

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0490.17
(392 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
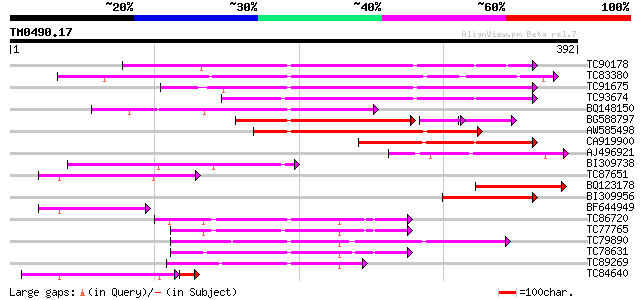
Score E
Sequences producing significant alignments: (bits) Value
TC90178 similar to GP|13958032|gb|AAK50769.1 polygalacturonase {... 184 5e-47
TC83380 weakly similar to GP|10176984|dbj|BAB10216. polygalactur... 176 1e-44
TC91675 similar to GP|6624205|dbj|BAA88472.1 polygalacturonase {... 170 8e-43
TC93674 similar to GP|15292729|gb|AAK92733.1 putative polygalact... 162 3e-40
BQ148150 weakly similar to GP|10185719|gb NTS1 protein {Nicotian... 149 2e-36
BG588797 weakly similar to PIR|T08215|T08 polygalacturonase (EC ... 122 2e-34
AW585498 weakly similar to GP|10177065|db polygalacturonase-like... 141 5e-34
CA919900 similar to GP|15292729|gb putative polygalacturonase PG... 95 4e-20
AJ496921 similar to GP|10185719|gb| NTS1 protein {Nicotiana taba... 89 3e-18
BI309738 weakly similar to GP|15028105|gb putative polygalacturo... 84 7e-17
TC87651 similar to GP|15292729|gb|AAK92733.1 putative polygalact... 78 7e-15
BQ123178 similar to GP|23495980|gb hypothetical protein {Plasmod... 70 1e-12
BI309956 weakly similar to GP|7381227|gb|A polygalacturonase {Ly... 65 3e-11
BF644949 similar to GP|15292729|gb putative polygalacturonase PG... 62 3e-10
TC86720 similar to PIR|T47941|T47941 hypothetical protein F2A19.... 62 4e-10
TC77765 similar to PIR|T47941|T47941 hypothetical protein F2A19.... 58 7e-09
TC79890 similar to GP|11762132|gb|AAG40344.1 AT3g62110 {Arabidop... 57 2e-08
TC78631 similar to PIR|T05388|T05388 hypothetical protein F16G20... 55 5e-08
TC89269 similar to PIR|T05614|T05614 hypothetical protein F9D16.... 54 8e-08
TC84640 similar to GP|10177065|dbj|BAB10507. polygalacturonase-l... 47 4e-07
>TC90178 similar to GP|13958032|gb|AAK50769.1 polygalacturonase {Pisum
sativum}, partial (94%)
Length = 1114
Score = 184 bits (467), Expect = 5e-47
Identities = 116/317 (36%), Positives = 159/317 (49%), Gaps = 30/317 (9%)
Frame = +3
Query: 79 KAPIEIQVDGTIQAPTDPNALN-GAFQWVTIGYVDYFTLSGNGTFDGRGASAWK------ 131
K I QVDGTI APT+ NA G QW+ + FT+ GNG FDGRG+ W+
Sbjct: 3 KPRIIFQVDGTIIAPTNSNAWGRGLLQWLDFTKLVGFTIQGNGIFDGRGSVWWQDTQYND 182
Query: 132 ----------------------LQKACGATC-SKRSMNIGFNFLKHSTVRSVTSKDSKFF 168
++ + G S + I F + TV +T ++S
Sbjct: 183 PLDDEEKLLVPLNNTIVSPPMQIESSLGGKMPSIKPTAIRFYGSINPTVTGITIQNSPQC 362
Query: 169 HVTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNK 228
H+ C I +PGDS NTDGIH+ S DV I S +A GDDC+S+ G
Sbjct: 363 HLKFDNCNGVLVHDVTISSPGDSP--NTDGIHLQNSRDVLIYKSTMACGDDCISIQTGCS 536
Query: 229 HITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPV 288
++ +HNV CGPGHG SIG LG + T V NI V++ + NT NGVRIKTW G+ V
Sbjct: 537 NVYVHNVDCGPGHGISIGGLGKDNTRACVSNITVRDVNMHNTMNGVRIKTW--QGGSGSV 710
Query: 289 TNLTFEDITMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILN 348
+ F +I + V P+IIDQ YC C Q+ + + ++ + ++ IKGT K V
Sbjct: 711 QGVLFSNIQVSQVQVPIIIDQFYCDKRNCKNQT-AAVALTGINYEGIKGTYTVKP-VHFA 884
Query: 349 CSSGVPCEGVELSDISL 365
CS +PC V L+ + L
Sbjct: 885 CSDSLPCIDVSLTSVEL 935
>TC83380 weakly similar to GP|10176984|dbj|BAB10216. polygalacturonase-like
protein {Arabidopsis thaliana}, partial (21%)
Length = 1220
Score = 176 bits (447), Expect = 1e-44
Identities = 110/352 (31%), Positives = 180/352 (50%), Gaps = 6/352 (1%)
Frame = +2
Query: 34 GAPNSDITQAFLNAWKQACASTSPSKIVISK--ATYKMNAVNVKGPCKAP-IEIQVDGTI 90
G +D ++AF++A+K C + +V+ + + +N GPC + I I++ G I
Sbjct: 119 GDGRTDDSKAFVDAFKALCGGRYGNTLVVPNGHSFFVRPTLNFSGPCYSKNINIKIMGNI 298
Query: 91 QAPTDPN-ALNGAFQWVTIGYVDYFTLSGNGTFDGRGASAWKLQKACGATCSKRSMNIGF 149
AP + + W+ + TL G+G +G G +K G C + + F
Sbjct: 299 LAPKRSDWGRECSLMWLHFFNISGLTLDGSGVINGNGEGWESREKGIGG-CPRIPTALQF 475
Query: 150 NFLKHSTVRSVTSKDSKFFHVTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKI 209
+ + +T + H+ ++ + T I +P S NTDGI + ++ V I
Sbjct: 476 DKCDGLQINGLTHINGPGAHIAVIDSQDITISHIHINSPKKSH--NTDGIDLTRTIGVNI 649
Query: 210 LNSNIATGDDCVSLGDGNKHITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTN 269
+ I +GDDC+++ G++ + + NV CGPGHG S+GSLG + +EE V++ VKNCT
Sbjct: 650 HDIQIESGDDCIAVKGGSQFVNVSNVTCGPGHGISVGSLGGHGSEEFVQHFSVKNCTFNG 829
Query: 270 TQNGVRIKTWPTAPGTSPVTNLTFEDITMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISN 329
+ V+IKTWP G + ++ FEDI + PV IDQ Y + + + +KISN
Sbjct: 830 ADSAVKIKTWPGGKGYA--KHIIFEDIIINQTNYPVFIDQHY----MRTPEQHQAVKISN 991
Query: 330 VTFKNIKGTSETKDGVILNCSSGVPCEGVELSDISLTF--NGAPAMAKCANV 379
+TF NI GT +D V+L+C+ + C + L+ I++T PA KC NV
Sbjct: 992 ITFSNIYGTCIGEDAVVLDCAK-IGCYNITLNQINITSINRKKPASVKCKNV 1144
>TC91675 similar to GP|6624205|dbj|BAA88472.1 polygalacturonase {Cucumis
sativus}, partial (65%)
Length = 1146
Score = 170 bits (431), Expect = 8e-43
Identities = 90/269 (33%), Positives = 145/269 (53%), Gaps = 8/269 (2%)
Frame = +1
Query: 105 WVTIGYVDYFTLSGNGTFDGRGASAWKLQKACGATCSKRSMN--------IGFNFLKHST 156
W+ + G+G DG G+ W A+C K N + +
Sbjct: 25 WLDFSKLKKAVFQGSGVIDGSGSKWW------AASCKKNKTNPCKGAPTALTIDTSSSIK 186
Query: 157 VRSVTSKDSKFFHVTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIAT 216
V+ +T ++++ H T+ C + G K+ +PGDS NTDGIHI++ST+V + + I T
Sbjct: 187 VKGLTIQNNQQMHFTISRCDSVRILGVKVASPGDSP--NTDGIHISESTNVIVQDCKIGT 360
Query: 217 GDDCVSLGDGNKHITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRI 276
GDDC+S+ + + +I + + CGPGHG SIGSLG + + V +I+ L T NG+RI
Sbjct: 361 GDDCISIVNASSNIKMKRIFCGPGHGISIGSLGKDNSTGVVTKVILDTAFLKGTTNGLRI 540
Query: 277 KTWPTAPGTSPVTNLTFEDITMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIK 336
KTW G V + F+++ + NV NP++IDQ YC S + S ++IS + ++N+
Sbjct: 541 KTW--QGGAGYVKGVRFQNVRVENVSNPILIDQFYCDSPTSCQNQSSALEISEIMYQNVS 714
Query: 337 GTSETKDGVILNCSSGVPCEGVELSDISL 365
GT+ + + +CS VPC + LS++ L
Sbjct: 715 GTTNSAKAIKFDCSDTVPCSNLVLSNVDL 801
>TC93674 similar to GP|15292729|gb|AAK92733.1 putative polygalacturonase PG1
{Arabidopsis thaliana}, partial (51%)
Length = 745
Score = 162 bits (409), Expect = 3e-40
Identities = 90/220 (40%), Positives = 126/220 (56%), Gaps = 1/220 (0%)
Frame = +3
Query: 147 IGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTD 206
I F + +R + ++S FHV GC D I AP +S NTDGIH+ + D
Sbjct: 15 IRFFMSSNLVLRGLKIQNSPQFHVKFDGCQGVLIDELSISAP--KLSPNTDGIHLGNTRD 188
Query: 207 VKILNSNIATGDDCVSLGDGNKHITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCT 266
V I NS I+ GDDC+S+G G ++ + V C P HG SIGSLG + + V N+ V+N
Sbjct: 189 VGIYNSLISNGDDCISIGPGCSNVNVDGVTCAPTHGISIGSLGVHNSHACVSNLTVRNSI 368
Query: 267 LTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVNVYNPVIIDQQYCPSNLCSKQSPSKIK 326
+ + NG+RIKTW GT VT LTF++I M NV N + IDQ YC S C Q+ S +
Sbjct: 369 IKESDNGLRIKTW--QGGTGSVTGLTFDNIQMENVRNCINIDQFYCLSKECMNQT-SAVY 539
Query: 327 ISNVTFKNIKGTSETKDGVI-LNCSSGVPCEGVELSDISL 365
++N++++ IKGT + + I CS V C + LS+I L
Sbjct: 540 VNNISYRKIKGTYDVRTPPIHFACSDTVACTNITLSEIEL 659
>BQ148150 weakly similar to GP|10185719|gb NTS1 protein {Nicotiana tabacum},
partial (31%)
Length = 669
Score = 149 bits (375), Expect = 2e-36
Identities = 79/203 (38%), Positives = 116/203 (56%), Gaps = 4/203 (1%)
Frame = +2
Query: 57 PSKIVISKATYKMNAVNVKGPCKAP--IEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYF 114
P+ ++I T+ GPC +P I +++ G QA TD + +W +D
Sbjct: 2 PATVLIPAGTFVTGQTLFAGPCTSPKPITVEIVGKAQATTDLSEYYSP-EWFQFENIDGL 178
Query: 115 TLSGNGTFDGRGASAWKLQ--KACGATCSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTL 172
L G+G FDG+G+ +W L K C+ ++ F+ +K++ V+ VTS +S FH L
Sbjct: 179 VLKGSGVFDGQGSISWPLNDCKQNKGNCASLPSSLKFDKIKNAIVQDVTSLNSMQFHFHL 358
Query: 173 LGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITI 232
GC+N +F I APG+S NTDG+HI+ S + + NS IATGDDC+S+G +ITI
Sbjct: 359 HGCSNISFTNLHITAPGNSP--NTDGMHISSSDFITVTNSVIATGDDCISVGHSTSNITI 532
Query: 233 HNVKCGPGHGFSIGSLGTNPTEE 255
+ CGPGHG S+GSLG P E+
Sbjct: 533 SGITCGPGHGISVGSLGKRPEEK 601
>BG588797 weakly similar to PIR|T08215|T08 polygalacturonase (EC 3.2.1.15) 3
precursor - muskmelon, partial (35%)
Length = 631
Score = 122 bits (307), Expect(3) = 2e-34
Identities = 58/124 (46%), Positives = 81/124 (64%)
Frame = +2
Query: 157 VRSVTSKDSKFFHVTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIAT 216
V+++ KDS+ HV C N + AP DS NTDGIH+A++ ++ I+N +I T
Sbjct: 8 VKNLRFKDSQQMHVVFERCFNVFVSNLIVRAPEDSP--NTDGIHVAETQNIDIINCDIGT 181
Query: 217 GDDCVSLGDGNKHITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRI 276
GDDC+S+ G+K++ ++ CGPGHG SIGSLG + +E V N++V LT T NGVRI
Sbjct: 182 GDDCISIVSGSKNVRAIDITCGPGHGISIGSLGADNSEAEVSNVVVNRAALTGTTNGVRI 361
Query: 277 KTWP 280
KTWP
Sbjct: 362 KTWP 373
Score = 30.8 bits (68), Expect(3) = 2e-34
Identities = 12/40 (30%), Positives = 23/40 (57%)
Frame = +1
Query: 311 YCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCS 350
YC ++ +++S V ++NI+GTS ++ + NCS
Sbjct: 460 YCDQTEPCQERNKAVQLSQVLYQNIRGTSASEIAIKFNCS 579
Score = 30.8 bits (68), Expect(3) = 2e-34
Identities = 16/32 (50%), Positives = 19/32 (59%)
Frame = +3
Query: 284 GTSPVTNLTFEDITMVNVYNPVIIDQQYCPSN 315
G+ N+ F +I M NV NPVIIDQ SN
Sbjct: 378 GSGYARNIKFMNIKMQNVTNPVIIDQYLL*SN 473
>AW585498 weakly similar to GP|10177065|db polygalacturonase-like protein
{Arabidopsis thaliana}, partial (26%)
Length = 583
Score = 141 bits (355), Expect = 5e-34
Identities = 73/159 (45%), Positives = 98/159 (60%)
Frame = +3
Query: 169 HVTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNK 228
H+TL C ++IAPG+S NTDGI I+ S D+++LNS IATGDDC+++ G+
Sbjct: 30 HITLTSCKKGIISNIRLIAPGESP--NTDGIDISASRDIQVLNSFIATGDDCIAISAGSS 203
Query: 229 HITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPV 288
I I + CGPGHG SIGSLG + VE++ VKNCTLT T GVRIKT G +
Sbjct: 204 VIKITAITCGPGHGISIGSLGARGDTDIVEDVHVKNCTLTETLTGVRIKTKQGGGGFA-- 377
Query: 289 TNLTFEDITMVNVYNPVIIDQQYCPSNLCSKQSPSKIKI 327
+TFE+I V +NP+ IDQ YC + + + IK+
Sbjct: 378 RRITFENIKFVRAHNPIWIDQFYCVNQMVCRNMTKAIKV 494
Score = 35.4 bits (80), Expect = 0.038
Identities = 23/63 (36%), Positives = 34/63 (53%)
Frame = +1
Query: 293 FEDITMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSG 352
FE IT + N +++ + + + L KQS K S+VT++ I GTS T + LNC
Sbjct: 409 FEPITPFGLTNFIVLIKWFAET*L--KQS----KWSDVTYRGISGTSLTDKAINLNCDQN 570
Query: 353 VPC 355
V C
Sbjct: 571 VGC 579
>CA919900 similar to GP|15292729|gb putative polygalacturonase PG1
{Arabidopsis thaliana}, partial (33%)
Length = 752
Score = 95.1 bits (235), Expect = 4e-20
Identities = 51/125 (40%), Positives = 78/125 (61%), Gaps = 1/125 (0%)
Frame = -2
Query: 242 GFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVNV 301
G SIGSLG + ++ V N+ V++ + + NG+RIKTW G+ V+NL FE+I M NV
Sbjct: 751 GISIGSLGVHNSQACVSNLTVRDTVIRESDNGLRIKTWQGGMGS--VSNLKFENIQMENV 578
Query: 302 YNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVI-LNCSSGVPCEGVEL 360
N ++IDQ YC + C Q+ S + +++V++KNIKGT + + I CS V C + L
Sbjct: 577 GNCILIDQYYCLTKECLNQT-SAVHVNDVSYKNIKGTYDVRTAPIHFACSDTVACTNITL 401
Query: 361 SDISL 365
S++ L
Sbjct: 400 SEVEL 386
>AJ496921 similar to GP|10185719|gb| NTS1 protein {Nicotiana tabacum},
partial (9%)
Length = 508
Score = 89.0 bits (219), Expect = 3e-18
Identities = 53/130 (40%), Positives = 71/130 (53%), Gaps = 6/130 (4%)
Frame = +3
Query: 263 KNCTLTNTQNGVRIKTWPTAPGTSPVT--NLTFEDITMVNVYNPVIIDQQYCPSNLCSKQ 320
+ CT + NG RIKTW GT+P N+ +ED+ M +V NP++IDQ Y
Sbjct: 18 QKCTFIGSTNGARIKTWI---GTAPAEAKNIVYEDLIMKDVQNPIVIDQSYGKKERVP-- 182
Query: 321 SPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVELSDISLTFNG----APAMAKC 376
S S KIS+ F+ IKGT+ + V L CSS PCE +E++D+ LT+ G P C
Sbjct: 183 STSVWKISDAHFRRIKGTTVSNIPVSLQCSSKNPCENIEVADVELTYVGPQKSIPFANSC 362
Query: 377 ANVKPIIIGK 386
N K I GK
Sbjct: 363 INAKAIFGGK 392
>BI309738 weakly similar to GP|15028105|gb putative polygalacturonase
{Arabidopsis thaliana}, partial (31%)
Length = 768
Score = 84.3 bits (207), Expect = 7e-17
Identities = 49/165 (29%), Positives = 83/165 (49%), Gaps = 5/165 (3%)
Frame = +2
Query: 41 TQAFLNAWKQACASTSPSKIVIS-KATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNAL 99
TQAFL W+ AC+ + IV + T+ + +++ GPC++ I +++ G I AP +P+
Sbjct: 278 TQAFLKVWEIACSLSGFINIVFPYQKTFLVTPIDIGGPCRSKITLRILGAIVAPRNPDVW 457
Query: 100 NG--AFQWVTIGYVDYFTLSGNGTFDGRGASAWKLQKACGAT--CSKRSMNIGFNFLKHS 155
+G +W+ V++ ++ G G DG G W T C + F+ K
Sbjct: 458 HGLNKRKWIYFHGVNHLSVEGGGRIDGMGQEWWSRSCKINTTNPCLPAPTALTFHKCKSL 637
Query: 156 TVRSVTSKDSKFFHVTLLGCTNFTFDGFKIIAPGDSISINTDGIH 200
VR++T +S+ H+ C K++AP ++S NTDGIH
Sbjct: 638 KVRNLTVLNSQKMHIAFTSCMRVVASRLKVLAP--ALSPNTDGIH 766
>TC87651 similar to GP|15292729|gb|AAK92733.1 putative polygalacturonase PG1
{Arabidopsis thaliana}, partial (35%)
Length = 822
Score = 77.8 bits (190), Expect = 7e-15
Identities = 41/117 (35%), Positives = 62/117 (52%), Gaps = 5/117 (4%)
Frame = +3
Query: 21 SAQSIVFDIRTFG--GAPNSDITQAFLNAWKQACASTSPSKIVISKATYKMNAVNVKGPC 78
S+ +FD+ +FG G ++D T AF AWK ACA S +V T+ + ++ GPC
Sbjct: 348 SSSDCIFDVTSFGAIGDGSADDTPAFKKAWKAACAVESGILLVPENYTFMITSIIFSGPC 527
Query: 79 KAPIEIQVDGTIQAPTDPNA---LNGAFQWVTIGYVDYFTLSGNGTFDGRGASAWKL 132
K + QVDG + AP P++ + QW+ +D +L+G GT +G G W L
Sbjct: 528 KPGLVFQVDGMLMAPDGPDSWPEADSRNQWLVFYKLDQMSLNGTGTIEGNGDQWWDL 698
>BQ123178 similar to GP|23495980|gb hypothetical protein {Plasmodium
falciparum 3D7}, partial (2%)
Length = 619
Score = 70.1 bits (170), Expect = 1e-12
Identities = 38/63 (60%), Positives = 44/63 (69%)
Frame = -3
Query: 323 SKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVELSDISLTFNGAPAMAKCANVKPI 382
SKIKIS VTFKNI G T++ V+L S+GVP E V LS+I LTFNG A CANVKP
Sbjct: 611 SKIKISKVTFKNIMGIFATQNCVVLIRSNGVPYEEVVLSNIDLTFNGVTVNATCANVKP* 432
Query: 383 IIG 385
+G
Sbjct: 431 FLG 423
>BI309956 weakly similar to GP|7381227|gb|A polygalacturonase {Lycopersicon
esculentum}, partial (21%)
Length = 512
Score = 65.5 bits (158), Expect = 3e-11
Identities = 30/66 (45%), Positives = 42/66 (63%)
Frame = +3
Query: 300 NVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVE 359
NV NP+IIDQ YC S K S +++ N++F NI+GTS T++ + CS PCEG+
Sbjct: 12 NVSNPIIIDQYYCDSRHPCKNQTSAVQVGNISFINIQGTSATEETIKFACSDASPCEGLY 191
Query: 360 LSDISL 365
L +I L
Sbjct: 192 LENIFL 209
>BF644949 similar to GP|15292729|gb putative polygalacturonase PG1
{Arabidopsis thaliana}, partial (22%)
Length = 673
Score = 62.4 bits (150), Expect = 3e-10
Identities = 32/79 (40%), Positives = 44/79 (55%), Gaps = 2/79 (2%)
Frame = +1
Query: 21 SAQSIVFDIRTFG--GAPNSDITQAFLNAWKQACASTSPSKIVISKATYKMNAVNVKGPC 78
S + VFD+R+FG G ++D T AF AWK ACA S + +K+ + GPC
Sbjct: 382 SPSNCVFDVRSFGAVGDGDADDTAAFRAAWKAACAVDSGVLLAPENYCFKITSTIFSGPC 561
Query: 79 KAPIEIQVDGTIQAPTDPN 97
K + Q+DGT+ AP PN
Sbjct: 562 KPGLVFQIDGTLMAPDGPN 618
>TC86720 similar to PIR|T47941|T47941 hypothetical protein F2A19.90 -
Arabidopsis thaliana, partial (85%)
Length = 1719
Score = 62.0 bits (149), Expect = 4e-10
Identities = 57/193 (29%), Positives = 89/193 (45%), Gaps = 15/193 (7%)
Frame = +2
Query: 101 GAFQWVTIG--YVDYFTLSGNGTFDGRGASAWKL--QKACGATCSKRSMNIGFNFLKHST 156
G F+ + G D GNGT DG+GA W+ +K T R I F
Sbjct: 548 GRFRSLIFGTNLTDVIVTGGNGTIDGQGAFWWQQFHRKKLKYT---RPYLIELMFSDSIQ 718
Query: 157 VRSVTSKDSKFFHVTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIAT 216
+ ++T +S ++V + +N G IIAP S NTDGI+ T+ KI + I +
Sbjct: 719 ISNLTLLNSPSWNVHPVYSSNIIIQGITIIAPISSP--NTDGINPDSCTNTKIEDCYIVS 892
Query: 217 GDDCVSLGDG-----------NKHITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNC 265
GDDCV++ G K + I + C +I +LG+ ++++ ++
Sbjct: 893 GDDCVAVKSGWDEYGIKFGWPTKQLVIRRLTCISPFSATI-ALGSE-MSGGIQDVRAEDI 1066
Query: 266 TLTNTQNGVRIKT 278
T T++GVRIKT
Sbjct: 1067TAIRTESGVRIKT 1105
>TC77765 similar to PIR|T47941|T47941 hypothetical protein F2A19.90 -
Arabidopsis thaliana, partial (61%)
Length = 1027
Score = 57.8 bits (138), Expect = 7e-09
Identities = 50/180 (27%), Positives = 84/180 (45%), Gaps = 13/180 (7%)
Frame = +3
Query: 112 DYFTLSGNGTFDGRGASAWKL--QKACGATCSKRSMNIGFNFLKHSTVRSVTSKDSKFFH 169
D NGT DG+G+ W+ K T R I F + + ++T DS ++
Sbjct: 501 DVIVTGDNGTIDGQGSFWWQQFHNKKLKYT---RPYLIELMFSDNIQISNLTLLDSPSWN 671
Query: 170 VTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDG--- 226
+ + +N G IIAP S NTDGI+ T+ KI + I +GDDCV++ G
Sbjct: 672 IHPVYSSNIIIKGITIIAP--IRSPNTDGINPDSCTNTKIEDCYIVSGDDCVAVKSGWDE 845
Query: 227 --------NKHITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKT 278
K + I + C + +I +LG+ ++++ ++ T +T++ +RIKT
Sbjct: 846 YGIKFGWPTKQLVIRRLTCISPYSATI-ALGSE-MSGGIQDVRAEDITAVHTESXIRIKT 1019
>TC79890 similar to GP|11762132|gb|AAG40344.1 AT3g62110 {Arabidopsis
thaliana}, partial (55%)
Length = 1151
Score = 56.6 bits (135), Expect = 2e-08
Identities = 60/246 (24%), Positives = 105/246 (42%), Gaps = 11/246 (4%)
Frame = +3
Query: 112 DYFTLSGNGTFDGRGASAWKLQKACGATCSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVT 171
D NGT DG+G+ W + ++ + N + + + T +S F+ +
Sbjct: 36 DVIITGNNGTIDGQGSIWWSKFRNKTLDHTRPHLVELINSTE-VLISNATFLNSPFWTIH 212
Query: 172 LLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDG----- 226
+ C+N T II P S NTDGI S +V I + I+TGDD +S+ G
Sbjct: 213 PVYCSNVTVQNVTIIVPFGSP--NTDGIDPDSSDNVCIEDCYISTGDDLISIKSGWDEYG 386
Query: 227 ------NKHITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWP 280
+ +I+IH + G S G + V + ++ + ++++ +RIKT P
Sbjct: 387 ISFGRPSTNISIHRL---TGRTTSAGIAIGSEMSGGVSEVYAEDIYIFDSKSAIRIKTSP 557
Query: 281 TAPGTSPVTNLTFEDITMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSE 340
G V N+ ++T++NV + Y S + I +T N+ G +
Sbjct: 558 GRGGY--VRNVYISNMTLINVDIAIRFTGLYGEHPDDSYDRDALPVIERITVVNVIGENI 731
Query: 341 TKDGVI 346
+ G+I
Sbjct: 732 KRAGLI 749
>TC78631 similar to PIR|T05388|T05388 hypothetical protein F16G20.200 -
Arabidopsis thaliana, partial (82%)
Length = 1613
Score = 55.1 bits (131), Expect = 5e-08
Identities = 48/179 (26%), Positives = 83/179 (45%), Gaps = 12/179 (6%)
Frame = +3
Query: 112 DYFTLSGNGTFDGRGASAW-KLQKACGATCSKRSMNIGFNFLKHSTVRSVTSKDSKFFHV 170
D NGT DG+G W K K G R I + + + ++T +S ++V
Sbjct: 507 DVVITGNNGTLDGQGELWWQKFHK--GKLTYTRPYLIEIMYSDNIQISNLTLVNSPSWNV 680
Query: 171 TLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDG---- 226
+ +N G I+AP +S NTDGI+ T+ +I + I +GDDCV++ G
Sbjct: 681 HPVYSSNIIVQGITILAPVNSP--NTDGINPDSCTNTRIEDCYIVSGDDCVAVKSGWDEY 854
Query: 227 -------NKHITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKT 278
K + I + C ++ +LG+ ++++ ++ N+++GVRIKT
Sbjct: 855 GIAYGMPTKQLVIRRLTC-ISPTSAVIALGSE-MSGGIQDVRAEDIVAINSESGVRIKT 1025
>TC89269 similar to PIR|T05614|T05614 hypothetical protein F9D16.290 -
Arabidopsis thaliana, partial (60%)
Length = 923
Score = 54.3 bits (129), Expect = 8e-08
Identities = 44/151 (29%), Positives = 73/151 (48%), Gaps = 12/151 (7%)
Frame = +1
Query: 109 GYVDYFTLSGNGTFDGRGASAWKLQKACGATCSKRSMNIGFNFLKHSTVRSVTSKDSKFF 168
G D NGT DG+G W + + ++ ++ + F ++ + +V K+S F+
Sbjct: 466 GVRDVIITGENGTIDGQGDVWWNMWRQRTLQFTRPNL-VEFLNSRNIIISNVIFKNSPFW 642
Query: 169 HVTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDG-- 226
++ + C+N I+AP DS NTDGI S++V I +S I+TGDD V++ G
Sbjct: 643 NIHPVYCSNVVVRYVTILAPRDSP--NTDGIDPDSSSNVCIEDSYISTGDDLVAVKSGWD 816
Query: 227 ---------NKHITIHNVK-CGPGHGFSIGS 247
+ ITI + P G ++GS
Sbjct: 817 AYGISYGRPSNDITIRRITGSSPFAGIALGS 909
>TC84640 similar to GP|10177065|dbj|BAB10507. polygalacturonase-like protein
{Arabidopsis thaliana}, partial (12%)
Length = 567
Score = 47.0 bits (110), Expect(2) = 4e-07
Identities = 32/116 (27%), Positives = 61/116 (52%), Gaps = 7/116 (6%)
Frame = +2
Query: 9 TAIVLFLLLLSGSAQSIVFDIRTFG--GAPNSDITQAFLNAWKQACASTS-PSKIVISKA 65
T +++ + G + + F++ +G G ++ + AFL AWK C S S S+++I A
Sbjct: 44 TLLMILSMFKLGLSITTTFNVLQYGAVGDGKTNDSPAFLKAWKDVCNSKSGTSRLIIPAA 223
Query: 66 -TYKMNAVNVKGPCKAP-IEIQVDGTIQAPTDPNALNGA--FQWVTIGYVDYFTLS 117
T+ + + +GPCK+ I I++ G I AP + +G+ W+ +V+ +S
Sbjct: 224 KTFLLRPIAFRGPCKSNYIYIELSGNIIAPKTKSEYSGSPINTWIGFSFVNGLIIS 391
Score = 24.6 bits (52), Expect(2) = 4e-07
Identities = 9/14 (64%), Positives = 10/14 (71%)
Frame = +3
Query: 118 GNGTFDGRGASAWK 131
G GT DGRG+ WK
Sbjct: 393 GKGTVDGRGSMWWK 434
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.133 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,440,878
Number of Sequences: 36976
Number of extensions: 171986
Number of successful extensions: 830
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 794
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 799
length of query: 392
length of database: 9,014,727
effective HSP length: 98
effective length of query: 294
effective length of database: 5,391,079
effective search space: 1584977226
effective search space used: 1584977226
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0490.17


