

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0544.10
(306 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
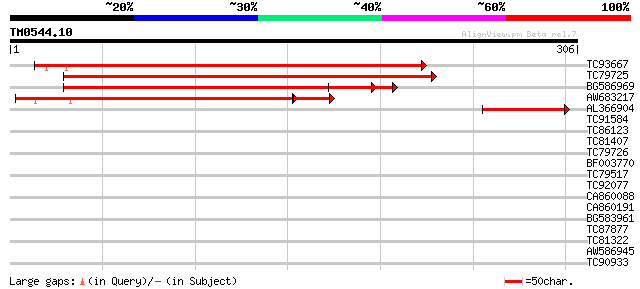
Score E
Sequences producing significant alignments: (bits) Value
TC93667 similar to GP|6729027|gb|AAF27023.1| unknown protein {Ar... 353 5e-98
TC79725 similar to GP|6729027|gb|AAF27023.1| unknown protein {Ar... 335 2e-92
BG586969 similar to GP|6729027|gb| unknown protein {Arabidopsis ... 231 3e-61
AW683217 similar to GP|6069661|dbj Similar to Sequence of BAC F7... 142 4e-39
AL366904 weakly similar to GP|6729027|gb|A unknown protein {Arab... 67 9e-12
TC91584 similar to GP|21592519|gb|AAM64469.1 unknown {Arabidopsi... 36 0.021
TC86123 homologue to PIR|S71770|S71770 calcium-dependent protein... 34 0.080
TC81407 homologue to GP|18650662|gb|AAL75809.1 ethylene response... 34 0.080
TC79726 similar to GP|17064808|gb|AAL32558.1 Unknown protein {Ar... 33 0.14
BF003770 similar to PIR|F96809|F96 protein F28K19.26 [imported] ... 32 0.23
TC79517 similar to PIR|T08874|T08874 calcium-dependent protein k... 32 0.23
TC92077 homologue to GP|23497650|gb|AAN37190.1 hypothetical prot... 32 0.23
CA860088 similar to GP|8248752|emb hypothetical protein MAL4P2.... 32 0.40
CA860191 homologue to GP|19697342|gb hypothetical protein {Dicty... 31 0.52
BG583961 similar to GP|14599408|em probable major surface glycop... 30 0.89
TC87877 similar to GP|21618275|gb|AAM67325.1 unknown {Arabidopsi... 30 1.5
TC81322 weakly similar to GP|17104669|gb|AAL34223.1 unknown prot... 29 2.0
AW586945 homologue to GP|23496297|gb hypothetical protein {Plasm... 29 2.0
TC90933 similar to SP|P27484|GRP2_NICSY Glycine-rich protein 2. ... 28 3.4
>TC93667 similar to GP|6729027|gb|AAF27023.1| unknown protein {Arabidopsis
thaliana}, partial (58%)
Length = 662
Score = 353 bits (906), Expect = 5e-98
Identities = 175/219 (79%), Positives = 195/219 (88%), Gaps = 7/219 (3%)
Frame = +2
Query: 14 NHHQQ--QIPNQNNHKPS----VLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGI 67
N H+Q QI K S VLPKRII+VRHGESQGNLDP AYT TPDHKI LTPQGI
Sbjct: 5 NRHKQRCQIAMHREEKSSSSSGVLPKRIIVVRHGESQGNLDPGAYTVTPDHKIPLTPQGI 184
Query: 68 TQARIAGARIRHVISSTSST-DWRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRI 126
+QA G+RIRHVISS+SS+ DWRVYFYVSPY RTRSTLRE+ +SFSK R++GVREECRI
Sbjct: 185 SQALSTGSRIRHVISSSSSSPDWRVYFYVSPYTRTRSTLRELAKSFSKKRVIGVREECRI 364
Query: 127 REQDFGNFQVQERINALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRL 186
REQDFGNFQVQER++A+KETRQRFGRFF+RFP+GESAADVFDRVSSFLES+WRDIDMNRL
Sbjct: 365 REQDFGNFQVQERMDAIKETRQRFGRFFYRFPEGESAADVFDRVSSFLESMWRDIDMNRL 544
Query: 187 NHDPSNDLNLIIVSHGLASRVFLMKWFKWTVEQFDLLNN 225
NH+PSNDLNLIIVSHGLASRVFLM+WF+WTVEQF+LLNN
Sbjct: 545 NHNPSNDLNLIIVSHGLASRVFLMRWFRWTVEQFELLNN 661
>TC79725 similar to GP|6729027|gb|AAF27023.1| unknown protein {Arabidopsis
thaliana}, partial (63%)
Length = 760
Score = 335 bits (858), Expect = 2e-92
Identities = 158/202 (78%), Positives = 179/202 (88%), Gaps = 1/202 (0%)
Frame = +3
Query: 30 VLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTS-STD 88
VLPKRIIL+RHGESQGNLD SAYTTTPDH I LTP GI QAR AGA + ++S S D
Sbjct: 120 VLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPSGIAQARHAGANLHRLVSGQGCSPD 299
Query: 89 WRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERINALKETRQ 148
WR+YFYVSPYARTRSTLRE+GRSFSK R++GVREE R+REQDFGNFQVQER+ +KETR+
Sbjct: 300 WRLYFYVSPYARTRSTLREVGRSFSKKRVIGVREESRVREQDFGNFQVQERMKIVKETRE 479
Query: 149 RFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVF 208
RFGRFF+RFP+GESAADVFDR+S F ESLWRDID+NRL+HDP+NDLNL+IVSHGL SR+F
Sbjct: 480 RFGRFFYRFPEGESAADVFDRISGFFESLWRDIDLNRLHHDPTNDLNLVIVSHGLTSRIF 659
Query: 209 LMKWFKWTVEQFDLLNNFGNGE 230
LMKWFKWTVEQF+ LNNFGN E
Sbjct: 660 LMKWFKWTVEQFEHLNNFGNCE 725
>BG586969 similar to GP|6729027|gb| unknown protein {Arabidopsis thaliana},
partial (56%)
Length = 788
Score = 231 bits (588), Expect = 3e-61
Identities = 116/170 (68%), Positives = 138/170 (80%), Gaps = 1/170 (0%)
Frame = +1
Query: 30 VLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTS-STD 88
VLPKRIIL+RHGESQGNLD SAYTTTPDH I LTP GI QAR AGA + ++S S D
Sbjct: 160 VLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPSGIAQARHAGANLHRLVSGQGCSPD 339
Query: 89 WRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERINALKETRQ 148
WR+YFYVSPYARTRSTLRE+GRSFSK R++GVREE R+REQDFGNFQVQER+ +KETR+
Sbjct: 340 WRLYFYVSPYARTRSTLREVGRSFSKKRVIGVREESRVREQDFGNFQVQERMKIVKETRE 519
Query: 149 RFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLII 198
RFGRFF+RFP+GESAADVFDR+S L+ + + +++H N LNL +
Sbjct: 520 RFGRFFYRFPEGESAADVFDRISGNC*LLFPSL*IIQIHHIDIN-LNLFL 666
Score = 70.9 bits (172), Expect = 6e-13
Identities = 30/37 (81%), Positives = 35/37 (94%)
Frame = +3
Query: 173 FLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVFL 209
F ESLWRDID+NRL+HDP+NDLNL+IVSHGL SR+FL
Sbjct: 678 FFESLWRDIDLNRLHHDPTNDLNLVIVSHGLTSRIFL 788
>AW683217 similar to GP|6069661|dbj Similar to Sequence of BAC F7G19 from
Arabidopsis thaliana chromosome 1; similar to
Saccharomyces, partial (29%)
Length = 664
Score = 142 bits (358), Expect(2) = 4e-39
Identities = 76/164 (46%), Positives = 100/164 (60%), Gaps = 12/164 (7%)
Frame = +2
Query: 4 HNNNDNDDN--NNHHQQQIPNQNNHKPSVL----------PKRIILVRHGESQGNLDPSA 51
H+ D D+ NN+ + P +N +L P+RIILVRHGES+GN+D S
Sbjct: 110 HSQTDPIDSLTNNNGHTRFPEKNPLINPLLASKVGAVPPRPRRIILVRHGESEGNVDESV 289
Query: 52 YTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWRVYFYVSPYARTRSTLREIGRS 111
YT PD KI LT +G QA G RI+++I S +W++YFYVSPY RT TL+ + R
Sbjct: 290 YTRVPDPKIGLTNRGRVQAEECGQRIKNMIEKDSDENWQLYFYVSPYRRTLETLQSLARP 469
Query: 112 FSKHRLLGVREECRIREQDFGNFQVQERINALKETRQRFGRFFF 155
F + R+ G REE RIREQDFGNFQ +E + K R +GRFF+
Sbjct: 470 FERSRIAGFREEPRIREQDFGNFQNRELMKVEKAQRHLYGRFFY 601
Score = 36.6 bits (83), Expect(2) = 4e-39
Identities = 15/22 (68%), Positives = 19/22 (86%)
Frame = +3
Query: 154 FFRFPDGESAADVFDRVSSFLE 175
F RFP+GESAADV+DR++ F E
Sbjct: 597 FTRFPNGESAADVYDRITGFRE 662
>AL366904 weakly similar to GP|6729027|gb|A unknown protein {Arabidopsis
thaliana}, partial (11%)
Length = 407
Score = 67.0 bits (162), Expect = 9e-12
Identities = 28/47 (59%), Positives = 36/47 (76%)
Frame = +3
Query: 256 WGLSPDMIADQKWRAHATKGAWHDQGPLSLDTFFDHLHDSDNEDNDD 302
WG+SPDMIADQKWRA A +GAW+DQ P LD FFD S+++D+ +
Sbjct: 3 WGMSPDMIADQKWRADAPRGAWNDQCPWYLDGFFDRFDASESDDDTE 143
>TC91584 similar to GP|21592519|gb|AAM64469.1 unknown {Arabidopsis
thaliana}, partial (71%)
Length = 590
Score = 35.8 bits (81), Expect = 0.021
Identities = 25/87 (28%), Positives = 41/87 (46%), Gaps = 3/87 (3%)
Frame = +1
Query: 30 VLPKRIILVRHGESQGN---LDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSS 86
VL R +VRHG+S N L S+ + L P+G+ QA++AG + + S +
Sbjct: 4 VLKNRYWIVRHGKSIPNERGLIVSSMENGILPEFQLAPEGVNQAQLAGYSFQKELESNNI 183
Query: 87 TDWRVYFYVSPYARTRSTLREIGRSFS 113
V SP++RT T + + +
Sbjct: 184 PLANVRICYSPFSRTTHTANVVATALN 264
>TC86123 homologue to PIR|S71770|S71770 calcium-dependent protein kinase (EC
2.7.1.-) - mung bean, complete
Length = 2448
Score = 33.9 bits (76), Expect = 0.080
Identities = 13/27 (48%), Positives = 21/27 (77%)
Frame = +3
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKP 28
+N+NNN+ND N++HH ++NN+KP
Sbjct: 348 NNNNNNNNDSNSDHHNH---HRNNNKP 419
>TC81407 homologue to GP|18650662|gb|AAL75809.1 ethylene response factor 1
{Lycopersicon esculentum}, partial (59%)
Length = 770
Score = 33.9 bits (76), Expect = 0.080
Identities = 14/40 (35%), Positives = 24/40 (60%)
Frame = +3
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKPSVLPKRIILVRHG 41
H + + ND NN+H ++ N+ +HK S+LP + L+ G
Sbjct: 321 HGFSFSSNDQTNNNHHKKSHNKFHHKVSILPLIVTLLSMG 440
>TC79726 similar to GP|17064808|gb|AAL32558.1 Unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 777
Score = 33.1 bits (74), Expect = 0.14
Identities = 53/206 (25%), Positives = 82/206 (39%), Gaps = 10/206 (4%)
Frame = +3
Query: 28 PSVLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSST 87
P + KR++LVRHG+S N + + D + LT +G +QA + +
Sbjct: 240 PLKVAKRVVLVRHGQSTWNAE-GRIQGSSDFSV-LTKKGESQAETSRQMLLE-------- 389
Query: 88 DWRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERINALKET- 146
D + SP AR++ T I S R + E +RE D +FQ LKE
Sbjct: 390 DNFDACFASPLARSKKTAEIIWGS----RQQQIIPEYDLREIDLYSFQ-----GLLKEEG 542
Query: 147 RQRFGRFF---------FRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLI 197
+ RFG F F D +++DR S W I L HD +++
Sbjct: 543 KARFGPAFHQWQVDAVNFVIDDHYPVRELWDRA----RSCWTKI----LAHD---SRSVL 689
Query: 198 IVSHGLASRVFLMKWFKWTVEQFDLL 223
+V+H ++ + E F L
Sbjct: 690 VVAHNAVNQALVATAIGLEAEYFRTL 767
>BF003770 similar to PIR|F96809|F96 protein F28K19.26 [imported] -
Arabidopsis thaliana, partial (15%)
Length = 570
Score = 32.3 bits (72), Expect = 0.23
Identities = 20/57 (35%), Positives = 34/57 (59%)
Frame = +3
Query: 24 NNHKPSVLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHV 80
++H P+ +IL+RHGES N + + +T D + LT +G+ +A AG RI ++
Sbjct: 219 SHHSPT--ESSLILIRHGESMWN-EKNLFTGCCD--VPLTRKGVEEAIEAGKRITYI 374
>TC79517 similar to PIR|T08874|T08874 calcium-dependent protein kinase (EC
2.7.1.-) gamma - soybean, partial (80%)
Length = 1930
Score = 32.3 bits (72), Expect = 0.23
Identities = 13/30 (43%), Positives = 20/30 (66%)
Frame = +3
Query: 1 MHNHNNNDNDDNNNHHQQQIPNQNNHKPSV 30
+HNHN+N N ++N++H N NH+P V
Sbjct: 423 IHNHNHNHNHNHNHNH-----NHKNHEPYV 497
Score = 31.6 bits (70), Expect = 0.40
Identities = 14/32 (43%), Positives = 20/32 (61%), Gaps = 1/32 (3%)
Frame = +3
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHK-PSVLP 32
HNHN+N N ++N++H+ P N K PS P
Sbjct: 432 HNHNHNHNHNHNHNHKNHEPYVNQSKTPSQQP 527
>TC92077 homologue to GP|23497650|gb|AAN37190.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (1%)
Length = 376
Score = 32.3 bits (72), Expect = 0.23
Identities = 12/24 (50%), Positives = 16/24 (66%)
Frame = +1
Query: 3 NHNNNDNDDNNNHHQQQIPNQNNH 26
N NNNDN++NNN++ N N H
Sbjct: 16 NDNNNDNNNNNNNNNHNNNNNNTH 87
Score = 30.4 bits (67), Expect = 0.89
Identities = 11/25 (44%), Positives = 18/25 (72%)
Frame = +1
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNH 26
+N N+N+ND+NNN++ N NN+
Sbjct: 7 NNDNDNNNDNNNNNNNNNHNNNNNN 81
Score = 29.3 bits (64), Expect = 2.0
Identities = 10/25 (40%), Positives = 19/25 (76%)
Frame = +1
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNH 26
+N++N++N+DNNN++ N NN+
Sbjct: 4 NNNDNDNNNDNNNNNNNNNHNNNNN 78
>CA860088 similar to GP|8248752|emb hypothetical protein MAL4P2.29
{Plasmodium falciparum 3D7}, partial (1%)
Length = 455
Score = 31.6 bits (70), Expect = 0.40
Identities = 11/22 (50%), Positives = 17/22 (77%)
Frame = +1
Query: 5 NNNDNDDNNNHHQQQIPNQNNH 26
NNND+D++NN+ + +I N N H
Sbjct: 232 NNNDDDNDNNNKESKIENTNEH 297
>CA860191 homologue to GP|19697342|gb hypothetical protein {Dictyostelium
discoideum}, partial (1%)
Length = 504
Score = 31.2 bits (69), Expect = 0.52
Identities = 11/31 (35%), Positives = 24/31 (76%)
Frame = +2
Query: 3 NHNNNDNDDNNNHHQQQIPNQNNHKPSVLPK 33
N+NNN++++NNN+++ N+ N + ++LP+
Sbjct: 311 NNNNNNSNNNNNNNEDNHHNKFNSEDAMLPQ 403
>BG583961 similar to GP|14599408|em probable major surface glycoprotein
{Pneumocystis carinii}, partial (4%)
Length = 803
Score = 30.4 bits (67), Expect = 0.89
Identities = 13/25 (52%), Positives = 16/25 (64%), Gaps = 1/25 (4%)
Frame = +1
Query: 3 NHNNNDND-DNNNHHQQQIPNQNNH 26
N N+NDND DN+N H N N+H
Sbjct: 625 NDNDNDNDNDNDNDHDNDNDNDNDH 699
Score = 29.6 bits (65), Expect = 1.5
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 3/41 (7%)
Frame = +1
Query: 3 NHNNNDND-DNNNHHQQQIPNQNNHKPSVLPK--RIILVRH 40
N N+NDND DN+N H N N + + P IIL+ H
Sbjct: 673 NDNDNDNDHDNDNDHDHVNVNDNVNVNDITPSYITIILIYH 795
Score = 26.9 bits (58), Expect = 9.8
Identities = 12/27 (44%), Positives = 16/27 (58%), Gaps = 3/27 (11%)
Frame = +1
Query: 3 NHNNNDND---DNNNHHQQQIPNQNNH 26
N N+NDND DN+N + N N+H
Sbjct: 637 NDNDNDNDNDHDNDNDNDNDHDNDNDH 717
>TC87877 similar to GP|21618275|gb|AAM67325.1 unknown {Arabidopsis
thaliana}, partial (79%)
Length = 1387
Score = 29.6 bits (65), Expect = 1.5
Identities = 42/212 (19%), Positives = 86/212 (39%), Gaps = 27/212 (12%)
Frame = +2
Query: 35 IILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHV-------------- 80
+IL+RHGES N + + +T D + L+ +GI +A AG RI +
Sbjct: 569 LILIRHGESLWN-EKNLFTGCVD--VPLSKKGIDEAIEAGKRISSIPVDLIFTSALIRAQ 739
Query: 81 ----ISSTSSTDWRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQV 136
++ T +V + + ++ +K + + V ++ E+ +G Q
Sbjct: 740 MTAMLAMTQHRRRKVPIVMHNESEQAKAWSQVFSEDTKKQSIPVIAAWQLNERMYGELQG 919
Query: 137 QERINALKETRQRFGR---------FFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLN 187
+ +ET R+G+ + P+GES +R ++ +++
Sbjct: 920 LNK----QETADRYGKEQVHEWRRSYDIPPPNGESLEMCAERAVAYFR--------DQIE 1063
Query: 188 HDPSNDLNLIIVSHGLASRVFLMKWFKWTVEQ 219
+ N++I +HG + R +M K T ++
Sbjct: 1064PQLLSGKNVMIAAHGNSLRSIIMYLDKLTSQE 1159
>TC81322 weakly similar to GP|17104669|gb|AAL34223.1 unknown protein
{Arabidopsis thaliana}, partial (13%)
Length = 788
Score = 29.3 bits (64), Expect = 2.0
Identities = 13/29 (44%), Positives = 17/29 (57%)
Frame = -3
Query: 1 MHNHNNNDNDDNNNHHQQQIPNQNNHKPS 29
M HNNN N N +Q PN ++HKP+
Sbjct: 204 MQTHNNNTN---NQSYQNINPNMSSHKPN 127
>AW586945 homologue to GP|23496297|gb hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 677
Score = 29.3 bits (64), Expect = 2.0
Identities = 11/25 (44%), Positives = 17/25 (68%)
Frame = +1
Query: 3 NHNNNDNDDNNNHHQQQIPNQNNHK 27
N+NNN+N++NNN + + N N K
Sbjct: 451 NNNNNNNNNNNNFNDKNNKNYKNDK 525
Score = 28.9 bits (63), Expect = 2.6
Identities = 14/49 (28%), Positives = 27/49 (54%)
Frame = +1
Query: 3 NHNNNDNDDNNNHHQQQIPNQNNHKPSVLPKRIILVRHGESQGNLDPSA 51
N +NN+N++NNN++ N N+K + K V+ +S ++ S+
Sbjct: 442 NDDNNNNNNNNNNNNFNDKNNKNYKNDKIDKSDRKVKRRKSLFSISSSS 588
>TC90933 similar to SP|P27484|GRP2_NICSY Glycine-rich protein 2. [Wood
tobacco] {Nicotiana sylvestris}, partial (44%)
Length = 368
Score = 28.5 bits (62), Expect = 3.4
Identities = 9/26 (34%), Positives = 17/26 (64%)
Frame = -3
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHK 27
HNH ++ ++ + HH+ IP Q +H+
Sbjct: 348 HNHLHHAHNHHRRHHRVSIPEQKHHQ 271
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,438,518
Number of Sequences: 36976
Number of extensions: 151210
Number of successful extensions: 1741
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 1297
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1564
length of query: 306
length of database: 9,014,727
effective HSP length: 96
effective length of query: 210
effective length of database: 5,465,031
effective search space: 1147656510
effective search space used: 1147656510
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0544.10


