjct: 541 IFQAMPEKNVISWTSMIVGFVRTGMHKKALSL 636
Score = 122 bits (306), Expect = 4e-28
Identities = 69/213 (32%), Positives = 110/213 (51%), Gaps = 2/213 (0%)
Frame = +1
Query: 161 VHAQAIKLGMGSHVCVQNAFLSLYVACRL--IRSARQVFDDISDRTLVSWNSMIVGYSKM 218
++ Q +K G H + L+ Y + + AR VFD IS V WN+MI YS
Sbjct: 34 IYVQFLKKGTIRHKLTVSRLLTTYASMEFSNLTYARMVFDRISSPNTVMWNTMIRAYSNS 213
Query: 219 GFSKEAILLFKEMQYLGVEADMFTLVGLLSVALRHGNLDLGRFVHLYIVVAGIEIDSIVT 278
+EA+LL+ +M + + + +T LL L +H+ I+ G + T
Sbjct: 214 NDPEEALLLYHQMLHHSIPHNAYTFPFLLKACSALSALAETHQIHVQIIKRGFGSEVYAT 393
Query: 279 NALIDMYAKCGHLQYAETVFDRMLDRNVVSWTCMINAYANHGLIDHALEIFDQMPVKNAV 338
N+L+ +YA G ++ A +FD + R++VSW MI+ Y G ++ A +IF MP KN +
Sbjct: 394 NSLLRVYAISGSIKSAHVLFDLLPSRDIVSWNTMIDGYIKCGNVEMAYKIFQAMPEKNVI 573
Query: 339 SWNSIISCHVQEGQHAEAMELFCRMCVSGVMPD 371
SW S+I V+ G H +A+ L +M V+ + PD
Sbjct: 574 SWTSMIVGFVRTGMHKKALSLLQQMLVARIKPD 672
Score = 77.4 bits (189), Expect = 2e-14
Identities = 59/225 (26%), Positives = 105/225 (46%), Gaps = 8/225 (3%)
Frame = +1
Query: 257 DLGRFVHLYI--VVAGIEIDSIVTNALIDMYAKC--GHLQYAETVFDRMLDRNVVSWTCM 312
++G +Y+ + G + + L+ YA +L YA VFDR+ N V W M
Sbjct: 13 NIGELKQIYVQFLKKGTIRHKLTVSRLLTTYASMEFSNLTYARMVFDRISSPNTVMWNTM 192
Query: 313 INAYANHGLIDHALEIFDQMP----VKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGV 368
I AY+N + AL ++ QM NA ++ ++ AE ++ ++ G
Sbjct: 193 IRAYSNSNDPEEALLLYHQMLHHSIPHNAYTFPFLLKACSALSALAETHQIHVQIIKRGF 372
Query: 369 MPDNATLVSILSSCSHMGDLALGKKAHDYICDNNITVSVTLCNSLMNMYAKCGALQTAMD 428
+ S+L + G + K AH + D + + N++++ Y KCG ++ A
Sbjct: 373 GSEVYATNSLLRVYAISGSI---KSAH-VLFDLLPSRDIVSWNTMIDGYIKCGNVEMAYK 540
Query: 429 IFFGMPQKNVVSWNVIIGALALHGSGKEAIEMFEKMRASGLFPDE 473
IF MP+KNV+SW +I G K+A+ + ++M + + PD+
Sbjct: 541 IFQAMPEKNVISWTSMIVGFVRTGMHKKALSLLQQMLVARIKPDK 675
Score = 70.1 bits (170), Expect = 3e-12
Identities = 46/208 (22%), Positives = 101/208 (48%), Gaps = 6/208 (2%)
Frame = +1
Query: 287 KCGHLQYAETVFDRMLDRNVV----SWTCMINAYANHGL--IDHALEIFDQMPVKNAVSW 340
+C ++ + ++ + L + + + + ++ YA+ + +A +FD++ N V W
Sbjct: 4 RCSNIGELKQIYVQFLKKGTIRHKLTVSRLLTTYASMEFSNLTYARMVFDRISSPNTVMW 183
Query: 341 NSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDYICD 400
N++I + EA+ L+ +M + + T +L +CS + LA + H I
Sbjct: 184 NTMIRAYSNSNDPEEALLLYHQMLHHSIPHNAYTFPFLLKACSALSALAETHQIHVQIIK 363
Query: 401 NNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKEAIEM 460
V NSL+ +YA G++++A +F +P +++VSWN +I G+ + A ++
Sbjct: 364 RGFGSEVYATNSLLRVYAISGSIKSAHVLFDLLPSRDIVSWNTMIDGYIKCGNVEMAYKI 543
Query: 461 FEKMRASGLFPDEITFTGLLSACSHSGL 488
F+ M + I++T ++ +G+
Sbjct: 544 FQAMPEKNV----ISWTSMIVGFVRTGM 615
Score = 48.5 bits (114), Expect = 8e-06
Identities = 49/192 (25%), Positives = 87/192 (44%), Gaps = 4/192 (2%)
Frame = +1
Query: 382 CSHMGDLALGKKAH-DYICDNNITVSVTLCNSLMNMYAKC--GALQTAMDIFFGMPQKNV 438
CS++G+L K+ + ++ I +T+ + L+ YA L A +F + N
Sbjct: 7 CSNIGEL---KQIYVQFLKKGTIRHKLTV-SRLLTTYASMEFSNLTYARMVFDRISSPNT 174
Query: 439 VSWNVIIGALALHGSGKEAIEMFEKMRASGLFPDEITFTGLLSACSH-SGLVEMGRYYFG 497
V WN +I A + +EA+ ++ +M + + TF LL ACS S L E + +
Sbjct: 175 VMWNTMIRAYSNSNDPEEALLLYHQMLHHSIPHNAYTFPFLLKACSALSALAETHQIHVQ 354
Query: 498 LMSSTFGISPDVAHYACMVDLLGRGGFLGEAMSLIQKLPMKPDAVVWGALLGACKTFGNL 557
++ FG +V ++ + G + A L LP + D V W ++ GN+
Sbjct: 355 IIKRGFG--SEVYATNSLLRVYAISGSIKSAHVLFDLLPSR-DIVSWNTMIDGYIKCGNV 525
Query: 558 EMGKQILKQLLE 569
EM +I + + E
Sbjct: 526 EMAYKIFQAMPE 561
>BF640739 weakly similar to GP|7363286|dbj Similar to Arabidopsis thaliana
DNA chromosome 4 BAC clone F4I10; vegetative storage
protein., partial (14%)
Length = 444
Score = 124 bits (311), Expect = 1e-28
Identities = 58/146 (39%), Positives = 94/146 (63%), Gaps = 1/146 (0%)
Frame = +1
Query: 355 EAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDYICDNNITVSVTLCNSLM 414
EA+ LFC M G+ PD+ T VS++++ + + K H N+ +V + +L+
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 415 NMYAKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKEAIEMFEKMR-ASGLFPDE 473
+MYAKCGA++TA ++F M +++V++WN +I HG GK A+++F+ M+ + L P++
Sbjct: 187 DMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPND 366
Query: 474 ITFTGLLSACSHSGLVEMGRYYFGLM 499
ITF ++SACSHSG VE G YYF +M
Sbjct: 367 ITFLSVISACSHSGFVEEGLYYFKIM 444
Score = 88.6 bits (218), Expect = 7e-18
Identities = 50/164 (30%), Positives = 82/164 (49%)
Frame = +1
Query: 223 EAILLFKEMQYLGVEADMFTLVGLLSVALRHGNLDLGRFVHLYIVVAGIEIDSIVTNALI 282
EA+ LF MQ G++ D FT V +++ +++H + ++ + V AL+
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 283 DMYAKCGHLQYAETVFDRMLDRNVVSWTCMINAYANHGLIDHALEIFDQMPVKNAVSWNS 342
DMYAKCG ++ A +FD M +R+V++W MI+ Y HGL AL++FD M +N S
Sbjct: 187 DMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDM--QNEAS--- 351
Query: 343 IISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMG 386
+ P++ T +S++S+CSH G
Sbjct: 352 -------------------------LKPNDITFLSVISACSHSG 408
Score = 76.6 bits (187), Expect = 3e-14
Identities = 43/142 (30%), Positives = 75/142 (52%), Gaps = 1/142 (0%)
Frame = +1
Query: 121 NKSLLLYRKMVCAGIMPNQFTIPFVLKACSAKPSCWDVAVVHAQAIKLGMGSHVCVQNAF 180
N++L L+ M GI P+ FT V+ A + +H AI+ M ++V V A
Sbjct: 4 NEALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATAL 183
Query: 181 LSLYVACRLIRSARQVFDDISDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQ-YLGVEAD 239
+ +Y C I +AR++FD + +R +++WN+MI GY G K A+ LF +MQ ++ +
Sbjct: 184 VDMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPN 363
Query: 240 MFTLVGLLSVALRHGNLDLGRF 261
T + ++S G ++ G +
Sbjct: 364 DITFLSVISACSHSGFVEEGLY 429
Score = 38.1 bits (87), Expect = 0.011
Identities = 31/117 (26%), Positives = 47/117 (39%), Gaps = 1/117 (0%)
Frame = +1
Query: 35 KLNSPTHQTLHCLLDQCSSLRQLKLLHAQIILHGLTGQILTLGKLLSFCVHLGDLRYAHL 94
K +S T ++ L S RQ K +H I + + L+ G + A
Sbjct: 49 KPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALVDMYAKCGAIETARE 228
Query: 95 LFDQTPQPNKFMYNHLIRGYSNSDDPNKSLLLYRKMVC-AGIMPNQFTIPFVLKACS 150
LFD + + +N +I GY +L L+ M A + PN T V+ ACS
Sbjct: 229 LFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPNDITFLSVISACS 399
Score = 32.3 bits (72), Expect = 0.60
Identities = 31/144 (21%), Positives = 55/144 (37%), Gaps = 34/144 (23%)
Frame = +1
Query: 456 EAIEMFEKMRASGLFPDEITFTGLLSACSHSGLVEMGRYYFGLMSST------FGISPDV 509
EA+ +F M++ G+ PD TF +++A + + ++ GL T F + V
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 510 AHYA------------------------CMVDLLGRGGFLGEAMSLIQKLP----MKPDA 541
YA M+D G G A+ L + +KP+
Sbjct: 187 DMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPND 366
Query: 542 VVWGALLGACKTFGNLEMGKQILK 565
+ + +++ AC G +E G K
Sbjct: 367 ITFLSVISACSHSGFVEEGLYYFK 438
>TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9O13.24
{Arabidopsis thaliana}, partial (55%)
Length = 1727
Score = 118 bits (295), Expect = 8e-27
Identities = 66/222 (29%), Positives = 109/222 (48%)
Frame = +2
Query: 349 QEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDYICDNNITVSVT 408
QEG+ EA+EL + G+ D + C + KK HDY +
Sbjct: 224 QEGKVKEALELMEK----GIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFK 391
Query: 409 LCNSLMNMYAKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKEAIEMFEKMRASG 468
+ N ++ MY C ++ A +F MP +N+ SW+++I A G E +++FE+M G
Sbjct: 392 MHNKVIEMYGNCKSMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELG 571
Query: 469 LFPDEITFTGLLSACSHSGLVEMGRYYFGLMSSTFGISPDVAHYACMVDLLGRGGFLGEA 528
L T +LSAC + VE Y M S +GI P V HY ++D+LG+ G+L EA
Sbjct: 572 LEITSETMLAVLSACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEA 751
Query: 529 MSLIQKLPMKPDAVVWGALLGACKTFGNLEMGKQILKQLLEL 570
I++LP +P V+ L + G++++ + + ++ L
Sbjct: 752 EEFIEQLPFEPTVTVFETLKNYARIHGDVDLEDHVEELIVSL 877
Score = 69.7 bits (169), Expect = 3e-12
Identities = 57/210 (27%), Positives = 96/210 (45%), Gaps = 8/210 (3%)
Frame = +2
Query: 138 NQFTIPFVLKACSAKPSCWDVAVVHAQAIKLGMGSHVCVQNAFLSLYVACRLIRSARQVF 197
N F I F L C S D VH ++ S + N + +Y C+ + AR+VF
Sbjct: 284 NCFEILFDL--CGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCKSMTDARRVF 457
Query: 198 DDISDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEADMFTLVGLLSVALRHGNLD 257
D + +R + SW+ MI GY+ E + LF++M LG+E T++ +LS G+ +
Sbjct: 458 DHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLSAC---GSAE 628
Query: 258 LGRFVHLYIVVA----GIEIDSIVTNALIDMYAKCGHLQYAETVFDRM-LDRNVVSWTCM 312
++Y+ GIE L+D+ + G+L+ AE +++ + V + +
Sbjct: 629 AVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEAEEFIEQLPFEPTVTVFETL 808
Query: 313 INAYANHG---LIDHALEIFDQMPVKNAVS 339
N HG L DH E+ + AV+
Sbjct: 809 KNYARIHGDVDLEDHVEELIVSLDPSKAVA 898
Score = 62.8 bits (151), Expect = 4e-10
Identities = 54/210 (25%), Positives = 94/210 (44%), Gaps = 5/210 (2%)
Frame = +2
Query: 215 YSKMGFSKEAILLFKEMQYLGVEADMFTLVGLLSVALRHGNLDLGRFVHLYIVVAGIEID 274
+ + G KEA+ L ++ G++AD L + + +++ + VH Y + + D
Sbjct: 218 FCQEGKVKEALELMEK----GIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSD 385
Query: 275 SIVTNALIDMYAKCGHLQYAETVFDRMLDRNVVSWTCMINAYANHGLIDHALEIFDQM-- 332
+ N +I+MY C + A VFD M +RN+ SW MI YAN + D L++F+QM
Sbjct: 386 FKMHNKVIEMYGNCKSMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNE 565
Query: 333 ---PVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLA 389
+ + + +C E + L G+ P + +L G L
Sbjct: 566 LGLEITSETMLAVLSACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYL- 742
Query: 390 LGKKAHDYICDNNITVSVTLCNSLMNMYAK 419
K+A ++I +VT+ +L N YA+
Sbjct: 743 --KEAEEFIEQLPFEPTVTVFETLKN-YAR 823
Score = 41.2 bits (95), Expect = 0.001
Identities = 31/137 (22%), Positives = 60/137 (43%)
Frame = +2
Query: 35 KLNSPTHQTLHCLLDQCSSLRQLKLLHAQIILHGLTGQILTLGKLLSFCVHLGDLRYAHL 94
K ++ + L L + S+ K +H + K++ + + A
Sbjct: 272 KADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCKSMTDARR 451
Query: 95 LFDQTPQPNKFMYNHLIRGYSNSDDPNKSLLLYRKMVCAGIMPNQFTIPFVLKACSAKPS 154
+FD P N ++ +IRGY+NS ++ L L+ +M G+ T+ VL AC + +
Sbjct: 452 VFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLSACGSAEA 631
Query: 155 CWDVAVVHAQAIKLGMG 171
D A ++ +++K G
Sbjct: 632 VED-AYIYLESMKSKYG 679
>BG645370 weakly similar to PIR|T00405|T004 hypothetical protein At2g44880
[imported] - Arabidopsis thaliana, partial (11%)
Length = 784
Score = 112 bits (281), Expect(2) = 1e-26
Identities = 43/126 (34%), Positives = 85/126 (67%)
Frame = -2
Query: 476 FTGLLSACSHSGLVEMGRYYFGLMSSTFGISPDVAHYACMVDLLGRGGFLGEAMSLIQKL 535
FT +L+AC+H+G ++ G YF M + +G+SPD+ YAC++DL R G L +A L++++
Sbjct: 783 FTAVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEM 604
Query: 536 PMKPDAVVWGALLGACKTFGNLEMGKQILKQLLELGRYDSGLYVLLSNMYSESQRWDDMK 595
P P+ ++W + L ACK +G++E+G++ QL+++ ++ Y+ L+++Y+ W++
Sbjct: 603 PYDPNCIIWSSFLSACKIYGDVELGREAAIQLIKMEPCNAAPYLTLAHIYTTKGLWNEAS 424
Query: 596 KIRKIM 601
++R +M
Sbjct: 423 EVRSLM 406
Score = 35.8 bits (81), Expect = 0.054
Identities = 30/119 (25%), Positives = 56/119 (46%), Gaps = 5/119 (4%)
Frame = -2
Query: 246 LLSVALRHGNLDLGR-FVHLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRM-LD 303
+L+ G +D G + + I G+ D + LID+YA+ G+L+ A + + M D
Sbjct: 774 VLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEMPYD 595
Query: 304 RNVVSWTCMINAYANHGLIDHALEIFDQ---MPVKNAVSWNSIISCHVQEGQHAEAMEL 359
N + W+ ++A +G ++ E Q M NA + ++ + +G EA E+
Sbjct: 594 PNCIIWSSFLSACKIYGDVELGREAAIQLIKMEPCNAAPYLTLAHIYTTKGLWNEASEV 418
Score = 31.2 bits (69), Expect = 1.3
Identities = 24/91 (26%), Positives = 42/91 (45%), Gaps = 5/91 (5%)
Frame = -2
Query: 309 WTCMINAYANHGLIDHALEIFDQMPVKNAVS-----WNSIISCHVQEGQHAEAMELFCRM 363
+T ++ A + G ID E F++M +S + +I + + G +A +L M
Sbjct: 783 FTAVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEM 604
Query: 364 CVSGVMPDNATLVSILSSCSHMGDLALGKKA 394
P+ S LS+C GD+ LG++A
Sbjct: 603 PYD---PNCIIWSSFLSACKIYGDVELGREA 520
Score = 25.4 bits (54), Expect(2) = 1e-26
Identities = 12/29 (41%), Positives = 17/29 (58%)
Frame = -3
Query: 611 AISSIEIDGCCYQFMVDDKNHEVSTSIYS 639
A S +E+D + F VDD H+ S IY+
Sbjct: 371 AWSWVEVDKHNHVFAVDDVTHQQSNEIYA 285
>BE202961 weakly similar to GP|6016735|gb|A hypothetical protein {Arabidopsis
thaliana}, partial (14%)
Length = 613
Score = 114 bits (285), Expect = 1e-25
Identities = 69/233 (29%), Positives = 115/233 (48%)
Frame = +2
Query: 227 LFKEMQYLGVEADMFTLVGLLSVALRHGNLDLGRFVHLYIVVAGIEIDSIVTNALIDMYA 286
L EM ++ + T+V +L NL+ G +H V G E+++ V+ AL+DMY
Sbjct: 8 LSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYM 187
Query: 287 KCGHLQYAETVFDRMLDRNVVSWTCMINAYANHGLIDHALEIFDQMPVKNAVSWNSIISC 346
KC + A +F+RM ++V++W + + YA++G++
Sbjct: 188 KCFSPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVH----------------------- 298
Query: 347 HVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDYICDNNITVS 406
E+M +F M SG PD LV IL++ S +G L H ++ N +
Sbjct: 299 --------ESMWVFRNMLSSGTRPDAIALVKILTTVSELGILQQAVCFHAFVIKNGFENN 454
Query: 407 VTLCNSLMNMYAKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKEAIE 459
+ SL+ +YAKC +++ A +F GM K+VV+W+ II A HG G+EA++
Sbjct: 455 QFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVTWSSIIAAYGFHGQGEEALK 613
Score = 102 bits (254), Expect = 5e-22
Identities = 56/202 (27%), Positives = 100/202 (48%)
Frame = +2
Query: 126 LYRKMVCAGIMPNQFTIPFVLKACSAKPSCWDVAVVHAQAIKLGMGSHVCVQNAFLSLYV 185
L +M+ I PN T+ VL+AC+ + + +H A+ G V A + +Y+
Sbjct: 8 LSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYM 187
Query: 186 ACRLIRSARQVFDDISDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEADMFTLVG 245
C A +F+ + + +++W + GY+ G E++ +F+ M G D LV
Sbjct: 188 KCFSPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIALVK 367
Query: 246 LLSVALRHGNLDLGRFVHLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRMLDRN 305
+L+ G L H +++ G E + + +LI++YAKC ++ A VF M ++
Sbjct: 368 ILTTVSELGILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYKD 547
Query: 306 VVSWTCMINAYANHGLIDHALE 327
VV+W+ +I AY HG + AL+
Sbjct: 548 VVTWSSIIAAYGFHGQGEEALK 613
Score = 90.1 bits (222), Expect = 2e-18
Identities = 54/209 (25%), Positives = 107/209 (50%)
Frame = +2
Query: 357 MELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDYICDNNITVSVTLCNSLMNM 416
++L M + P+ T+VS+L +C+ + +L G K H+ + + T+ +LM+M
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDM 181
Query: 417 YAKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKEAIEMFEKMRASGLFPDEITF 476
Y KC + + A+DIF MP+K+V++W V+ A +G E++ +F M +SG PD I
Sbjct: 182 YMKCFSPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIAL 361
Query: 477 TGLLSACSHSGLVEMGRYYFGLMSSTFGISPDVAHYACMVDLLGRGGFLGEAMSLIQKLP 536
+L+ S G+++ + + G + A ++++ + + +A + + +
Sbjct: 362 VKILTTVSELGILQQAVCFHAFVIKN-GFENNQFIGASLIEVYAKCSSIEDANKVFKGMT 538
Query: 537 MKPDAVVWGALLGACKTFGNLEMGKQILK 565
K D V W +++ A +G G++ LK
Sbjct: 539 YK-DVVTWSSIIAA---YGFHGQGEEALK 613
>AW687696 weakly similar to PIR|T49965|T499 hypothetical protein F8M21.190 -
Arabidopsis thaliana, partial (16%)
Length = 289
Score = 109 bits (272), Expect = 4e-24
Identities = 47/91 (51%), Positives = 65/91 (70%)
Frame = +2
Query: 471 PDEITFTGLLSACSHSGLVEMGRYYFGLMSSTFGISPDVAHYACMVDLLGRGGFLGEAMS 530
PDEITF +L ACSH GLV+ GR YF +M+ + I P + HY CMVDLLGR G EA
Sbjct: 17 PDEITFLCVLCACSHGGLVDEGRRYFEIMNRDYNIKPTIKHYGCMVDLLGRAGLFVEAYE 196
Query: 531 LIQKLPMKPDAVVWGALLGACKTFGNLEMGK 561
LI+ +P++ +A++W LL AC+ +GN+E+G+
Sbjct: 197 LIKSMPVECNAIIWRTLLAACRNYGNVELGE 289
Score = 37.0 bits (84), Expect = 0.024
Identities = 20/86 (23%), Positives = 43/86 (49%), Gaps = 2/86 (2%)
Frame = +2
Query: 370 PDNATLVSILSSCSHMGDLALGKKAHDYI-CDNNITVSVTLCNSLMNMYAKCGALQTAMD 428
PD T + +L +CSH G + G++ + + D NI ++ ++++ + G A +
Sbjct: 17 PDEITFLCVLCACSHGGLVDEGRRYFEIMNRDYNIKPTIKHYGCMVDLLGRAGLFVEAYE 196
Query: 429 IFFGMP-QKNVVSWNVIIGALALHGS 453
+ MP + N + W ++ A +G+
Sbjct: 197 LIKSMPVECNAIIWRTLLAACRNYGN 274
Score = 29.3 bits (64), Expect = 5.1
Identities = 13/48 (27%), Positives = 25/48 (52%), Gaps = 1/48 (2%)
Frame = +2
Query: 299 DRMLDRNVVSWTCMINAYANHGLIDHALEIFDQMPVK-NAVSWNSIIS 345
D + + + CM++ GL A E+ MPV+ NA+ W ++++
Sbjct: 110 DYNIKPTIKHYGCMVDLLGRAGLFVEAYELIKSMPVECNAIIWRTLLA 253
>TC83657 weakly similar to GP|11994382|dbj|BAB02341.
gb|AAB82628.1~gene_id:MIL23.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1299
Score = 106 bits (264), Expect = 3e-23
Identities = 62/174 (35%), Positives = 95/174 (53%), Gaps = 1/174 (0%)
Frame = +3
Query: 479 LLSACSHSGLVEMGRYYFGLMSSTFGISPDVAHYACMVDLLGRGGFLGEAMSLIQKLPMK 538
+LSAC+H GL+ M +GI + HY CMVDLLGR G L EA LI+++PMK
Sbjct: 84 VLSACAHGGLMSEALEVISKMEE-YGIEMGIRHYGCMVDLLGRAGKLKEAYELIKRMPMK 260
Query: 539 PDAVVWGALLGACKTFGNLEMGKQILKQL-LELGRYDSGLYVLLSNMYSESQRWDDMKKI 597
P+ V GA++GAC +++M +Q++K + + + VLLSN+Y+ S++W+ + I
Sbjct: 261 PNETVLGAMIGACWIHSDMKMAEQVMKMIGADSAACVNSHNVLLSNIYAASEKWEKAEMI 440
Query: 598 RKIMNDSGIKKCRAISSIEIDGCCYQFMVDDKNHEVSTSIYSMVDQLMDHLKSV 651
R M D G +K SSI + N V SI + + M+ L ++
Sbjct: 441 RSSMVDGGSEKIPGYSSIIL-----------SNSAVDLSISKEISRQMNGLSAI 569
>BF646834 weakly similar to PIR|A84784|A847 hypothetical protein At2g36730
[imported] - Arabidopsis thaliana, partial (10%)
Length = 602
Score = 103 bits (256), Expect = 3e-22
Identities = 60/172 (34%), Positives = 96/172 (54%), Gaps = 3/172 (1%)
Frame = +2
Query: 47 LLDQCSSLRQLKLLHAQIILHGLTGQILTLGKLLSFCVHLGDLRYAH---LLFDQTPQPN 103
LL+ S+ +L L AQI L+ L L +L+ F +H L+F + P+
Sbjct: 83 LLNSLHSITKLHQLQAQIHLNSLHNDTHILSQLVYFFSLSPFKNLSHARKLVFHFSNNPS 262
Query: 104 KFMYNHLIRGYSNSDDPNKSLLLYRKMVCAGIMPNQFTIPFVLKACSAKPSCWDVAVVHA 163
+N LIRGY++SD P +S+ +++KM G+ PN+ T PF+ K+C+ + VHA
Sbjct: 263 PISWNILIRGYASSDSPIESIWVFKKMRENGVKPNKLTYPFIFKSCAMALVLCEGKQVHA 442
Query: 164 QAIKLGMGSHVCVQNAFLSLYVACRLIRSARQVFDDISDRTLVSWNSMIVGY 215
+K G+ S V V N ++ Y C+ I AR+VFD++ T+VSWNS++ +
Sbjct: 443 DLVKFGLDSDVYVCNNMINFYGXCKKIVYARKVFDEMCVXTIVSWNSVMTAW 598
Score = 68.2 bits (165), Expect = 1e-11
Identities = 42/137 (30%), Positives = 68/137 (48%)
Frame = +2
Query: 180 FLSLYVACRLIRSARQVFDDISDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEAD 239
F SL L + + VF ++ + +SWN +I GY+ E+I +FK+M+ GV+ +
Sbjct: 188 FFSLSPFKNLSHARKLVFHFSNNPSPISWNILIRGYASSDSPIESIWVFKKMRENGVKPN 367
Query: 240 MFTLVGLLSVALRHGNLDLGRFVHLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFD 299
T + L G+ VH +V G++ D V N +I+ Y C + YA VFD
Sbjct: 368 KLTYPFIFKSCAMALVLCEGKQVHADLVKFGLDSDVYVCNNMINFYGXCKKIVYARKVFD 547
Query: 300 RMLDRNVVSWTCMINAY 316
M +VSW ++ A+
Sbjct: 548 EMCVXTIVSWNSVMTAW 598
Score = 60.1 bits (144), Expect = 3e-09
Identities = 32/110 (29%), Positives = 54/110 (49%)
Frame = +2
Query: 338 VSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDY 397
+SWN +I + E++ +F +M +GV P+ T I SC+ L GK+ H
Sbjct: 266 ISWNILIRGYASSDSPIESIWVFKKMRENGVKPNKLTYPFIFKSCAMALVLCEGKQVHAD 445
Query: 398 ICDNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQKNVVSWNVIIGA 447
+ + V +CN+++N Y C + A +F M +VSWN ++ A
Sbjct: 446 LVKFGLDSDVYVCNNMINFYGXCKKIVYARKVFDEMCVXTIVSWNSVMTA 595
Score = 42.4 bits (98), Expect = 6e-04
Identities = 25/84 (29%), Positives = 48/84 (56%), Gaps = 2/84 (2%)
Frame = +2
Query: 439 VSWNVIIGALALHGSGKEAIEMFEKMRASGLFPDEITFTGLLSACSHSGLVEMGRYYFGL 498
+SWN++I A S E+I +F+KMR +G+ P+++T+ + +C+ + ++ G+
Sbjct: 266 ISWNILIRGYASSDSPIESIWVFKKMRENGVKPNKLTYPFIFKSCAMALVLCEGKQVHAD 445
Query: 499 MSSTFGISPDVAHYAC--MVDLLG 520
+ FG+ DV Y C M++ G
Sbjct: 446 LVK-FGLDSDV--YVCNNMINFYG 508
>TC84835 weakly similar to PIR|T02328|T02328 hypothetical protein At2g34400
[imported] - Arabidopsis thaliana, partial (5%)
Length = 647
Score = 102 bits (253), Expect = 6e-22
Identities = 61/200 (30%), Positives = 104/200 (51%), Gaps = 4/200 (2%)
Frame = +1
Query: 78 KLLSFCVHLGDLRYAHL---LFDQTPQPNKFMYNHLIRGYSNSDDPNKSLLLYRKMVCAG 134
KL S +HL + Y H +F+ T P+ F+YN LI+ + + + L+ ++ G
Sbjct: 13 KLFSVSIHLNNNNYFHYSLSIFNHTLHPSLFLYNLLIKSFFKRNSFQTLISLFNQLRLNG 192
Query: 135 IMPNQFTIPFVLKACSAKPSCWDVAVVHAQAIKLGMGSHVCVQNAFLSLYVACRLIRSAR 194
+ P+ +T PFVLKA + +HA K G+ S V N+F+ +Y I R
Sbjct: 193 LYPDNYTYPFVLKAVAFIADFRQGTKIHAFVFKTGLDSDYYVSNSFMDMYAELGRIDFVR 372
Query: 195 QVFDDISDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVE-ADMFTLVGLLSVALRH 253
++FD+IS+R VSWN MI G K +EA+ +F+ M+ E T+V L+
Sbjct: 373 KLFDEISERDSVSWNVMISGCVKCRRFEEAVEVFQRMRVDSNEKISEATVVSSLTACAAS 552
Query: 254 GNLDLGRFVHLYIVVAGIEI 273
N+++G+ +H +I+ +++
Sbjct: 553 RNVEVGKEIHGFIIEKELDL 612
Score = 83.2 bits (204), Expect = 3e-16
Identities = 49/183 (26%), Positives = 90/183 (48%), Gaps = 1/183 (0%)
Frame = +1
Query: 318 NHGLIDHALEIFDQMPVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVS 377
N+ ++L IF+ + +N +I + + LF ++ ++G+ PDN T
Sbjct: 43 NNNYFHYSLSIFNHTLHPSLFLYNLLIKSFFKRNSFQTLISLFNQLRLNGLYPDNYTYPF 222
Query: 378 ILSSCSHMGDLALGKKAHDYICDNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQKN 437
+L + + + D G K H ++ + + NS M+MYA+ G + +F + +++
Sbjct: 223 VLKAVAFIADFRQGTKIHAFVFKTGLDSDYYVSNSFMDMYAELGRIDFVRKLFDEISERD 402
Query: 438 VVSWNVIIGALALHGSGKEAIEMFEKMRA-SGLFPDEITFTGLLSACSHSGLVEMGRYYF 496
VSWNV+I +EA+E+F++MR S E T L+AC+ S VE+G+
Sbjct: 403 SVSWNVMISGCVKCRRFEEAVEVFQRMRVDSNEKISEATVVSSLTACAASRNVEVGKEIH 582
Query: 497 GLM 499
G +
Sbjct: 583 GFI 591
Score = 76.3 bits (186), Expect = 4e-14
Identities = 49/208 (23%), Positives = 95/208 (45%)
Frame = +1
Query: 196 VFDDISDRTLVSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEADMFTLVGLLSVALRHGN 255
+F+ +L +N +I + K + I LF +++ G+ D +T +L +
Sbjct: 73 IFNHTLHPSLFLYNLLIKSFFKRNSFQTLISLFNQLRLNGLYPDNYTYPFVLKAVAFIAD 252
Query: 256 LDLGRFVHLYIVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRMLDRNVVSWTCMINA 315
G +H ++ G++ D V+N+ +DMYA+ G + + +FD + +R+ VSW MI+
Sbjct: 253 FRQGTKIHAFVFKTGLDSDYYVSNSFMDMYAELGRIDFVRKLFDEISERDSVSWNVMISG 432
Query: 316 YANHGLIDHALEIFDQMPVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATL 375
+ A+E+F +M V + N IS AT+
Sbjct: 433 CVKCRRFEEAVEVFQRMRVDS----NEKIS--------------------------EATV 522
Query: 376 VSILSSCSHMGDLALGKKAHDYICDNNI 403
VS L++C+ ++ +GK+ H +I + +
Sbjct: 523 VSSLTACAASRNVEVGKEIHGFIIEKEL 606
Score = 30.8 bits (68), Expect = 1.7
Identities = 29/133 (21%), Positives = 58/133 (42%), Gaps = 6/133 (4%)
Frame = +1
Query: 25 QTQITEITKTKLNS--PTHQTLHCLLDQCSSL---RQLKLLHAQIILHGLTGQILTLGKL 79
QT I+ + +LN P + T +L + + RQ +HA + GL
Sbjct: 151 QTLISLFNQLRLNGLYPDNYTYPFVLKAVAFIADFRQGTKIHAFVFKTGLDSDYYVSNSF 330
Query: 80 LSFCVHLGDLRYAHLLFDQTPQPNKFMYNHLIRGYSNSDDPNKSLLLYRKM-VCAGIMPN 138
+ LG + + LFD+ + + +N +I G +++ ++++M V + +
Sbjct: 331 MDMYAELGRIDFVRKLFDEISERDSVSWNVMISGCVKCRRFEEAVEVFQRMRVDSNEKIS 510
Query: 139 QFTIPFVLKACSA 151
+ T+ L AC+A
Sbjct: 511 EATVVSSLTACAA 549
>BF642497 weakly similar to PIR|H96713|H96 hypothetical protein T6L1.11
[imported] - Arabidopsis thaliana, partial (20%)
Length = 461
Score = 101 bits (251), Expect = 1e-21
Identities = 58/183 (31%), Positives = 93/183 (50%)
Frame = +3
Query: 206 VSWNSMIVGYSKMGFSKEAILLFKEMQYLGVEADMFTLVGLLSVALRHGNLDLGRFVHLY 265
+SW SMI G+++ G ++AI +F+EM+ ++ D +T +L+ L G+ VH Y
Sbjct: 6 ISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAY 185
Query: 266 IVVAGIEIDSIVTNALIDMYAKCGHLQYAETVFDRMLDRNVVSWTCMINAYANHGLIDHA 325
I+ + + V +AL+DMY KC +++ AE VF +M +NVVSWT M+ Y
Sbjct: 186 IIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYG-------- 341
Query: 326 LEIFDQMPVKNAVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHM 385
Q G EA++ F M G+ PD+ TL S++SSC+++
Sbjct: 342 -----------------------QNGYSEEAVKTFSDMQKYGIEPDDFTLGSVISSCANL 452
Query: 386 GDL 388
L
Sbjct: 453 ASL 461
Score = 94.4 bits (233), Expect = 1e-19
Identities = 45/149 (30%), Positives = 86/149 (57%)
Frame = +3
Query: 337 AVSWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHD 396
++SW S+I+ Q G +A+++F M + + D T S+L++C + L GK+ H
Sbjct: 3 SISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHA 182
Query: 397 YICDNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKE 456
YI + ++ + ++L++MY KC +++A +F M KNVVSW ++ +G +E
Sbjct: 183 YIIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEE 362
Query: 457 AIEMFEKMRASGLFPDEITFTGLLSACSH 485
A++ F M+ G+ PD+ T ++S+C++
Sbjct: 363 AVKTFSDMQKYGIEPDDFTLGSVISSCAN 449
Score = 89.7 bits (221), Expect = 3e-18
Identities = 44/142 (30%), Positives = 81/142 (56%)
Frame = +3
Query: 107 YNHLIRGYSNSDDPNKSLLLYRKMVCAGIMPNQFTIPFVLKACSAKPSCWDVAVVHAQAI 166
+ +I G++ + ++ ++R+M + +Q+T VL AC + + VHA I
Sbjct: 12 WTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAYII 191
Query: 167 KLGMGSHVCVQNAFLSLYVACRLIRSARQVFDDISDRTLVSWNSMIVGYSKMGFSKEAIL 226
+ ++ V +A + +Y C+ I+SA VF ++ + +VSW +M+VGY + G+S+EA+
Sbjct: 192 RTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEEAVK 371
Query: 227 LFKEMQYLGVEADMFTLVGLLS 248
F +MQ G+E D FTL ++S
Sbjct: 372 TFSDMQKYGIEPDDFTLGSVIS 437
Score = 35.0 bits (79), Expect = 0.093
Identities = 21/98 (21%), Positives = 44/98 (44%)
Frame = +3
Query: 53 SLRQLKLLHAQIILHGLTGQILTLGKLLSFCVHLGDLRYAHLLFDQTPQPNKFMYNHLIR 112
+L++ K +HA II I L+ +++ A +F + N + ++
Sbjct: 153 ALQEGKQVHAYIIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLV 332
Query: 113 GYSNSDDPNKSLLLYRKMVCAGIMPNQFTIPFVLKACS 150
GY + +++ + M GI P+ FT+ V+ +C+
Sbjct: 333 GYGQNGYSEEAVKTFSDMQKYGIEPDDFTLGSVISSCA 446
>BQ147515 weakly similar to PIR|T05355|T05 hypothetical protein F8B4.150 -
Arabidopsis thaliana, partial (23%)
Length = 427
Score = 100 bits (248), Expect = 2e-21
Identities = 46/142 (32%), Positives = 85/142 (59%)
Frame = +2
Query: 397 YICDNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQKNVVSWNVIIGALALHGSGKE 456
Y+ + + V+ CN ++ MY +CG++ A+++F M ++++ + ++I LA +G ++
Sbjct: 2 YVLQHLSPLKVSTCNGILEMYFQCGSVDDAVNVFKNMTERDLTTICIMIKQLAKNGFAED 181
Query: 457 AIEMFEKMRASGLFPDEITFTGLLSACSHSGLVEMGRYYFGLMSSTFGISPDVAHYACMV 516
+I++F + + SGL PD F G+ AC G + G +F MS + I P + HY +V
Sbjct: 182 SIDLFTQFKRSGLKPDGQMFIGVFGACXMLGDIVEGMLHFESMSRDYDIVPTMEHYVSLV 361
Query: 517 DLLGRGGFLGEAMSLIQKLPMK 538
D++G G L EA+ I+K+PM+
Sbjct: 362 DMIGSIGHLDEALEFIEKMPME 427
Score = 49.7 bits (117), Expect = 4e-06
Identities = 36/159 (22%), Positives = 67/159 (41%), Gaps = 1/159 (0%)
Frame = +2
Query: 279 NALIDMYAKCGHLQYAETVFDRMLDRNVVSWTCMINAYANHGLIDHALEIFDQMPVKNAV 338
N +++MY +CG + A VF M +R++ + MI A +G + ++++F Q
Sbjct: 44 NGILEMYFQCGSVDDAVNVFKNMTERDLTTICIMIKQLAKNGFAEDSIDLFTQ------- 202
Query: 339 SWNSIISCHVQEGQHAEAMELFCRMCVSGVMPDNATLVSILSSCSHMGDLALGKKAHDYI 398
F R SG+ PD + + +C +GD+ G + +
Sbjct: 203 ---------------------FKR---SGLKPDGQMFIGVFGACXMLGDIVEGMLHFESM 310
Query: 399 C-DNNITVSVTLCNSLMNMYAKCGALQTAMDIFFGMPQK 436
D +I ++ SL++M G L A++ MP +
Sbjct: 311 SRDYDIVPTMEHYVSLVDMIGSIGHLDEALEFIEKMPME 427
>BG646586 weakly similar to GP|10177683|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (9%)
Length = 687
Score = 100 bits (248), Expect = 2e-21
Identities = 48/112 (42%), Positives = 72/112 (63%)
Frame = -3
Query: 505 ISPDVAHYACMVDLLGRGGFLGEAMSLIQKLPMKPDAVVWGALLGACKTFGNLEMGKQIL 564
I P HY MVDLLGR G L +A SLI++ DA WG+LL AC+ +GN+E+G+
Sbjct: 664 IEPLPEHYTGMVDLLGRAGQLEKAYSLIKENSTSADATTWGSLLAACRVYGNVELGEIAA 485
Query: 565 KQLLELGRYDSGLYVLLSNMYSESQRWDDMKKIRKIMNDSGIKKCRAISSIE 616
+ L E+ DSG YVLL+N Y+ + +W+ ++++K+M+ G+KK S I+
Sbjct: 484 RHLFEIDPTDSGNYVLLANTYASNDKWERAEEVKKLMSKKGMKKPSGYSWIQ 329
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,261,888
Number of Sequences: 36976
Number of extensions: 340800
Number of successful extensions: 2373
Number of sequences better than 10.0: 126
Number of HSP's better than 10.0 without gapping: 2092
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2294
length of query: 664
length of database: 9,014,727
effective HSP length: 102
effective length of query: 562
effective length of database: 5,243,175
effective search space: 2946664350
effective search space used: 2946664350
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0559.15


