

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0544.5
(471 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
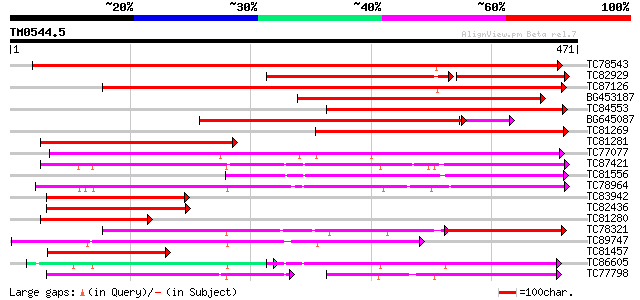
Score E
Sequences producing significant alignments: (bits) Value
TC78543 similar to PIR|T14545|T14545 probable sugar transporter ... 415 e-116
TC82929 similar to PIR|D96589|D96589 hypothetical protein T22H22... 219 e-102
TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {... 364 e-101
BG453187 weakly similar to GP|17381265|gb AT5g18840/F17K4_90 {Ar... 363 e-100
TC84553 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90 ... 243 2e-64
BG645087 similar to GP|17381265|gb AT5g18840/F17K4_90 {Arabidops... 228 3e-64
TC81269 similar to GP|23504385|emb|CAC00697. putative sugar tran... 240 1e-63
TC81281 similar to PIR|D96589|D96589 hypothetical protein T22H22... 233 9e-62
TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter p... 192 2e-49
TC87421 sugar transporter 176 2e-44
TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar tran... 163 2e-40
TC78964 weakly similar to PIR|T00450|T00450 probable monosacchar... 160 1e-39
TC83942 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90 ... 151 5e-37
TC82436 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90 ... 150 1e-36
TC81280 similar to PIR|D96589|D96589 hypothetical protein T22H22... 139 3e-33
TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 138 5e-33
TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported]... 135 3e-32
TC81457 similar to GP|20197560|gb|AAD13706.2 putative sugar tran... 121 5e-28
TC86605 similar to PIR|T01853|T01853 probable hexose transport p... 108 6e-24
TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 108 6e-24
>TC78543 similar to PIR|T14545|T14545 probable sugar transporter protein -
beet, partial (93%)
Length = 1653
Score = 415 bits (1067), Expect = e-116
Identities = 208/446 (46%), Positives = 304/446 (67%), Gaps = 6/446 (1%)
Frame = +1
Query: 20 QQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGA 79
Q R + ++ L+ LG FG GY+SPTQSAI+ DL L+V++FS+FGS+ +GA
Sbjct: 121 QAIRDSSISVIACVLIVALGPIQFGFTAGYTSPTQSAIITDLGLSVSEFSLFGSLSNVGA 300
Query: 80 MIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYV 139
M+GAI SG++A+Y GR+ ++ + + I+GWL+I+F+ + +LY+GRLL G G+G++SY
Sbjct: 301 MVGAIASGQIAEYMGRKGSLMIASIPNIIGWLMISFANDSSFLYMGRLLEGFGVGIISYT 480
Query: 140 VPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLP 199
VPVY+AEI+P+NLRG +++QL + G+ L YL+G FV WR LA++G IPC + L
Sbjct: 481 VPVYIAEISPQNLRGSLVSVNQLSVTLGIMLAYLLGLFVEWRFLAILGIIPCTLLIPGLF 660
Query: 200 FIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQW 259
FIP+SPRWLAK+G +E +++LQ LRG D+ E EI+ + R+T L Q
Sbjct: 661 FIPESPRWLAKMGMTEEFENSLQVLRGFETDISVEVNEIKTAVASANRRTTVRFSELKQR 840
Query: 260 KYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSES-IGTIAMVAVKIPVTVLGVL 318
+Y L IG+GL++LQQ GING++FY+++IF +AG S S + T + AV++ T L +
Sbjct: 841 RYWLPLMIGIGLLVLQQLSGINGVLFYSSTIFQNAGISSSDVATFGVGAVQVLATTLTLW 1020
Query: 319 LMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLH-----EWKGVSPILALVGVLVYVGS 373
L DKSGRR LL++S++ L + +++F L+ + G+ +L++ GV+V V +
Sbjct: 1021LADKSGRRLLLIVSSSAMTLSLLVVSISFYLKDYYISADSSLYGILSLLSVAGVVVMVIA 1200
Query: 374 YSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFS 433
+SLGMGA+PW+IMSEI PIN+KG AGS T NW SW+++ N L+ WSS GTF I++
Sbjct: 1201FSLGMGAMPWIIMSEILPINIKGLAGSFATLANWFFSWLITLTANLLLDWSSGGTFTIYT 1380
Query: 434 SICGFTVLFVAKLVPETKGRTLEEIQ 459
+C FTV FVA VPETKG+TLEEIQ
Sbjct: 1381VVCAFTVGFVAIWVPETKGKTLEEIQ 1458
>TC82929 similar to PIR|D96589|D96589 hypothetical protein T22H22.15
[imported] - Arabidopsis thaliana, partial (30%)
Length = 779
Score = 219 bits (558), Expect(2) = e-102
Identities = 111/155 (71%), Positives = 132/155 (84%)
Frame = +2
Query: 214 LKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMI 273
L+ S+S LQ LRGKN D+ +EA EIR++TE Q+QTEA+I GLFQ +Y+ SLT+GVGL+I
Sbjct: 5 LERSESTLQHLRGKNVDISEEATEIREFTEASQQQTEANIFGLFQLQYLKSLTVGVGLII 184
Query: 274 LQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSA 333
LQQFGG+N I FYA+SIFVSAGFS SIGTIAMV V+IP+T LGV+LMDKSGRRPLLL+SA
Sbjct: 185 LQQFGGVNAIAFYASSIFVSAGFSRSIGTIAMVVVQIPMTALGVILMDKSGRRPLLLISA 364
Query: 334 AGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVL 368
+GTCLGCFL +L+F LQ LH K SPILALVGVL
Sbjct: 365 SGTCLGCFLVSLSFYLQDLH--KEFSPILALVGVL 463
Score = 172 bits (436), Expect(2) = e-102
Identities = 80/94 (85%), Positives = 88/94 (93%)
Frame = +1
Query: 372 GSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFI 431
GS+SLGMG IPWVIMSEIFPINVKGSAGS VTF +WLCSWIVSYAFNFLMSW+S GTFFI
Sbjct: 463 GSFSLGMGGIPWVIMSEIFPINVKGSAGSFVTFVHWLCSWIVSYAFNFLMSWNSAGTFFI 642
Query: 432 FSSICGFTVLFVAKLVPETKGRTLEEIQASLSSH 465
FS+ICG T+LFVAKLVPETKGRTLEE+QASL+ +
Sbjct: 643 FSTICGLTILFVAKLVPETKGRTLEEVQASLNPY 744
>TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {Arabidopsis
thaliana}, partial (79%)
Length = 1524
Score = 364 bits (935), Expect = e-101
Identities = 177/390 (45%), Positives = 272/390 (69%), Gaps = 5/390 (1%)
Frame = +3
Query: 78 GAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLS 137
GAM+GAI SG++A+Y GR+ ++ + + I+GWL I+F+K + +L++GRLL G G+G++S
Sbjct: 6 GAMVGAIASGQIAEYVGRKGSLMIASIPNIIGWLAISFAKDSSFLFMGRLLEGFGVGIIS 185
Query: 138 YVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLS 197
YVVPVY+AEI P+N+RG +++QL + G+ L YL+G F NWR+LA++G +PC +
Sbjct: 186 YVVPVYIAEIAPENMRGSLGSVNQLSVTIGIMLAYLLGLFANWRVLAILGILPCTVLIPG 365
Query: 198 LPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLF 257
L FIP+SPRWLAK+G ++E +++LQ LRG + D+ E EI+ + ++ L
Sbjct: 366 LFFIPESPRWLAKMGMMEEFETSLQVLRGFDTDISVEVHEIKKAVASNGKRATIRFADLQ 545
Query: 258 QWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSES-IGTIAMVAVKIPVTVLG 316
+ +Y L++G+GL++LQQ GING++FY+ SIF +AG S S T+ + A+++ T +
Sbjct: 546 RKRYWFPLSVGIGLLVLQQLSGINGVLFYSTSIFANAGISSSNAATVGLGAIQVIATGVA 725
Query: 317 VLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHE----WKGVSPILALVGVLVYVG 372
L+DKSGRR LL++S++ + ++AF L+G+ E + + I+++VG++V V
Sbjct: 726 TWLVDKSGRRVLLIISSSLMTASLLVVSIAFYLEGVVEKDSQYFSILGIISVVGLVVMVI 905
Query: 373 SYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIF 432
+SLG+G IPW+IMSEI P+N+KG AGS T NWL +WI++ N L++WSS GTF I+
Sbjct: 906 GFSLGLGPIPWLIMSEILPVNIKGLAGSTATMANWLVAWIITMTANLLLTWSSGGTFLIY 1085
Query: 433 SSICGFTVLFVAKLVPETKGRTLEEIQASL 462
+ + FTV+F + VPETKGRTLEEIQ SL
Sbjct: 1086TVVAAFTVVFTSLWVPETKGRTLEEIQFSL 1175
>BG453187 weakly similar to GP|17381265|gb AT5g18840/F17K4_90 {Arabidopsis
thaliana}, partial (29%)
Length = 623
Score = 363 bits (931), Expect = e-100
Identities = 178/206 (86%), Positives = 193/206 (93%)
Frame = +2
Query: 240 DYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSES 299
DYTE LQ+QTEA+IIGLFQ +Y+ SLT+G+GLMILQQFGGINGIVFYANSIF+SAG SES
Sbjct: 5 DYTEALQQQTEANIIGLFQLQYLKSLTVGLGLMILQQFGGINGIVFYANSIFISAGLSES 184
Query: 300 IGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVS 359
IGTIAMVAVKIP+T LGV LMDKSGRRPLLLLSA GTCLGCFLAAL+F LQ +H+WK VS
Sbjct: 185 IGTIAMVAVKIPMTTLGVFLMDKSGRRPLLLLSAVGTCLGCFLAALSFFLQDIHKWKEVS 364
Query: 360 PILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNF 419
PILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVT NWLCSWI+SYAFNF
Sbjct: 365 PILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTLVNWLCSWIISYAFNF 544
Query: 420 LMSWSSTGTFFIFSSICGFTVLFVAK 445
LM+WSSTGTFF F++ICGFTVLFVAK
Sbjct: 545 LMTWSSTGTFFGFAAICGFTVLFVAK 622
>TC84553 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90
{Arabidopsis thaliana}, partial (41%)
Length = 827
Score = 243 bits (619), Expect = 2e-64
Identities = 118/201 (58%), Positives = 153/201 (75%), Gaps = 1/201 (0%)
Frame = +2
Query: 264 SLTIGVGLMILQQFGGINGIVFYANSIFVSAGF-SESIGTIAMVAVKIPVTVLGVLLMDK 322
S+ +GVGLM+ QQ GI GI FY + FV+A F S IGTIA +++P T+LG +LMDK
Sbjct: 5 SVVLGVGLMVCQQSVGIMGIGFYTSETFVAADFLSGKIGTIAYACMQVPFTILGAILMDK 184
Query: 323 SGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMGAIP 382
SGRRPL+ SA+GT LGCF+ +AF L+ + + PILA+ G+L+YV ++S+GMG +P
Sbjct: 185 SGRRPLITASASGTFLGCFMTGIAFFLKDQNLLLELVPILAVAGILIYVAAFSIGMGPVP 364
Query: 383 WVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLF 442
WVIMSEIFPI+VKG+AGSLV NWL +W+VSY FNF +SWSS GT F+++ T+LF
Sbjct: 365 WVIMSEIFPIHVKGTAGSLVVLINWLGAWVVSYTFNFFLSWSSPGTLFLYAGCSLLTILF 544
Query: 443 VAKLVPETKGRTLEEIQASLS 463
VAKLVPETKG+TLEEIQA L+
Sbjct: 545 VAKLVPETKGKTLEEIQACLN 607
>BG645087 similar to GP|17381265|gb AT5g18840/F17K4_90 {Arabidopsis
thaliana}, partial (46%)
Length = 805
Score = 228 bits (582), Expect(2) = 3e-64
Identities = 115/223 (51%), Positives = 153/223 (68%), Gaps = 1/223 (0%)
Frame = +2
Query: 158 TMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKES 217
T +QLMI G S+++LIG+ +NWR LAL G +PC L+ L FIP+SPRWLAKVG KE
Sbjct: 11 TTNQLMIVIGSSMSFLIGSIINWRQLALAGLVPCICLLVGLCFIPESPRWLAKVGREKEF 190
Query: 218 DSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQF 277
AL++LRGK+ D+ EA EI D ETLQ + + LFQ KY+ S+ IGVGLM QQ
Sbjct: 191 QLALRKLRGKDIDISDEANEILDNIETLQSLPKTKFLDLFQSKYVRSVIIGVGLMAFQQS 370
Query: 278 GGINGIVFYANSIFVSAGFSES-IGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGT 336
GINGI FY FV+AG S + GTIA +++P T+LG +LMDKSGR+PL+ +SA+GT
Sbjct: 371 VGINGIGFYTAETFVAAGLSSAKAGTIAYACIQVPFTLLGAILMDKSGRKPLITVSASGT 550
Query: 337 CLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMG 379
LGCF+ +AF + W P LA+ GVL+Y+ ++S+G+G
Sbjct: 551 FLGCFITGVAFFFKNQSLWLEWVPTLAVAGVLIYIAAFSIGLG 679
Score = 35.0 bits (79), Expect(2) = 3e-64
Identities = 18/47 (38%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Frame = +3
Query: 374 YSLGMGAIPWVIMSEIFPINVKGSAGSLVTFF-NWLCSWIVSYAFNF 419
+ LG G++PWV+MS + + + F+ L +WIVSY FNF
Sbjct: 663 FQLGWGSVPWVMMSGGVSYKCEKNCWKVW*FW*RGLGAWIVSYTFNF 803
>TC81269 similar to GP|23504385|emb|CAC00697. putative sugar transporter
{Lycopersicon esculentum}, partial (42%)
Length = 966
Score = 240 bits (612), Expect = 1e-63
Identities = 110/210 (52%), Positives = 155/210 (73%)
Frame = +2
Query: 255 GLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTV 314
G +Y+ SLTIGVGLM+ QQ GGING+ FY +SIF AGF + G+I ++I +T
Sbjct: 11 GFVSRRYLRSLTIGVGLMVCQQLGGINGVCFYTSSIFDLAGFPSATGSIIYAILQIVITG 190
Query: 315 LGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSY 374
+G L+D++GR+PLLL+S +G GC A+AF L+ G P LA+ G+LVY+GS+
Sbjct: 191 VGAALIDRAGRKPLLLVSGSGLVAGCIFTAVAFYLKVHDVAVGAVPALAVTGILVYIGSF 370
Query: 375 SLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSS 434
S+GMGAIPWV+MSEIFP+N+KG AGS+ T NW +W+ SY FNFLMSWSS GTF ++++
Sbjct: 371 SIGMGAIPWVVMSEIFPVNIKGQAGSIATLVNWFGAWLCSYTFNFLMSWSSYGTFVLYAA 550
Query: 435 ICGFTVLFVAKLVPETKGRTLEEIQASLSS 464
I +LF+A +VPETKG++LE++QA++++
Sbjct: 551 INALAILFIAVVVPETKGKSLEQLQAAINA 640
>TC81281 similar to PIR|D96589|D96589 hypothetical protein T22H22.15
[imported] - Arabidopsis thaliana, partial (24%)
Length = 585
Score = 233 bits (595), Expect = 9e-62
Identities = 115/165 (69%), Positives = 136/165 (81%), Gaps = 1/165 (0%)
Frame = +2
Query: 26 TATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIV 85
T+ L L+TL AV GSY FG+A+GYSSP QS I DLNL VA++S+FGSILTIGAM+GAIV
Sbjct: 83 TSILILTTLAAVSGSYVFGSAVGYSSPAQSGITDDLNLGVAEYSLFGSILTIGAMVGAIV 262
Query: 86 SGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVA 145
SG +ADYAGRR AMGFS+LFCILGWL I SKVAWWLYVGRLL+GCG+G+LSYVVP+Y+A
Sbjct: 263 SGSLADYAGRRAAMGFSELFCILGWLAIAVSKVAWWLYVGRLLLGCGMGILSYVVPIYIA 442
Query: 146 -EITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAI 189
T + RG +LMIC G+SLTYLIGAF+NWR+LA+IG I
Sbjct: 443 *NNTQGSSRGIHLQFIKLMICFGVSLTYLIGAFLNWRLLAIIGTI 577
>TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter protein
{Arabidopsis thaliana}, partial (85%)
Length = 1752
Score = 192 bits (489), Expect = 2e-49
Identities = 132/456 (28%), Positives = 223/456 (47%), Gaps = 28/456 (6%)
Frame = +1
Query: 34 LVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVADYA 93
++A + S G IG S I DL ++ + + I+ I ++IG+ ++G+ +D+
Sbjct: 124 MLASMTSILLGYDIGVMSGAAIYIKRDLKVSDGKIEVLLGIINIYSLIGSCLAGRTSDWI 303
Query: 94 GRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPKNLR 153
GRR + F+ +G L++ FS +L GR + G GIG + PVY AE++P + R
Sbjct: 304 GRRYTIVFAGAIFFVGALLMGFSPNYNFLMFGRFVAGVGIGYALMIAPVYTAEVSPASSR 483
Query: 154 GGFTTMHQLMICCGMSLTYL-------IGAFVNWRILALIGAIPCFAQLLSLPFIPDSPR 206
G T+ ++ I G+ L Y+ + + WR++ +GAIP + + +P+SPR
Sbjct: 484 GFLTSFPEVFINSGILLGYISNYAFSKLSLKLGWRMMLGVGAIPSVILAVGVLAMPESPR 663
Query: 207 WLAKVGHLKESDSALQRLRGKNADVHQEAAEIR-----------DYTETLQRQTEASI-- 253
WL G L ++ L + + AEI+ D E + T +
Sbjct: 664 WLVMRGRLGDAIKVLNKTSDSKEEAQLRLAEIKQAAGIPEDCNDDVVEVKVKNTGEGVWK 843
Query: 254 -IGLFQWKYMTSLTI-GVGLMILQQFGGINGIVFYANSIFVSAGFSES----IGTIAMVA 307
+ L+ + + I +G+ QQ G++ +V Y+ +IF AG + I TIA+
Sbjct: 844 ELFLYPTPAVRHIVIAALGIHFFQQASGVDAVVLYSPTIFKKAGINGDTHLLIATIAVGF 1023
Query: 308 VKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPI-LALVG 366
VK ++ ++D+ GRRPLLL S G + A++ + K I L++
Sbjct: 1024VKTLFILVATFMLDRYGRRPLLLTSVGGMVVSLLTLAVSLTIIDHSNTKLNWAIGLSIAT 1203
Query: 367 VLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSST 426
VL YV ++S+G G I WV SEIFP+ ++ + N + S ++S F L +
Sbjct: 1204VLSYVATFSIGAGPITWVYSSEIFPLRLRAQGAACGVVVNRVTSGVISMTFLSLSKGITI 1383
Query: 427 -GTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQAS 461
G FF+F I +F ++PET+G+TLEE++AS
Sbjct: 1384GGAFFLFGGIATIGWIFFYIMLPETQGKTLEEMEAS 1491
>TC87421 sugar transporter
Length = 1753
Score = 176 bits (446), Expect = 2e-44
Identities = 140/480 (29%), Positives = 228/480 (47%), Gaps = 40/480 (8%)
Frame = +2
Query: 26 TATLTLSTLVAVLGSYAFGAAIGYSSPTQS----------AIMPDLNLTVAQ-------- 67
T +T++ +VA +G FG IG S S A+ N +
Sbjct: 86 TPFVTITCIVAAMGGLIFGYDIGISGGVTSMDPFLKKFFPAVYRKKNKDKSTNQYCQYDS 265
Query: 68 --FSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVG 125
++F S L + A++ ++V+ + GR+++M F L ++G L+ F+ W L VG
Sbjct: 266 QTLTMFTSSLYLAALLSSLVASTITRRFGRKLSMLFGGLLFLVGALINGFANHVWMLIVG 445
Query: 126 RLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFV------- 178
R+L+G GIG + VP+Y++E+ P RG QL I G+ + ++ F
Sbjct: 446 RILLGFGIGFANQPVPLYLSEMAPYKYRGALNIGFQLSITIGILVANVLNYFFAKIKGGW 625
Query: 179 NWRILALIGA-IPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAE 237
WR L+L GA +P + +PD+P + + G + + L+R+RG DV +E +
Sbjct: 626 GWR-LSLGGAMVPALIITIGSLVLPDTPNSMIERGDRDGAKAQLKRIRGIE-DVDEEFND 799
Query: 238 IRDYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFS 297
+ +E Q E L Q KY LT+ V + QQF GIN I+FYA +F S GF
Sbjct: 800 LVAASEA-SMQVENPWRNLLQRKYRPQLTMAVLIPFFQQFTGINVIMFYAPVLFNSIGFK 976
Query: 298 ESIGTIAMV---AVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALA-----FLL 349
+ ++ V V + T + + +DK GRR L L A L C +A A F
Sbjct: 977 DDASLMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGGA-QMLICQVAVAAAIGAKFGT 1153
Query: 350 QG----LHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFF 405
G L EW I+ ++ + +YV ++ G + W++ SEIFP+ ++ +A S+
Sbjct: 1154SGNPGNLPEWYA---IVVVLFICIYVAGFAWSWGPLGWLVPSEIFPLEIRSAAQSVNVSV 1324
Query: 406 NWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSH 465
N L +++V+ F ++ G F F+ ++V L+PETKG +EE+ SH
Sbjct: 1325NMLFTFLVAQVFLIMLCHMKFGLFLFFAFFVLVMSIYVFFLLPETKGIPIEEMDRVWKSH 1504
>TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar transporter
{Prunus armeniaca}, partial (59%)
Length = 1074
Score = 163 bits (412), Expect = 2e-40
Identities = 95/287 (33%), Positives = 160/287 (55%), Gaps = 2/287 (0%)
Frame = +1
Query: 180 WRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIR 239
WR + I +P L + P+SPRWL + G + E++ A++ L GK V ++R
Sbjct: 19 WRTMFGIAVVPSILLALGMAISPESPRWLFQQGKIAEAEKAIKTLYGKER-VATVMYDLR 195
Query: 240 DYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSES 299
++ + EA LF +Y +++G L + QQ GIN +V+Y+ S+F SAG +
Sbjct: 196 TASQG-SSEPEAGWFDLFSSRYWKVVSVGAALFLFQQLAGINAVVYYSTSVFRSAGIASD 372
Query: 300 IGTIAMV-AVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGV 358
+ A+V A + T + LMDK GR+ LL+ S +G L +L+F + L + G
Sbjct: 373 VAASALVGASNVIGTAIASSLMDKQGRKSLLITSFSGMAASMLLLSLSFTWKVLAPYSGT 552
Query: 359 SPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVS-YAF 417
LA++G ++YV S+SLG G +P +++ EIF ++ A SL +W+ ++++ Y
Sbjct: 553 ---LAVLGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAVSLSLGTHWISNFVIGLYFL 723
Query: 418 NFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSS 464
+ + + + + FS++C VL++A V ETKGR+LEEI+ +LSS
Sbjct: 724 SVVNKFGISSVYLGFSAVCVLAVLYIAGNVVETKGRSLEEIERALSS 864
>TC78964 weakly similar to PIR|T00450|T00450 probable monosaccharide
transport protein T14N5.7 - Arabidopsis thaliana,
partial (98%)
Length = 1771
Score = 160 bits (405), Expect = 1e-39
Identities = 129/480 (26%), Positives = 218/480 (44%), Gaps = 36/480 (7%)
Frame = +1
Query: 22 QRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSA------IMPDL------NLTVAQF- 68
+ FTA + +V LG FG +G S S PD+ +L +
Sbjct: 85 EHKFTAYFAFTCVVGALGGSLFGYDLGVSGGVTSMDDFLEKFFPDVYRKKHAHLKETDYC 264
Query: 69 -------SIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWW 121
++F S L A++ + + GR+ + L ++G ++ ++
Sbjct: 265 KYDNQVLTLFTSSLYFSALVMTFFASYLTRNKGRKATIIVGALSFLIGAILNAAAQNIPT 444
Query: 122 LYVGRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVN-- 179
L +GR+ +G GIG + VP+Y++E+ P + RG + Q C G+ + L+ F +
Sbjct: 445 LIIGRVFLGGGIGFGNQAVPLYLSEMAPASSRGAVNQLFQFTTCAGILIANLVNYFTDKI 624
Query: 180 ----WRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRG-KNADVHQE 234
WRI + IP LL F ++P L + G L E+ L+++RG KN D E
Sbjct: 625 HPHGWRISLGLAGIPAVLMLLGGIFCAETPNSLVEQGRLDEARKVLEKVRGTKNVDAEFE 804
Query: 235 AAEIRDYTETLQRQTEASIIGLFQWKYMTSLTIG-VGLMILQQFGGINGIVFYANSIFVS 293
+++D +E L + ++ L + KY L IG +G QQ G N I+FYA IF S
Sbjct: 805 --DLKDASE-LAQAVKSPFKVLLKRKYRPQLIIGALGTPAFQQLTGNNSILFYAPVIFQS 975
Query: 294 AGFSESIGTIAMVAVK---IPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLL- 349
+GF + + + TV+ + L+DK G R L AG + C + A +L
Sbjct: 976 SGFGSNAALFSSFITNGALLVATVISMFLVDKFGTRKFFL--EAGFEMICCMIITAVVLA 1149
Query: 350 ----QGLHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFF 405
G KG+S L ++ + +V +Y G + W++ SE+FP+ ++ +A S+
Sbjct: 1150VEFGHGKELSKGISAFLVIM-IFWFVLAYGRSWGPLGWLVPSELFPLEIRSAAQSIAVCV 1326
Query: 406 NWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSH 465
N + + +V+ F + G F +F + +FV L+PETK +EEI +H
Sbjct: 1327NMIFTALVAQLFLLSLCHLKYGIFLLFGGLIVVMSVFVFFLLPETKQVPIEEIYLLFENH 1506
>TC83942 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90
{Arabidopsis thaliana}, partial (26%)
Length = 531
Score = 151 bits (382), Expect = 5e-37
Identities = 72/119 (60%), Positives = 94/119 (78%)
Frame = +2
Query: 31 LSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVA 90
LST VAV GS++FG +GYS+PTQ+AI DLNL++A+FS+FGS++TIGAM+GAI SG+V
Sbjct: 173 LSTFVAVCGSFSFGTCVGYSAPTQAAIRADLNLSLAEFSMFGSLVTIGAMLGAITSGRVT 352
Query: 91 DYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITP 149
D GR+ AM S FCI+GWL + FSK ++ L +GR G GIG++SYVVPVY+AEI P
Sbjct: 353 DIIGRKGAMRISTGFCIIGWLAVFFSKSSYTLDLGRFFTGYGIGVISYVVPVYIAEIAP 529
>TC82436 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90
{Arabidopsis thaliana}, partial (25%)
Length = 656
Score = 150 bits (378), Expect = 1e-36
Identities = 71/120 (59%), Positives = 93/120 (77%)
Frame = +2
Query: 31 LSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVA 90
LST VAV GS++FG +GYSSPTQ+AI DLNL++++FS+FGS++TIGAM+GAI SG++
Sbjct: 215 LSTFVAVCGSFSFGTCVGYSSPTQAAIRADLNLSISEFSMFGSLVTIGAMLGAITSGRIT 394
Query: 91 DYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPK 150
D+ GR+ AM S FCI GWL + FSK + L +GR G GIG++SYVVPVY+AEI K
Sbjct: 395 DFIGRKGAMRLSTGFCITGWLAVFFSKDPYSLDIGRFFTGYGIGVISYVVPVYIAEIPQK 574
>TC81280 similar to PIR|D96589|D96589 hypothetical protein T22H22.15
[imported] - Arabidopsis thaliana, partial (18%)
Length = 650
Score = 139 bits (349), Expect = 3e-33
Identities = 68/93 (73%), Positives = 78/93 (83%)
Frame = +3
Query: 26 TATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIV 85
T+ L L+TL AV GSY FG+A+GYSSP QS I DLNL VA++S+FGSILTIGAM+GAIV
Sbjct: 129 TSILILTTLAAVSGSYVFGSAVGYSSPAQSGITDDLNLGVAEYSLFGSILTIGAMVGAIV 308
Query: 86 SGKVADYAGRRVAMGFSQLFCILGWLVITFSKV 118
SG +ADYAGRR AMGFS+LFCILGWL I SKV
Sbjct: 309 SGSLADYAGRRAAMGFSELFCILGWLAIAVSKV 407
Score = 40.0 bits (92), Expect = 0.002
Identities = 21/37 (56%), Positives = 24/37 (64%)
Frame = +3
Query: 103 QLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYV 139
Q C L LV K AW LYVGRLL+ G+G+LSYV
Sbjct: 531 QSSCYLMGLVAFCLKXAWXLYVGRLLLXXGMGILSYV 641
>TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 2093
Score = 138 bits (347), Expect = 5e-33
Identities = 91/301 (30%), Positives = 158/301 (52%), Gaps = 13/301 (4%)
Frame = +2
Query: 78 GAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLS 137
GA+IGA + G + D GR+V++ + +LG +++ + L VGR+ VG G+G+ S
Sbjct: 377 GAIIGAAIGGWINDRFGRKVSIIVADTLFLLGSIILAAAPNPATLIVGRVFVGLGVGMAS 556
Query: 138 YVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIG-AFV----NWRILALIGAIPCF 192
P+Y++E +P +RG +++ +I G L+YLI AF WR + + A P
Sbjct: 557 MASPLYISEASPTRVRGALVSLNSFLITGGQFLSYLINLAFTKAPGTWRWMLGVAAAPAV 736
Query: 193 AQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEAS 252
Q++ + +P+SPRWL + G +E+ L+++ + D E +++ E ++TE
Sbjct: 737 IQIVLMLSLPESPRWLYRKGKEEEAKVILKKIY-EVEDYDNEIQALKESVEMELKETEK- 910
Query: 253 IIGLFQWKYMTS----LTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAV 308
I + Q TS L GVGL QQF GIN +++Y+ SI AGF+ + + +
Sbjct: 911 -ISIMQLVKTTSVRRGLYAGVGLAFFQQFTGINTVMYYSPSIVQLAGFASKRTALLLSLI 1087
Query: 309 KIPV----TVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILAL 364
+ ++L + +DK+GR+ L L+S G L L + F +H +P+++L
Sbjct: 1088TSGLNAFGSILSIYFIDKTGRKKLALISLTGVVLSLTLLTVTFRESEIH-----APMVSL 1252
Query: 365 V 365
V
Sbjct: 1253V 1255
Score = 74.3 bits (181), Expect = 9e-14
Identities = 35/102 (34%), Positives = 62/102 (60%), Gaps = 1/102 (0%)
Frame = +2
Query: 362 LALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAF-NFL 420
+A++ + +Y+ +S GMG +PWV+ SEI+P+ +G G + + W+ + +VS +F +
Sbjct: 1490 IAILALALYIIFFSPGMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQSFLSLT 1669
Query: 421 MSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASL 462
++ TF IF+ I + FV VPETKG +EE+++ L
Sbjct: 1670 VAIGPAWTFMIFAIIAIVAIFFVIIFVPETKGVPMEEVESML 1795
>TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1372
Score = 135 bits (340), Expect = 3e-32
Identities = 99/354 (27%), Positives = 164/354 (45%), Gaps = 11/354 (3%)
Frame = +3
Query: 2 MVKTSEISIPLVFNDEEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDL 61
M + E ++ L+ N +++ P LVA + S+ FG +G + +I DL
Sbjct: 291 MEEDVEENLDLLDNFIDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDL 470
Query: 62 NL---TVAQFSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKV 118
T+A+ + SI GA+ G ++SG +AD GRR A L I+G + +
Sbjct: 471 GFNGNTLAE-GLVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMSAATNN 647
Query: 119 AWWLYVGRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFV 178
+ + VGRL VG G+GL V +YV E++P +RG + + Q+ C G+ + IG V
Sbjct: 648 LFGMLVGRLFVGTGLGLGPPVAALYVTEVSPAFVRGTYGALIQIATCFGILGSLFIGIPV 827
Query: 179 N-----WRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQ 233
WR+ + IP L++ F +SP WL K G E+++ +RL G +
Sbjct: 828 KEISGWWRVCFWVSTIPAAILALAMDFCAESPHWLYKQGRTAEAEAEFERLLGVS----- 992
Query: 234 EAAEIRDYTETLQRQTEASII---GLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSI 290
EA + R + + L + + IG L LQQ GIN + ++++++
Sbjct: 993 EAKFAMSQLSKVDRGEDTDTVKFSELLHGHHSKVVFIGSTLFALQQLSGINAVFYFSSTV 1172
Query: 291 FVSAGFSESIGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAA 344
F SAG + + + +++ LMDK GR+ LL S G + + A
Sbjct: 1173FKSAGVPSDFANVCIGVANLTGSIISTGLMDKLGRKVLLFWSFFGMAISMIIQA 1334
>TC81457 similar to GP|20197560|gb|AAD13706.2 putative sugar transporter
{Arabidopsis thaliana}, partial (23%)
Length = 507
Score = 121 bits (304), Expect = 5e-28
Identities = 61/102 (59%), Positives = 75/102 (72%)
Frame = +1
Query: 32 STLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVAD 91
+T +AV GSY FGA GYSSPTQ AI DL+L++A++S+FGSILT GAMIGAI SG +AD
Sbjct: 202 TTFIAVCGSYEFGACAGYSSPTQEAIRKDLSLSLAEYSLFGSILTFGAMIGAITSGPIAD 381
Query: 92 YAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGI 133
+ GR+ AM S FCI GWLVI FS+ L +GRL G G+
Sbjct: 382 FVGRKGAMRVSSAFCIAGWLVIYFSEGPXPLDIGRLATGYGM 507
>TC86605 similar to PIR|T01853|T01853 probable hexose transport protein
F9D12.17 - Arabidopsis thaliana, partial (91%)
Length = 3111
Score = 108 bits (269), Expect = 6e-24
Identities = 72/251 (28%), Positives = 128/251 (50%), Gaps = 6/251 (2%)
Frame = +3
Query: 214 LKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMI 273
L + + L+++RG + ++ E E+ + + + ++ + L + L I + LMI
Sbjct: 1992 LDKGKAVLRKIRGTD-NIEPEFLELVEASR-VAKEVKHPFRNLLKRNNRPQLVISIALMI 2165
Query: 274 LQQFGGINGIVFYANSIFVSAGFSESIGTIAMV---AVKIPVTVLGVLLMDKSGRRPLLL 330
QQF GIN I+FYA +F + GF + V A+ + T++ + +DK GRR LLL
Sbjct: 2166 FQQFTGINAIMFYAPVLFNTLGFKNDAALYSAVITGAINVISTIVSIYSVDKLGRRKLLL 2345
Query: 331 LSAAGTCLGCFLAALAFLLQGLHEWKGVSP---ILALVGVLVYVGSYSLGMGAIPWVIMS 387
+ L + A+ ++ + +S L +V V ++V +++ G + W+I S
Sbjct: 2346 EAGVQMLLSQMVIAIVLGIKVKDHSEELSKGYAALVVVMVCIFVSAFAWSWGPLAWLIPS 2525
Query: 388 EIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAKLV 447
EIFP+ + + S+ N+L + +++ AF ++ + G FF FS F FV LV
Sbjct: 2526 EIFPLETRSAGQSVTVCVNFLFTAVIAQAFLSMLCYFKFGIFFFFSGWILFMSTFVFFLV 2705
Query: 448 PETKGRTLEEI 458
PETK +EE+
Sbjct: 2706 PETKNVPIEEM 2738
Score = 78.2 bits (191), Expect = 6e-15
Identities = 61/237 (25%), Positives = 96/237 (39%), Gaps = 28/237 (11%)
Frame = +2
Query: 15 NDEEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSS------PTQSAIMPDLNLTVAQ- 67
ND E + + T + +S ++A G FG +G S P P + +
Sbjct: 1316 NDREFEAK--ITPIIIISCIMAATGGLMFGYDVGVSGGVASMPPFLKKFFPTVLRQTTES 1489
Query: 68 --------------FSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVI 113
+F S L + + + GRR+ M + F I G +
Sbjct: 1490 DGSESNYCKYDNQGLQLFTSSLYLAGLTVTFFASYTTRVLGRRLTMLIAGFFFIAGVSLN 1669
Query: 114 TFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYL 173
++ L VGR+L+GCGIG + VPV+++EI P +RG + QL I G+ L
Sbjct: 1670 ASAQNLLMLIVGRVLLGCGIGFANQAVPVFLSEIAPSRIRGALNILFQLDITLGILYANL 1849
Query: 174 IGAFVN-------WRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQR 223
+ N WRI +G IP L + D+P L + GHL + S ++
Sbjct: 1850 VNYATNKIKGHWGWRISLGLGGIPALLLTLGAYLVVDTPNSLIERGHLGQRKSCSEK 2020
>TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (88%)
Length = 2467
Score = 108 bits (269), Expect = 6e-24
Identities = 76/214 (35%), Positives = 114/214 (52%), Gaps = 8/214 (3%)
Frame = +3
Query: 31 LSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVA 90
L + A +G++ G + + I DL L + ++ IGA + SG ++
Sbjct: 126 LVAIAASIGNFLQGWDNATIAGSILYIKKDLALQTTMEGLVVAMSLIGATVITTCSGPIS 305
Query: 91 DYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPK 150
D+ GRR M S + LG LV+ +S + L + RLL G GIGL +VPVY++E P
Sbjct: 306 DWLGRRPMMIISSVLYFLGSLVMLWSPNVYVLCLARLLDGFGIGLAVTLVPVYISETAPS 485
Query: 151 NLRGGFTTMHQLMICCGMSLTYLIGAFV-------NWRILALIGAIP-CFAQLLSLPFIP 202
++RG T+ Q GM L+Y + FV +WRI+ + +IP F LL++ F+P
Sbjct: 486 DIRGSLNTLPQFSGSGGMFLSYCM-VFVMSLSPSPSWRIMLGVLSIPSLFYFLLTVFFLP 662
Query: 203 DSPRWLAKVGHLKESDSALQRLRGKNADVHQEAA 236
+SPRWL G + E+ LQRLRG++ DV E A
Sbjct: 663 ESPRWLVSKGKMLEAKKVLQRLRGQD-DVSGEMA 761
Score = 98.2 bits (243), Expect = 6e-21
Identities = 65/211 (30%), Positives = 107/211 (49%), Gaps = 16/211 (7%)
Frame = +3
Query: 264 SLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAM-------------VAVKI 310
+L +G+G+ +LQQF GING+++Y I AG + + + + + +
Sbjct: 1632 ALIVGIGIQLLQQFSGINGVLYYTPQILEEAGVAVLLADLGLSSTSSSFLISAVTTLLML 1811
Query: 311 PVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQG--LHEWKGVSPILALVGVL 368
P L + LMD +GRR LLL++ + + +L L+ G + V ++ V V+
Sbjct: 1812 PSIGLAMRLMDVTGRRQLLLVT-----IPVLIVSLVILVLGSVIDFGSVVHAAISTVCVV 1976
Query: 369 VYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFN-FLMSWSSTG 427
VY + +G G IP ++ SEIFP V+G ++ W+ IV+Y+ L S G
Sbjct: 1977 VYFCFFVMGYGPIPNILCSEIFPTRVRGLCIAICALVFWIGDIIVTYSLPVMLSSLGLAG 2156
Query: 428 TFFIFSSICGFTVLFVAKLVPETKGRTLEEI 458
F +++ +C + +FV VPETKG LE I
Sbjct: 2157 VFGVYAIVCCISWVFVYLKVPETKGMPLEVI 2249
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,156,985
Number of Sequences: 36976
Number of extensions: 226748
Number of successful extensions: 1936
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 1832
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1867
length of query: 471
length of database: 9,014,727
effective HSP length: 100
effective length of query: 371
effective length of database: 5,317,127
effective search space: 1972654117
effective search space used: 1972654117
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0544.5


