

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0550.4
(888 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
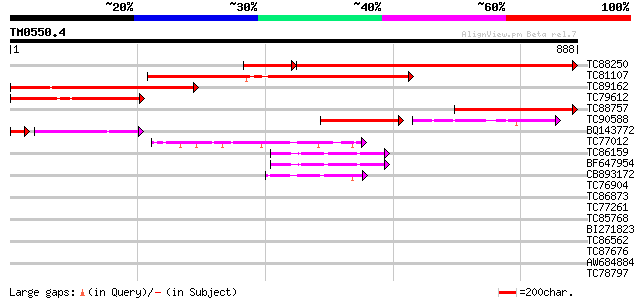
Score E
Sequences producing significant alignments: (bits) Value
TC88250 similar to SP|O64948|LON1_ARATH Lon protease homolog 1 ... 590 0.0
TC81107 similar to SP|O64948|LON1_ARATH Lon protease homolog 1 ... 597 e-171
TC89162 similar to SP|O04979|LON1_SPIOL Lon protease homolog 1 ... 478 e-135
TC79612 similar to SP|O04979|LON1_SPIOL Lon protease homolog 1 ... 352 4e-97
TC88757 similar to GP|4666311|dbj|BAA77218.1 LON protease homolo... 325 4e-89
TC90588 similar to SP|P93648|LON2_MAIZE Lon protease homolog 2 ... 131 1e-30
BQ143772 similar to SP|O04979|LON1_ Lon protease homolog 1 mito... 92 1e-26
TC77012 homologue to SP|P54776|PRSA_LYCES 26S protease regulator... 49 8e-06
TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AA... 44 2e-04
BF647954 homologue to GP|11094192|db 26S proteasome regulatory p... 44 2e-04
CB893172 homologue to PIR|T45642|T45 FtsH metalloproteinase-like... 42 8e-04
TC76904 homologue to GP|4325041|gb|AAD17230.1| FtsH-like protein... 40 0.003
TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protei... 40 0.004
TC77261 homologue to GP|8777330|dbj|BAA96920.1 26S proteasome AA... 38 0.015
TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle pr... 37 0.025
BI271823 similar to GP|21592745|gb cell division protein FtsH-li... 37 0.025
TC86562 similar to GP|21553404|gb|AAM62497.1 26S proteasome regu... 36 0.074
TC87676 PIR|G96577|G96577 26S proteasome ATPase subunit [importe... 36 0.074
AW684884 similar to GP|6069665|dbj EST D15969(C1695) corresponds... 36 0.074
TC78797 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 36 0.074
>TC88250 similar to SP|O64948|LON1_ARATH Lon protease homolog 1
mitochondrial precursor (EC 3.4.21.-). [Mouse-ear
cress], partial (55%)
Length = 2128
Score = 590 bits (1521), Expect(2) = 0.0
Identities = 314/441 (71%), Positives = 348/441 (78%), Gaps = 2/441 (0%)
Frame = +1
Query: 450 TYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHY 509
TY+GSMPGRLIDGLKRVAVCNPVMLLDE+DKTGSDVRGDPASALLEVLDPEQNKTFNDHY
Sbjct: 256 TYVGSMPGRLIDGLKRVAVCNPVMLLDEVDKTGSDVRGDPASALLEVLDPEQNKTFNDHY 435
Query: 510 LNVPFDLSKVIFVATANRAQQIPPPLLDRMEVIELPGYTPEEKLKIAMKHLIPRVLDQHG 569
LNVPFDLSKVIFVATANR Q IPPPLLDRMEVIELPGYTPEEKL+IAM+HLIPRVLDQHG
Sbjct: 436 LNVPFDLSKVIFVATANRMQPIPPPLLDRMEVIELPGYTPEEKLQIAMQHLIPRVLDQHG 615
Query: 570 LSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLASLARAAAVRVAEQEQVVPLNKEVQGL 629
LSSE++QIPE MV+LVIQRYTREAGVRNLERNLA+LARAAAVR+ EQEQVVPL K +QGL
Sbjct: 616 LSSEFIQIPEAMVKLVIQRYTREAGVRNLERNLAALARAAAVRIVEQEQVVPLTKGMQGL 795
Query: 630 TTPLLENRLTDGAEVEMEVIPMDVNSRDISNTFRITSPLVVNEAILEKVLGPPKFDGREA 689
+TPLLENRL DG EVEM+V+PM VNSR ISNTFRI SPLVV+E +LEKVLGPP+FD EA
Sbjct: 796 STPLLENRLADGTEVEMDVMPMSVNSRVISNTFRIASPLVVDETMLEKVLGPPRFDDSEA 975
Query: 690 AERVAAPGICVGLVWTTFGGEVQFVEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRAR 749
AERVA+PG+ VGLVWT FGGEVQFVEAT MVGKG+LHLTGQLGDVIKESAQ +
Sbjct: 976 AERVASPGVSVGLVWTAFGGEVQFVEATAMVGKGELHLTGQLGDVIKESAQNCTNMG*GQ 1155
Query: 750 A-ADLRLASAEGINLLEGRDIHIHFPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAM 808
LR A+GI G F G + + F ++ +
Sbjct: 1156GQLILRWMLAKGI*SFGGP*CTYSFSCGCCSERWALGRCNFSHIIGITFHSEKCEIRYSY 1335
Query: 809 TGEMTLRGLVLPVGGI-KDKILAAHRCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKR 867
+ G+ + + + AAHR GIKRVI+PERNLKDL EVP+SVL NLEILLAKR
Sbjct: 1336DWRDDVEGISSTCWRVLRIRYYAAHRYGIKRVILPERNLKDLAEVPASVLANLEILLAKR 1515
Query: 868 MEDVLEQALEGGCPWRHQSKL 888
MEDVLE A +GGCPWR SKL
Sbjct: 1516MEDVLEHAFDGGCPWRQHSKL 1578
Score = 158 bits (400), Expect(2) = 0.0
Identities = 78/84 (92%), Positives = 82/84 (96%)
Frame = +3
Query: 366 LKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIAAA 425
L++AQ+RLDSDHYGL KVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIAAA
Sbjct: 3 LRAAQKRLDSDHYGLVKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIAAA 182
Query: 426 LDRKFVRISLGGVKDEADIRGHRR 449
L RKF+RISLGGVKDEADIRGHRR
Sbjct: 183 LGRKFIRISLGGVKDEADIRGHRR 254
>TC81107 similar to SP|O64948|LON1_ARATH Lon protease homolog 1
mitochondrial precursor (EC 3.4.21.-). [Mouse-ear
cress], partial (45%)
Length = 1266
Score = 597 bits (1540), Expect = e-171
Identities = 330/430 (76%), Positives = 354/430 (81%), Gaps = 13/430 (3%)
Frame = +2
Query: 216 KLADIFVASFEISFEEQLSMLDSVDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSK 275
++ADIFVASFEISFEEQLSMLDSVDAK+RLSKATELVDRHLQSIRVAEKITQKVEGQLSK
Sbjct: 11 QVADIFVASFEISFEEQLSMLDSVDAKVRLSKATELVDRHLQSIRVAEKITQKVEGQLSK 190
Query: 276 SQKEFLLRQQMRAIKEELGDNDDDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQP 335
SQKEFLLRQQM+AIKEELGDNDD+EDDLAALERKMQSAGMPQ++WK + RELR LKKMQP
Sbjct: 191 SQKEFLLRQQMKAIKEELGDNDDEEDDLAALERKMQSAGMPQNVWKLSLRELRTLKKMQP 370
Query: 336 QQPGYSSSRAYLDLLADLPWQKASEEHEMDLKSA-----------QERLDSDHYGLAKVK 384
QQPG + + LKS +E +DH +
Sbjct: 371 QQPGV*QFTGLPGTSC*SSMAEGQQRA*TGLKSCTGTPGQ*SLWFREGQAADH----*IP 538
Query: 385 QRIIEYLAVRKLKPDARGPVLC--FVGPPGVGKTSLASSIAAALDRKFVRISLGGVKDEA 442
R +++ LC FVGPPGVGKTSLASSIAAALDRKFVRISLGGVKDEA
Sbjct: 539 SRS*A*ARCKRV--------LCWCFVGPPGVGKTSLASSIAAALDRKFVRISLGGVKDEA 694
Query: 443 DIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQN 502
DIRGHRRTYIGSMPGRLIDGLK+VAVCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQN
Sbjct: 695 DIRGHRRTYIGSMPGRLIDGLKKVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQN 874
Query: 503 KTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDRMEVIELPGYTPEEKLKIAMKHLIP 562
KTFNDHYLNVPFDLSKVIFVATANRAQ IPPPLLDRMEVIELPGYTPEEKLKIAMKHLIP
Sbjct: 875 KTFNDHYLNVPFDLSKVIFVATANRAQPIPPPLLDRMEVIELPGYTPEEKLKIAMKHLIP 1054
Query: 563 RVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLASLARAAAVRVAEQEQVVPL 622
RVLDQHGLSSE+LQI QLVIQRYTREAGVR+LER+LA+LARAAA +VA+QEQVVPL
Sbjct: 1055RVLDQHGLSSEFLQIXGSDGQLVIQRYTREAGVRSLERSLAALARAAAXKVAKQEQVVPL 1234
Query: 623 NKEVQGLTTP 632
K + GLTTP
Sbjct: 1235XKXLXGLTTP 1264
>TC89162 similar to SP|O04979|LON1_SPIOL Lon protease homolog 1
mitochondrial precursor (EC 3.4.21.-). [Spinach]
{Spinacia oleracea}, partial (32%)
Length = 905
Score = 478 bits (1230), Expect = e-135
Identities = 253/297 (85%), Positives = 266/297 (89%), Gaps = 2/297 (0%)
Frame = +3
Query: 1 MAESVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRD 60
MAESVELPSR+AILPFRNKVLLPGAIIRIRCTSP+SVKLVEQELWQ+E+KGLIGILPVRD
Sbjct: 21 MAESVELPSRIAILPFRNKVLLPGAIIRIRCTSPSSVKLVEQELWQKEEKGLIGILPVRD 200
Query: 61 AAETEVKPVGPTISQDG--DTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALH 118
A E + GPT SQ D L+QSSK+ SSDS K D K QND+VHWHNRGVAAR LH
Sbjct: 201 ALE--INSAGPTASQGAGTDILNQSSKLQDGSSDSIKPDTKRQNDIVHWHNRGVAARPLH 374
Query: 119 LSRGVEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFI 178
LSRGVEKPSGRVTY VVLEGLCRFSV ELSTRG YHTA+ISSLEM K EMEQ+E DPD+I
Sbjct: 375 LSRGVEKPSGRVTYTVVLEGLCRFSVLELSTRGIYHTAKISSLEMTKIEMEQLEQDPDYI 554
Query: 179 MLSRQFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDS 238
MLSRQFKATA ELISVLE KQKTGGRTKVLL+ +PVHKLADIFVASFEISFEEQLSMLDS
Sbjct: 555 MLSRQFKATATELISVLELKQKTGGRTKVLLDNIPVHKLADIFVASFEISFEEQLSMLDS 734
Query: 239 VDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGD 295
V KLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGD
Sbjct: 735 VGPKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGD 905
>TC79612 similar to SP|O04979|LON1_SPIOL Lon protease homolog 1
mitochondrial precursor (EC 3.4.21.-). [Spinach]
{Spinacia oleracea}, partial (19%)
Length = 768
Score = 352 bits (903), Expect = 4e-97
Identities = 184/212 (86%), Positives = 195/212 (91%), Gaps = 1/212 (0%)
Frame = +3
Query: 1 MAESVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRD 60
MA+SVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQRE+KG+IGILPVRD
Sbjct: 150 MADSVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREEKGMIGILPVRD 329
Query: 61 -AAETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHL 119
AA E K +G T+S D++DQ+SKVHG SSDS+ AK QNDVVHWHNRGVAARALHL
Sbjct: 330 TAASAETKQLGSTVS---DSIDQTSKVHGGSSDSN---AKVQNDVVHWHNRGVAARALHL 491
Query: 120 SRGVEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIM 179
SRGVEKPSGRVTYIVVLEGLCRF+VQEL+TRGTYHTARISSLEM KTEMEQVEHDPDFIM
Sbjct: 492 SRGVEKPSGRVTYIVVLEGLCRFNVQELNTRGTYHTARISSLEMTKTEMEQVEHDPDFIM 671
Query: 180 LSRQFKATAMELISVLEQKQKTGGRTKVLLET 211
LSRQFKATAMELISVLE KQ TGGRTKVLLET
Sbjct: 672 LSRQFKATAMELISVLEXKQSTGGRTKVLLET 767
>TC88757 similar to GP|4666311|dbj|BAA77218.1 LON protease homologue
{Lithospermum erythrorhizon}, partial (84%)
Length = 1104
Score = 325 bits (834), Expect = 4e-89
Identities = 164/192 (85%), Positives = 176/192 (91%)
Frame = +3
Query: 697 GICVGLVWTTFGGEVQFVEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLA 756
G LVWT+ GGEVQFVEA+ MVGKG+LHLTGQLGDVIKESAQIALTWVRARA DL+LA
Sbjct: 15 GYLSXLVWTSVGGEVQFVEASTMVGKGELHLTGQLGDVIKESAQIALTWVRARATDLKLA 194
Query: 757 SAEGINLLEGRDIHIHFPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRG 816
+AE I+LLEGRDIHIHFPAGA+PKDGPSAGVTLVTALVSLFS+++VRSDTAMTGEMTLRG
Sbjct: 195 AAESISLLEGRDIHIHFPAGAVPKDGPSAGVTLVTALVSLFSQRKVRSDTAMTGEMTLRG 374
Query: 817 LVLPVGGIKDKILAAHRCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQAL 876
LVLPVGGIKDKILAAHR GIKRVI+PERNLKDLVEVP SVL NLEILLAKRMEDVLE A
Sbjct: 375 LVLPVGGIKDKILAAHRYGIKRVILPERNLKDLVEVPPSVLANLEILLAKRMEDVLEHAF 554
Query: 877 EGGCPWRHQSKL 888
+ GCPWR SKL
Sbjct: 555 DSGCPWRQHSKL 590
>TC90588 similar to SP|P93648|LON2_MAIZE Lon protease homolog 2
mitochondrial precursor (EC 3.4.21.-). [Maize] {Zea
mays}, partial (34%)
Length = 1540
Score = 131 bits (330), Expect = 1e-30
Identities = 63/130 (48%), Positives = 90/130 (68%)
Frame = +2
Query: 487 GDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDRMEVIELPG 546
GDPASALLE+LDPEQN F DHYL+VP DLSKV+FV TAN + IP PLLDRMEV+ + G
Sbjct: 11 GDPASALLELLDPEQNANFLDHYLDVPIDLSKVLFVCTANVVEMIPNPLLDRMEVVTIAG 190
Query: 547 YTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLASLA 606
Y +EK+ IA +L + G+ E +++ + + +I+ Y RE+GVRNL++++ +
Sbjct: 191 YIADEKMHIARDYLEKTTREACGIKPEQVEVTDAALLALIENYCRESGVRNLQKHIEKIY 370
Query: 607 RAAAVRVAEQ 616
R A+++ Q
Sbjct: 371 RKIALQLVRQ 400
Score = 90.5 bits (223), Expect = 3e-18
Identities = 70/247 (28%), Positives = 109/247 (43%), Gaps = 16/247 (6%)
Frame = +1
Query: 632 PLLENRLTDGAEVEMEVIPMDVNSRDISNTFRITSPLVVNEAILEKVLGPPKFDGREAAE 691
P ++ D +ME + + S T ++++++ L+ +G P F +
Sbjct: 565 PADQSLRVDNKSTDMEGTKEEEDKETESKT---VEKVLIDKSNLDDFVGKPVFHAERIYD 735
Query: 692 RVAAPGICVGLVWTTFGGEVQFVEAT---EMVGKGDLHLTGQLGDVIKESAQIALTWVRA 748
+ A G+ +GL WT GG ++E E GKG LH+TGQLGDV+KESAQ+A T R
Sbjct: 736 QTPA-GVVMGLAWTAMGGSTLYIETKFVEEGDGKGALHITGQLGDVMKESAQLAHTIAR- 909
Query: 749 RAADLRLASAEGINLLEGRDIHIHFPAGAIPKDGPSAGVTLVTA-------------LVS 795
++L G HF KD P T +V
Sbjct: 910 -------------DILRGERTRKHFLCQY--KDSPPCSCRGYTKGWTKCWLHNDHFYVVP 1044
Query: 796 LFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKRVIIPERNLKDLVEVPSS 855
+ + G+ L G +LP+GG+K+K +AA R +K +I P N +D E+ +
Sbjct: 1045CLEEACEEGSSNDWGK*HLLGKILPIGGVKEKTIAARRSEVKTIIFPSANKRDFDELAPN 1224
Query: 856 VLVNLEI 862
V LE+
Sbjct: 1225VKEGLEV 1245
>BQ143772 similar to SP|O04979|LON1_ Lon protease homolog 1 mitochondrial
precursor (EC 3.4.21.-). [Spinach] {Spinacia oleracea},
partial (8%)
Length = 682
Score = 91.7 bits (226), Expect(2) = 1e-26
Identities = 70/175 (40%), Positives = 88/175 (50%), Gaps = 4/175 (2%)
Frame = +2
Query: 39 LVEQELW-QREDKGLIGILPVRDAAETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKLD 97
LVEQ+LW QR + P DA E + + S D L+Q+S +H S+
Sbjct: 149 LVEQDLWHQRREGDDEAFFPCTDALEIDSPAPNASQSAGTDILNQNSTLHNGISNPINPA 328
Query: 98 AKAQNDVVHWHNRGVAARALHLSRGVEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTAR 157
K ND+ HWH RGV A LH+SRGV+KP GRVTY V +EG F V +L TR R
Sbjct: 329 TKRPNDLDHWHFRGVGALPLHISRGVDKPCGRVTYTVTVEG*YTFVVLKLRTRRNI-PYR 505
Query: 158 ISSLEMNKTEMEQVEHDPDFIML--SRQFKATAMELISVLEQKQKT-GGRTKVLL 209
I + + + QFKAT+ ELISV + T GGRTKV L
Sbjct: 506 IDLFA*DDYN*HGTIRAGLLLRYC*TPQFKATSTELISVPGTEINTPGGRTKVPL 670
Score = 47.4 bits (111), Expect(2) = 1e-26
Identities = 23/30 (76%), Positives = 27/30 (89%)
Frame = +1
Query: 1 MAESVELPSRLAILPFRNKVLLPGAIIRIR 30
MAES ELP+R+ ILPF NKVLLPGAII+I+
Sbjct: 31 MAESGELPTRIPILPFINKVLLPGAIIKIQ 120
>TC77012 homologue to SP|P54776|PRSA_LYCES 26S protease regulatory subunit
6A homolog (TAT-binding protein homolog 1) (TBP-1)
(Mg(2+), complete
Length = 1793
Score = 48.9 bits (115), Expect = 8e-06
Identities = 86/375 (22%), Positives = 162/375 (42%), Gaps = 38/375 (10%)
Frame = +1
Query: 223 ASFEISFEEQLSMLDSVDAKLRLSKATELVDRHLQSIRVAEKIT----QKVEGQLSKSQK 278
A+FE ++QL+ + + D + +A+ L+D ++ ++ + T + + ++ ++Q+
Sbjct: 157 ANFE---DDQLANMSTDD----IVRASRLLDNEIRILKEESQRTNLELESYKEKIKENQE 315
Query: 279 EFLLRQQMRAIKE------ELGDNDDDEDDLAALERKMQSAGMPQSIWKHAHRELRRLK- 331
+ L +Q+ + E+ D+ E+D A ++ Q G + K + R+ L
Sbjct: 316 KIKLNKQLPYLVGNIVEILEMNPEDEAEEDGANIDLDSQRKGKCV-VLKTSTRQTIFLPV 492
Query: 332 -------KMQPQQPGYSSSRAYLDLLADLPWQKASEEHEMDLKSAQERLDSDHYGLAKVK 384
K++P + +YL +L LP + S M++ +D GL K
Sbjct: 493 VGLVDPDKLKPGDLVGVNKDSYL-ILDTLPSEYDSRVKAMEVDEKPTEDYNDIGGLEKQI 669
Query: 385 QRIIEYLAV------RKLKPDARGPV-LCFVGPPGVGKTSLASSIAAALDRKFVRISLGG 437
Q ++E + + R K R P + GPPG GKT +A + AA + F++++
Sbjct: 670 QELVEAIVLPMTHKERFQKLGIRPPKGVLLYGPPGTGKTLMARACAAQTNATFLKLAGPQ 849
Query: 438 VKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNP-VMLLDEIDKTG-----SDVRGD--P 489
+ + +IG + D + +P ++ +DEID G S+V GD
Sbjct: 850 LV---------QMFIGDGAKLVRDAFQLAKEKSPCIIFIDEIDAIGTKRFDSEVSGDREV 1002
Query: 490 ASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPL-----LDRMEVIEL 544
+LE+L+ + +D ++ +A NRA + P L LDR IE
Sbjct: 1003QRTMLELLNQLDGFSSDD----------RIKVIAATNRADILDPALMRSGRLDRK--IEF 1146
Query: 545 PGYTPEEKLKIAMKH 559
P T E + +I H
Sbjct: 1147PHPTEEARARILQIH 1191
>TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AAA-ATPase
subunit RPT4a-like protein {Arabidopsis thaliana},
complete
Length = 1640
Score = 44.3 bits (103), Expect = 2e-04
Identities = 54/197 (27%), Positives = 91/197 (45%), Gaps = 10/197 (5%)
Frame = +2
Query: 409 GPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAV 468
GPPG GKT LA +IA+ +D F+++ + D+ YIG RLI + A
Sbjct: 608 GPPGTGKTLLARAIASNIDANFLKVVSSAIIDK---------YIGE-SARLIREMFGYAR 757
Query: 469 CNP--VMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFND--HYLNVPFDLSKVIFVAT 524
+ ++ +DEID G + SA D E +T + + L+ L KV +
Sbjct: 758 DHQPCIIFMDEIDAIGGRRFSEGTSA-----DREIQRTLMELLNQLDGFDQLGKVKMIMA 922
Query: 525 ANRAQQIPPPLL--DRME-VIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEY---LQIP 578
NR + P LL R++ IE+P P E+ ++ + + + +HG +Y +++
Sbjct: 923 TNRPDVLDPALLRPGRLDRKIEIP--LPNEQSRMEILKIHAAGIAKHG-EIDYEAVVKLA 1093
Query: 579 EGMVQLVIQRYTREAGV 595
EG ++ EAG+
Sbjct: 1094EGFNGADLRNVCTEAGM 1144
>BF647954 homologue to GP|11094192|db 26S proteasome regulatory particle
triple-A ATPase subunit4 {Oryza sativa (japonica
cultivar-group)}, partial (53%)
Length = 649
Score = 44.3 bits (103), Expect = 2e-04
Identities = 54/197 (27%), Positives = 91/197 (45%), Gaps = 10/197 (5%)
Frame = +2
Query: 409 GPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAV 468
GPPG GKT LA +IA+ +D F+++ + D+ YIG RLI + A
Sbjct: 62 GPPGTGKTLLARAIASNIDANFLKVVSSAIIDK---------YIGE-SSRLIREMFGYAR 211
Query: 469 CNP--VMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFND--HYLNVPFDLSKVIFVAT 524
+ ++ +DEID G + SA D E +T + + L+ L KV +
Sbjct: 212 DHQPCIIFMDEIDAIGGRRFSEGTSA-----DREIQRTLMELLNQLDGFDQLGKVKIIMA 376
Query: 525 ANRAQQIPPPLL--DRME-VIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEY---LQIP 578
NR + P LL R++ IE+P P E+ ++ + + + +HG +Y +++
Sbjct: 377 TNRPDVLDPALLRPGRLDRKIEIP--LPNEQSRMEILKIHAAGIAKHG-EIDYEAVVKLA 547
Query: 579 EGMVQLVIQRYTREAGV 595
EG ++ EAG+
Sbjct: 548 EGFNGADLRNICTEAGM 598
>CB893172 homologue to PIR|T45642|T45 FtsH metalloproteinase-like protein -
Arabidopsis thaliana, partial (25%)
Length = 661
Score = 42.4 bits (98), Expect = 8e-04
Identities = 50/171 (29%), Positives = 72/171 (41%), Gaps = 11/171 (6%)
Frame = +3
Query: 401 RGPVLCFVGPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLI 460
RG +L VG PG GKT LA ++A D F+ S + Y+G R+
Sbjct: 87 RGVLL--VGLPGTGKTLLAKAVAGEADVPFISCSASEFVE---------LYVGMGASRVR 233
Query: 461 DGLKRVAVCNP-VMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVP-FDL-S 517
D R P ++ +DEID G V + E+ +T N + FD S
Sbjct: 234 DLFARAKKEAPSIIFIDEIDAVAKSRDG----KFRIVGNDEREQTLNQLLTEMDGFDSNS 401
Query: 518 KVIFVATANRAQQIPPPL-----LDRMEVIELPGYTPEE---KLKIAMKHL 560
VI +A NRA + P L DR+ ++E P E K+ ++ K L
Sbjct: 402 PVIVLAATNRADVLDPALRRPGRFDRIVMVETPDRIGRESILKVHVSKKEL 554
>TC76904 homologue to GP|4325041|gb|AAD17230.1| FtsH-like protein Pftf
precursor {Nicotiana tabacum}, partial (91%)
Length = 2667
Score = 40.4 bits (93), Expect = 0.003
Identities = 98/394 (24%), Positives = 150/394 (37%), Gaps = 43/394 (10%)
Frame = +1
Query: 195 LEQKQKTGGR--TKVLLETVPVHK----------LADIFVASFEISFEEQLSMLDSVDAK 242
LEQ Q+ G R K+LL V V AD +S +S+ L LD K
Sbjct: 295 LEQNQQEGRRGILKLLLGNVGVGLPALLGNGTAYAADEQGSSSRMSYSRFLEYLD----K 462
Query: 243 LRLSKATELVDRHLQSIRVAEKIT-------QKVEGQLSKSQKEFLLRQQMRAIKEELGD 295
R++K +L + +I + E ++ Q+V QL +E L Q+ R +
Sbjct: 463 DRVTKV-DLYENG--TIAIVEAVSPELGNRLQRVRVQLPGLSQELL--QKFREKNIDFAA 627
Query: 296 NDDDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQ-PQQPGYSSSRAYLDLLADLP 354
++ ED + L + + P ++ RR M P PG+ P
Sbjct: 628 HNAQEDSGSLLFNLIGNLAFPAAVIGVLFLLSRRSGGMGGPGGPGF-------------P 768
Query: 355 WQKASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPD------ARGPV-LCF 407
Q + + ++ D G+ + KQ +E + K KP+ AR P +
Sbjct: 769 MQFGQSKAKFQMEPNTGVTFDDVAGVDEAKQDFMEVVEFLK-KPERFTSVGARIPKGVLL 945
Query: 408 VGPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVA 467
VGPPG GKT LA +IA F IS + ++G R+ D K+
Sbjct: 946 VGPPGTGKTLLAKAIAGEAGVPFFSISGSEFVE---------MFVGIGASRVRDLFKKAK 1098
Query: 468 VCNP-VMLLDEIDKT----------GSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDL 516
P ++ +DEID G+D R + LL +D + T
Sbjct: 1099ENAPCIVFVDEIDAVGRQRGTGIGGGNDEREQTLNQLLTEMDGFEGNT------------ 1242
Query: 517 SKVIFVATANRAQQIPPPLL-----DRMEVIELP 545
VI VA NRA + LL DR +++P
Sbjct: 1243-GVIVVAATNRADILDSALLRPGRFDRQVSVDVP 1341
>TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protein {Glycine
max}, partial (53%)
Length = 1475
Score = 40.0 bits (92), Expect = 0.004
Identities = 32/99 (32%), Positives = 52/99 (52%), Gaps = 7/99 (7%)
Frame = +2
Query: 407 FVGPPGVGKTSLASSIAAAL---DRKFVRISLGGVKDEADIR---GHRRTYIG-SMPGRL 459
F+GP GVGKT LA ++A L + + VRI + ++ + G Y+G G+L
Sbjct: 713 FLGPTGVGKTELAKALAEQLFDDENQLVRIDMSEYMEQHSVSRLIGAPPGYVGHEEGGQL 892
Query: 460 IDGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLD 498
+ ++R V+L DE++K + V + LL+VLD
Sbjct: 893 TEAVRRRPY--SVVLFDEVEKAHTSV----FNTLLQVLD 991
>TC77261 homologue to GP|8777330|dbj|BAA96920.1 26S proteasome AAA-ATPase
subunit RPT3 {Arabidopsis thaliana}, partial (94%)
Length = 1575
Score = 38.1 bits (87), Expect = 0.015
Identities = 57/207 (27%), Positives = 91/207 (43%), Gaps = 14/207 (6%)
Frame = +2
Query: 409 GPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAV 468
GPPG GKT LA ++A F+R+ V E + Y+G P R++ + R+A
Sbjct: 698 GPPGTGKTMLAKAVANHTTAAFIRV----VGSE-----FVQKYLGEGP-RMVRDVFRLAK 847
Query: 469 CN--PVMLLDEID---------KTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLS 517
N ++ +DE+D +TG+D + L+E+L+ Q F D +NV
Sbjct: 848 ENAPAIIFIDEVDAIATARFDAQTGAD--REVQRILMELLN--QMDGF-DQTVNV----- 997
Query: 518 KVIFVATANRAQQIPPPLL--DRME-VIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEY 574
KVI NRA + P LL R++ IE P +K + L +Y
Sbjct: 998 KVIM--ATNRADTLDPALLRPGRLDRKIEFPLPDRRQKRLVFQVCTAKMNLSDEVDLEDY 1171
Query: 575 LQIPEGMVQLVIQRYTREAGVRNLERN 601
+ P+ + I +EAG+ + +N
Sbjct: 1172VSRPDKISAAEISAICQEAGMHAVRKN 1252
>TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle protein 48
homolog (Valosin containing protein homolog) (VCP).
[Soybean], partial (95%)
Length = 2785
Score = 37.4 bits (85), Expect = 0.025
Identities = 60/231 (25%), Positives = 93/231 (39%), Gaps = 18/231 (7%)
Frame = +2
Query: 407 FVGPPGVGKTSLASSIAAALDRKFVRI------SLGGVKDEADIRGHRRTYIGSMPGRLI 460
F GPPG GKT LA +IA F+ I ++ + EA++R +
Sbjct: 1652 FYGPPGCGKTLLAKAIANECQANFISIKGPELLTMWFGESEANVR------------EIF 1795
Query: 461 DGLKRVAVCNPVMLLDEID----KTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDL 516
D + A C V+ DE+D + GS V GD A VL+ Q T D
Sbjct: 1796 DKARGSAPC--VLFFDELDSIATQRGSSV-GDAGGAADRVLN--QLLTEMD-----GMSA 1945
Query: 517 SKVIFVATA-NRAQQIPPPL-----LDRMEVIELPGYTPEEKLKIA--MKHLIPRVLDQH 568
K +F+ A NR I P L LD++ I LP ++ A K I + +D
Sbjct: 1946 KKTVFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDEDSRHQIFKACLRKSPISKDVDIR 2125
Query: 569 GLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLASLARAAAVRVAEQEQV 619
L+ + + QR + A N+E+++ + + A +E +
Sbjct: 2126 ALAKYTQGFSGADITEICQRACKYAIRENIEKDIEKERKRSENPEAMEEDI 2278
>BI271823 similar to GP|21592745|gb cell division protein FtsH-like protein
{Arabidopsis thaliana}, partial (33%)
Length = 714
Score = 37.4 bits (85), Expect = 0.025
Identities = 37/138 (26%), Positives = 59/138 (41%), Gaps = 3/138 (2%)
Frame = +3
Query: 401 RGPVLCFVGPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLI 460
RG +L VGPPG GKT LA ++A F +S + ++G R+
Sbjct: 201 RGVLL--VGPPGTGKTLLARAVAGEAGVPFFTVSASEFVE---------MFVGRGAARIR 347
Query: 461 DGLKRVAVCNP-VMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLS-- 517
D R P ++ +DE+D G RG + E+++T N + S
Sbjct: 348 DLFSRARKFAPSIIFIDELDAVGGK-RG-------RGFNEERDQTLNQLLTEMDGFESEI 503
Query: 518 KVIFVATANRAQQIPPPL 535
+V+ +A NR + + P L
Sbjct: 504 RVVVIAATNRPEALDPAL 557
>TC86562 similar to GP|21553404|gb|AAM62497.1 26S proteasome regulatory
particle chain RPT6-like protein {Arabidopsis thaliana},
partial (77%)
Length = 1564
Score = 35.8 bits (81), Expect = 0.074
Identities = 46/196 (23%), Positives = 82/196 (41%), Gaps = 16/196 (8%)
Frame = +1
Query: 379 GLAKVKQRIIEYLAVRKLKPD--ARGPVL------CFVGPPGVGKTSLASSIAAALDRKF 430
GL +KQ + E + + +PD + G +L GPPG GKT LA +IA F
Sbjct: 418 GLETIKQTLFELVILPLQRPDLFSHGKLLGPQKGVLLYGPPGTGKTMLAKAIAKESGAVF 597
Query: 431 VRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVA--VCNPVMLLDEIDKTGSDVRGD 488
+ + + + + + G +L+ + +A + ++ +DE+D R
Sbjct: 598 INVRISNLMSK---------WFGDAQ-KLVAAVFSLAHKLQPSIIFIDEVDSFLGQRRSS 747
Query: 489 PASALLEVLDPEQNKTFNDHYLNVPFDLS-KVIFVATANRAQQIPPPLLDRMEVIELPGY 547
A+L + F + D S +V+ +A NR ++ +L R+ GY
Sbjct: 748 DHEAVLNM-----KTEFMALWDGFATDQSARVMVLAATNRPSELDEAILRRLPQAFEIGY 912
Query: 548 TPEEK-----LKIAMK 558
P+ K LK+ +K
Sbjct: 913 -PDRKERADILKVILK 957
>TC87676 PIR|G96577|G96577 26S proteasome ATPase subunit [imported] -
Arabidopsis thaliana, partial (56%)
Length = 963
Score = 35.8 bits (81), Expect = 0.074
Identities = 19/39 (48%), Positives = 24/39 (60%)
Frame = +2
Query: 395 KLKPDARGPVLCFVGPPGVGKTSLASSIAAALDRKFVRI 433
KL D VLC+ GPPG GKT LA ++A D F+R+
Sbjct: 35 KLGIDPPKGVLCY-GPPGTGKTLLARAVANRTDACFIRV 148
>AW684884 similar to GP|6069665|dbj EST D15969(C1695) corresponds to a region
of the predicted gene.~similar to valosin-containing,
partial (18%)
Length = 561
Score = 35.8 bits (81), Expect = 0.074
Identities = 38/127 (29%), Positives = 50/127 (38%)
Frame = +3
Query: 409 GPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAV 468
GPPG GKT LA +IA F RIS V + + G YI L D KR A
Sbjct: 147 GPPGCGKTRLAHAIANETGLPFHRISATEV--VSGVSGASEEYI----RELFDKAKRTA- 305
Query: 469 CNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRA 528
++ +DEID S + + +E Q T D V+ + NR
Sbjct: 306 -PSIVFIDEIDAIAS--KREDLQREMEKRIVTQLMTSMDEPETSDESRGYVLVIGATNRP 476
Query: 529 QQIPPPL 535
+ P L
Sbjct: 477 DSLDPAL 497
>TC78797 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22 {Arabidopsis
thaliana}, partial (74%)
Length = 2270
Score = 35.8 bits (81), Expect = 0.074
Identities = 26/86 (30%), Positives = 41/86 (47%), Gaps = 15/86 (17%)
Frame = +1
Query: 375 SDHYGLAKVKQRIIEYLAVRKLKPD---------ARGPVLCFVGPPGVGKTSLASSIAAA 425
SD L + K+ + E + + +PD RG +L GPPG GKT LA +IA
Sbjct: 1714 SDIGALEETKESLQELVMLPLRRPDLFTGGLLKPCRGILL--FGPPGTGKTMLAKAIAKE 1887
Query: 426 LDRKFVRISLGGV------KDEADIR 445
F+ +S+ + +DE ++R
Sbjct: 1888 AGASFINVSMSTITSKWFGEDEKNVR 1965
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,537,081
Number of Sequences: 36976
Number of extensions: 251212
Number of successful extensions: 1362
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 1308
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1353
length of query: 888
length of database: 9,014,727
effective HSP length: 105
effective length of query: 783
effective length of database: 5,132,247
effective search space: 4018549401
effective search space used: 4018549401
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0550.4


