

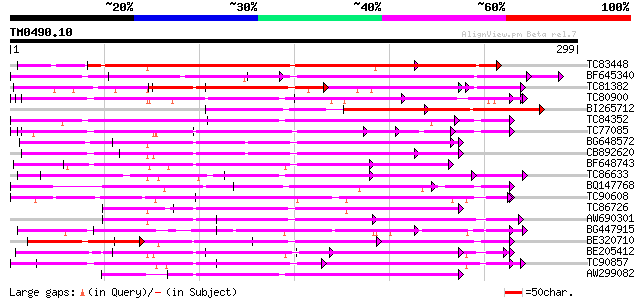
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0490.10
(299 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83448 weakly similar to GP|14269077|gb|AAK58011.1 verticillium... 259 1e-69
BF645340 weakly similar to GP|6016693|gb|A putative disease resi... 139 1e-33
TC81382 weakly similar to GP|14269077|gb|AAK58011.1 verticillium... 134 3e-32
TC80900 weakly similar to PIR|G84652|G84652 probable receptor-li... 133 7e-32
BI265712 weakly similar to GP|4235646|gb|A SC0A {Lycopersicon es... 110 7e-25
TC84352 weakly similar to GP|6979333|gb|AAF34426.1| leucine rich... 106 9e-24
TC77085 similar to GP|21536600|gb|AAM60932.1 putative disease re... 104 5e-23
BG648572 weakly similar to GP|9802748|gb Unknown protein {Arabid... 100 5e-22
CB892620 weakly similar to GP|22655012|g At1g73080/F3N23_28 {Ara... 100 1e-21
BF648743 weakly similar to PIR|H84632|H846 probable receptor-lik... 97 6e-21
TC86633 weakly similar to SP|Q00874|D100_ARATH DNA-damage-repair... 97 1e-20
BQ147768 weakly similar to PIR|H84632|H84 probable receptor-like... 96 1e-20
TC90608 weakly similar to GP|10716599|gb|AAG21897.1 putative dis... 95 4e-20
TC86726 similar to GP|14626935|gb|AAK70805.1 leucine-rich repeat... 94 5e-20
AW690301 weakly similar to GP|18175662|gb putative receptor prot... 92 3e-19
BG447915 similar to GP|7268809|emb leucine rich repeat-like prot... 91 7e-19
BE320710 weakly similar to GP|16924044|gb| Putative protein kina... 91 7e-19
BE205412 weakly similar to GP|14495542|gb receptor-like protein ... 90 9e-19
TC90857 weakly similar to GP|16930691|gb|AAL32011.1 AT4g26540/M3... 89 2e-18
AW299082 weakly similar to GP|10177183|d receptor protein kinase... 89 3e-18
>TC83448 weakly similar to GP|14269077|gb|AAK58011.1 verticillium wilt
disease resistance protein Ve2 {Lycopersicon
esculentum}, partial (13%)
Length = 774
Score = 259 bits (661), Expect = 1e-69
Identities = 133/218 (61%), Positives = 161/218 (73%)
Frame = +3
Query: 42 DLEGPLQKLNNVSSLHYLDLHNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSS 101
D EGPLQ N S L LDLHNNQL+GPIP+FP Y+DYS N+F SVIPQDI +Y++
Sbjct: 60 DFEGPLQ--NITSKLIALDLHNNQLKGPIPVFPEFASYLDYSMNKFDSVIPQDISNYLAF 233
Query: 102 AIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDN 161
FLSLS+N G+IP SLCNA+ LQVLD+SIN SG IPSCLM M + L VLNL+ N
Sbjct: 234 TTFLSLSNNTLQGSIPHSLCNASNLQVLDISINRISGAIPSCLMKMT--QTLVVLNLKMN 407
Query: 162 NLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKN 221
NL GTIPD+FP SC L TL+L+ N LHG IPKSL +CS LEVLDL +N+I FPC LKN
Sbjct: 408 NLIGTIPDVFPPSCVLRTLDLQKNNLHGQIPKSLVKCSALEVLDLAQNNIIDIFPCLLKN 587
Query: 222 ISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAF 259
IST+RV++LR N+F G +GC + + W LQI+D+AF
Sbjct: 588 ISTIRVIVLRKNKFYGRIGCPKTHG-TWPRLQIVDLAF 698
Score = 83.2 bits (204), Expect = 1e-16
Identities = 70/217 (32%), Positives = 99/217 (45%), Gaps = 5/217 (2%)
Frame = +3
Query: 5 STLTYLDLSKNQIHGVLPNWLWELNLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNN 64
S L LDL NQ+ G +P ++ YL+ S N D P N ++ +L L NN
Sbjct: 90 SKLIALDLHNNQLKGPIP--VFPEFASYLDYSMNKF-DSVIPQDISNYLAFTTFLSLSNN 260
Query: 65 QLQGPIP---IFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLC 121
LQG IP N+ +D S NR S IP + + + L+L N G IPD
Sbjct: 261 TLQGSIPHSLCNASNLQVLDISINRISGAIPSCLMKMTQTLVVLNLKMNNLIGTIPDVFP 440
Query: 122 NATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLN 181
+ L+ LDL NN G IP L+ K LEVL+L NN+ P + + +
Sbjct: 441 PSCVLRTLDLQKNNLHGQIPKSLV---KCSALEVLDLAQNNIIDIFPCLLKNISTIRVIV 611
Query: 182 LRGNQLHGPI--PKSLAQCSTLEVLDLGKNHITGGFP 216
LR N+ +G I PK+ L+++DL +P
Sbjct: 612 LRKNKFYGRIGCPKTHGTWPRLQIVDLAFTTSVENYP 722
Score = 33.9 bits (76), Expect = 0.078
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Frame = +2
Query: 243 QVNDEPWKILQIMDI--AFNNFSGTLKGKYFKNWETMMHD 280
+++ PW + Q D + NFSG L GK F WE M D
Sbjct: 641 RMSKNPWNLAQTSDS*SSLYNFSGKLPGKCFTTWEAMRAD 760
>BF645340 weakly similar to GP|6016693|gb|A putative disease resistance
protein {Arabidopsis thaliana}, partial (7%)
Length = 655
Score = 139 bits (351), Expect = 1e-33
Identities = 85/224 (37%), Positives = 127/224 (55%), Gaps = 1/224 (0%)
Frame = +2
Query: 53 VSSLHYLDLHNNQLQGPIPIFPVNVVY-VDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNK 111
+ L LDL NN+L G + + + ++ S+N F+S+ I L LS N
Sbjct: 2 LGKLESLDLSNNKLNGTVSNWLLETSRSLNLSQNLFTSI--DQISRNSDQLGDLDLSFNL 175
Query: 112 FHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMF 171
GN+ S+CN + L+ L+L NNF+G IP CL + +L++L+L+ NN GT+P+ F
Sbjct: 176 LVGNLSVSICNLSSLEFLNLGHNNFTGNIPQCLANLP---SLQILDLQMNNFYGTLPNNF 346
Query: 172 PASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILR 231
S L TLNL NQL G PKSL+ C L+VL+L N + FP +L+ + L+VL+LR
Sbjct: 347 SKSSKLITLNLNDNQLEGYFPKSLSHCENLQVLNLRNNKMEDKFPVWLQTLQYLKVLVLR 526
Query: 232 NNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKYFKNWE 275
+N+ G + ++ P+ L I DI+ NNF+G L Y K +E
Sbjct: 527 DNKLHGHIANLKIR-HPFPSLVIFDISSNNFTGPLPKAYLKYFE 655
Score = 58.5 bits (140), Expect = 3e-09
Identities = 47/167 (28%), Positives = 73/167 (43%)
Frame = +2
Query: 126 LQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGN 185
L+ LDLS N +GT+ + L+ ++ NL NL +I + S L L+L N
Sbjct: 11 LESLDLSNNKLNGTVSNWLLETSRSLNLS------QNLFTSIDQISRNSDQLGDLDLSFN 172
Query: 186 QLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVN 245
L G + S+ S+LE L+LG N+ TG P L N+ +L
Sbjct: 173 LLVGNLSVSICNLSSLEFLNLGHNNFTGNIPQCLANLPSL-------------------- 292
Query: 246 DEPWKILQIMDIAFNNFSGTLKGKYFKNWETMMHDEEDQYVSNFIHK 292
QI+D+ NNF GTL + K+ + + + D + + K
Sbjct: 293 -------QILDLQMNNFYGTLPNNFSKSSKLITLNLNDNQLEGYFPK 412
Score = 51.6 bits (122), Expect = 4e-07
Identities = 43/149 (28%), Positives = 66/149 (43%), Gaps = 4/149 (2%)
Frame = +2
Query: 1 LRNHSTLTYLDLSKNQIHGVLP-NWLWELNLHYLNISSNLLTDLEGPLQK-LNNVSSLHY 58
L N +L LDL N +G LP N+ L LN++ N LEG K L++ +L
Sbjct: 272 LANLPSLQILDLQMNNFYGTLPNNFSKSSKLITLNLNDN---QLEGYFPKSLSHCENLQV 442
Query: 59 LDLHNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPD 118
L+L NN+++ P++ + Y+ L L DNK HG+I +
Sbjct: 443 LNLRNNKMEDKFPVWLQTLQYLK----------------------VLVLRDNKLHGHIAN 556
Query: 119 SLCN--ATGLQVLDLSINNFSGTIPSCLM 145
L + D+S NNF+G +P +
Sbjct: 557 LKIRHPFPSLVIFDISSNNFTGPLPKAYL 643
>TC81382 weakly similar to GP|14269077|gb|AAK58011.1 verticillium wilt
disease resistance protein Ve2 {Lycopersicon
esculentum}, partial (16%)
Length = 1974
Score = 134 bits (338), Expect = 3e-32
Identities = 106/301 (35%), Positives = 157/301 (51%), Gaps = 31/301 (10%)
Frame = +3
Query: 3 NHSTLTYLDLSKNQIHGVLP---------------NWLWELNLHY---LNISSNLLTDLE 44
N + LT L LS N + G + +W +L+L++ +N S L +LE
Sbjct: 156 NLTNLTNLILSLNDLSGFVNFQHFSKLTNLRFLSLSWNTQLSLNFESNVNHSVFYLDELE 335
Query: 45 GPLQKLNNVS---------SLHYLDLHNNQLQGPIP--IFPVNVV-YVDYSRNRFSSVIP 92
L +N + +L YLDL NN+L G +P ++ N + +++ S+N F S I
Sbjct: 336 --LSSVNLIKFPKLQGKFPNLDYLDLSNNKLDGRMPNWLYEKNSLKFLNLSQNYFMS-ID 506
Query: 93 QDIGDYMSSAIF-LSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPE 151
Q I S+ + L LSDN IP +CN + L+ L+L NN +G IP CL A+
Sbjct: 507 QWINVNRSNGLSGLDLSDNLLDDEIPLVVCNISSLEFLNLGYNNLTGIIPQCL---AEST 677
Query: 152 NLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHI 211
+L+VLNL+ N GT+P F + +LNL GN+L G PKSL +C LE L+LG N I
Sbjct: 678 SLQVLNLQMNRFHGTLPSNFSKHSKIVSLNLYGNELEGRFPKSLFRCKKLEFLNLGVNKI 857
Query: 212 TGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKYF 271
FP +L+ + L+VL+LR+N+ GSL ++ ++ L I DI+ NN F
Sbjct: 858 EDNFPDWLQTMQDLKVLVLRDNKLHGSLVNLKIK-HSFQSLIIFDISGNNLRWVSTKSLF 1034
Query: 272 K 272
K
Sbjct: 1035K 1037
Score = 71.6 bits (174), Expect = 3e-13
Identities = 41/95 (43%), Positives = 60/95 (63%)
Frame = +1
Query: 74 PVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSI 133
P+ V +D+SRN+F IP IG+ + + L+LS N+ G+IP S+ N T L+ LDLS+
Sbjct: 1186 PIKFVSIDFSRNKFEGEIPNAIGE-LHALKGLNLSHNRLTGHIPKSIGNLTYLESLDLSL 1362
Query: 134 NNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIP 168
N +G IP+ L + LEV+NL +N+L G IP
Sbjct: 1363 NMLTGVIPAELTNL---NFLEVMNLSNNHLVGEIP 1458
Score = 58.2 bits (139), Expect = 4e-09
Identities = 45/141 (31%), Positives = 63/141 (43%), Gaps = 1/141 (0%)
Frame = +1
Query: 104 FLSLSDNKFHGNIPDSLCNAT-GLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNN 162
+L S KF DS+ AT G+Q+ + I P ++ N
Sbjct: 1093 YLHKSYEKFDAGYSDSVTVATKGIQMKLVKI----------------PIKFVSIDFSRNK 1224
Query: 163 LKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNI 222
+G IP+ L LNL N+L G IPKS+ + LE LDL N +TG P L N+
Sbjct: 1225 FEGEIPNAIGELHALKGLNLSHNRLTGHIPKSIGNLTYLESLDLSLNMLTGVIPAELTNL 1404
Query: 223 STLRVLILRNNRFQGSLGCGQ 243
+ L V+ L NN G + G+
Sbjct: 1405 NFLEVMNLSNNHLVGEIPRGK 1467
Score = 56.2 bits (134), Expect = 1e-08
Identities = 64/238 (26%), Positives = 91/238 (37%), Gaps = 74/238 (31%)
Frame = +3
Query: 76 NVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINN 135
++VY+ S NR + I + S L+L +NK GNIP+S+ N T L L LS+N+
Sbjct: 27 SLVYLYLSNNRLTGSISATSSYSLES---LNLYNNKLQGNIPESIFNLTNLTNLILSLND 197
Query: 136 FSGTIPSCLMTMAKPENLEVL--------------------------------------- 156
SG + +K NL L
Sbjct: 198 LSGFVN--FQHFSKLTNLRFLSLSWNTQLSLNFESNVNHSVFYLDELELSSVNLIKFPKL 371
Query: 157 ----------NLRDNNLKGTIPDMFPASCFLSTLNL---------------RGNQLHG-- 189
+L +N L G +P+ L LNL R N L G
Sbjct: 372 QGKFPNLDYLDLSNNKLDGRMPNWLYEKNSLKFLNLSQNYFMSIDQWINVNRSNGLSGLD 551
Query: 190 --------PIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSL 239
IP + S+LE L+LG N++TG P L ++L+VL L+ NRF G+L
Sbjct: 552 LSDNLLDDEIPLVVCNISSLEFLNLGYNNLTGIIPQCLAESTSLQVLNLQMNRFHGTL 725
Score = 29.3 bits (64), Expect = 1.9
Identities = 18/39 (46%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Frame = +1
Query: 3 NHSTLTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLL 40
N + L LDLS N + GV+P L LN L +N+S+N L
Sbjct: 1327 NLTYLESLDLSLNMLTGVIPAELTNLNFLEVMNLSNNHL 1443
>TC80900 weakly similar to PIR|G84652|G84652 probable receptor-like protein
kinase [imported] - Arabidopsis thaliana, partial (24%)
Length = 1054
Score = 133 bits (335), Expect = 7e-32
Identities = 97/274 (35%), Positives = 147/274 (53%), Gaps = 7/274 (2%)
Frame = +2
Query: 7 LTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPL-QKLNNVSSLHYLDLHNN 64
L ++ L N + +P + L +L++LN+ N LT GP+ + L N+++L YL L+ N
Sbjct: 20 LKWIYLGYNNLSREIPKNIGNLVSLNHLNLVYNNLT---GPIPESLGNLTNLQYLFLYLN 190
Query: 65 QLQGPIP--IFPV-NVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLC 121
+L GPIP IF + N++ +D S N S I + + I L L N F G IP+++
Sbjct: 191 KLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVNLQKLEI-LHLFSNNFTGKIPNTIT 367
Query: 122 NATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLN 181
+ LQVL L N +G IP T+ NL +L+L NNL G IP+ AS L +
Sbjct: 368 SLPHLQVLQLWSNKLTGEIPQ---TLGIHNNLTILDLSSNNLTGKIPNSLCASKNLHKII 538
Query: 182 LRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSLGC 241
L N L G IPK L C TLE + L N+++G P + + + +L + N+F
Sbjct: 539 LFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQLPQIYLLDISGNKFS----- 703
Query: 242 GQVNDEPWKI--LQIMDIAFNNFSGTLKGKYFKN 273
G++ND W + LQ++++A NNFSG L + N
Sbjct: 704 GKINDRKWNMPSLQMLNLANNNFSGDLPNSFGGN 805
Score = 94.0 bits (232), Expect = 6e-20
Identities = 77/212 (36%), Positives = 113/212 (52%), Gaps = 6/212 (2%)
Frame = +2
Query: 4 HSTLTYLDLSKNQIHGVLPNWLW-ELNLHYLNISSNLLTDLEGPLQK-LNNVSSLHYLDL 61
H+ LT LDLS N + G +PN L NLH + + SN L+G + K L + +L + L
Sbjct: 443 HNNLTILDLSSNNLTGKIPNSLCASKNLHKIILFSN---SLKGEIPKGLTSCKTLERVRL 613
Query: 62 HNNQLQGPIPI----FPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIP 117
+N L G +P+ P + +D S N+FS I D M S L+L++N F G++P
Sbjct: 614 QDNNLSGKLPLEITQLP-QIYLLDISGNKFSGKI-NDRKWNMPSLQMLNLANNNFSGDLP 787
Query: 118 DSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFL 177
+S ++ LDLS N FSG I + PE L L L +NNL G P+ L
Sbjct: 788 NSF-GGNKVEGLDLSQNQFSGYIQIGFKNL--PE-LVQLKLNNNNLFGKFPEELFQCNKL 955
Query: 178 STLNLRGNQLHGPIPKSLAQCSTLEVLDLGKN 209
+L+L N+L+G IP+ LA+ L +LD+ +N
Sbjct: 956 VSLDLSHNRLNGEIPEKLAKMPVLGLLDISEN 1051
Score = 88.6 bits (218), Expect = 3e-18
Identities = 88/306 (28%), Positives = 133/306 (42%), Gaps = 34/306 (11%)
Frame = +2
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQKLN-NVSSLHY 58
L N + L YL L N++ G +P ++ L NL L++S N L+ G + L N+ L
Sbjct: 146 LGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLS---GEISNLVVNLQKLEI 316
Query: 59 LDLHNNQLQGPIPIFPVNVVYVDYSR---NRFSSVIPQDIGDYMSSAIFLSLSDNKFHGN 115
L L +N G IP ++ ++ + N+ + IPQ +G + + I L LS N G
Sbjct: 317 LHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTI-LDLSSNNLTGK 493
Query: 116 IPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIP------- 168
IP+SLC + L + L N+ G IP L + + LE + L+DNNL G +P
Sbjct: 494 IPNSLCASKNLHKIILFSNSLKGEIPKGLTSC---KTLERVRLQDNNLSGKLPLEITQLP 664
Query: 169 ---------DMFPASC--------FLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHI 211
+ F L LNL N G +P S + +E LDL +N
Sbjct: 665 QIYLLDISGNKFSGKINDRKWNMPSLQMLNLANNNFSGDLPNSFGG-NKVEGLDLSQNQF 841
Query: 212 TGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQI-----MDIAFNNFSGTL 266
+G KN+ L L L NN G P ++ Q +D++ N +G +
Sbjct: 842 SGYIQIGFKNLPELVQLKLNNNNLFGKF--------PEELFQCNKLVSLDLSHNRLNGEI 997
Query: 267 KGKYFK 272
K K
Sbjct: 998 PEKLAK 1015
Score = 55.1 bits (131), Expect = 3e-08
Identities = 40/116 (34%), Positives = 58/116 (49%)
Frame = +2
Query: 151 ENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNH 210
+ L+ + L NNL IP L+ LNL N L GPIP+SL + L+ L L N
Sbjct: 14 KRLKWIYLGYNNLSREIPKNIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNK 193
Query: 211 ITGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTL 266
+TG P + N+ L L L +N G + VN + L+I+ + NNF+G +
Sbjct: 194 LTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVNLQK---LEILHLFSNNFTGKI 352
>BI265712 weakly similar to GP|4235646|gb|A SC0A {Lycopersicon esculentum},
partial (7%)
Length = 654
Score = 110 bits (275), Expect = 7e-25
Identities = 52/106 (49%), Positives = 71/106 (66%)
Frame = +1
Query: 177 LSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQ 236
L ++L N L GPIPKSL C L+VL+LGKN +TG FPCFL I TLR+++LR+N+
Sbjct: 1 LRFVDLNDNLLGGPIPKSLINCKELQVLNLGKNALTGRFPCFLSKIPTLRIMVLRSNKLH 180
Query: 237 GSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKYFKNWETMMHDEE 282
GS+ C + WK+L I+D+A NNFSG + +W+ MM DE+
Sbjct: 181 GSIRCPN-STGYWKMLHIVDLARNNFSGMISSALLNSWQAMMRDED 315
Score = 46.6 bits (109), Expect = 1e-05
Identities = 36/118 (30%), Positives = 49/118 (41%)
Frame = +1
Query: 104 FLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNL 163
F+ L+DN G IP SL N LQVL+L N +G P L +K L ++ LR N L
Sbjct: 7 FVDLNDNLLGGPIPKSLINCKELQVLNLGKNALTGRFPCFL---SKIPTLRIMVLRSNKL 177
Query: 164 KGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKN 221
G+I P S L ++DL +N+ +G L N
Sbjct: 178 HGSI----------------------RCPNSTGYWKMLHIVDLARNNFSGMISSALLN 285
>TC84352 weakly similar to GP|6979333|gb|AAF34426.1| leucine rich repeat
containing protein kinase {Oryza sativa}, partial (7%)
Length = 794
Score = 106 bits (265), Expect = 9e-24
Identities = 78/243 (32%), Positives = 118/243 (48%), Gaps = 6/243 (2%)
Frame = +1
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLW----ELNLHYLNISSNLLTDLEGPLQKLNNVSSL 56
++ L LD+SK I +P W W + +NIS+N +L+G + L +
Sbjct: 82 IQTQKYLLDLDISKAGISDNVPEWFWAKLSSQECNSINISNN---NLKGSIPNLQVKNHC 252
Query: 57 HYLDLHNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIF-LSLSDNKFHGN 115
L L +N+ +GPIP F +D S+N+FS P + ++ + +S+N+ G
Sbjct: 253 SLLSLSSNEFEGPIPAFLQGSALIDLSKNKFSDSRPFLCANGINEILAQFDVSNNQLSGR 432
Query: 116 IPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASC 175
IPD N L +DLS NNFSG IP+ +M L L LR+NNL G IP
Sbjct: 433 IPDCWSNFKSLVYVDLSHNNFSGKIPT---SMGSLVILRALLLRNNNLTGEIPFSLMNCT 603
Query: 176 FLSTLNLRGNQLHGPIPKSL-AQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNR 234
L L++R N+L G IP + ++ L+VL L N+ G P L ++ ++ L N
Sbjct: 604 QLVMLDMRDNRLEGHIPYWIGSELKELQVLSLKGNYFFGSLPLELCHLQFIQFFDLSLNS 783
Query: 235 FQG 237
G
Sbjct: 784 LSG 792
Score = 55.8 bits (133), Expect = 2e-08
Identities = 48/164 (29%), Positives = 72/164 (43%), Gaps = 2/164 (1%)
Frame = +1
Query: 105 LSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLK 164
+ L +K P + L LD+S S +P + +N+ +NNLK
Sbjct: 37 IGLRSSKLGLTFPKWIQTQKYLLDLDISKAGISDNVPEWFWAKLSSQECNSINISNNNLK 216
Query: 165 GTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKN--I 222
G+IP++ S L+L N+ GPIP L Q S L +DL KN + P N
Sbjct: 217 GSIPNL-QVKNHCSLLSLSSNEFEGPIPAFL-QGSAL--IDLSKNKFSDSRPFLCANGIN 384
Query: 223 STLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTL 266
L + NN+ G + N +K L +D++ NNFSG +
Sbjct: 385 EILAQFDVSNNQLSGRIPDCWSN---FKSLVYVDLSHNNFSGKI 507
>TC77085 similar to GP|21536600|gb|AAM60932.1 putative disease resistance
protein {Arabidopsis thaliana}, partial (88%)
Length = 1662
Score = 104 bits (259), Expect = 5e-23
Identities = 78/240 (32%), Positives = 119/240 (49%), Gaps = 9/240 (3%)
Frame = +2
Query: 5 STLTYLD----LSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPL-QKLNNVSSLHY 58
S L +LD ++ +I G P++L++L NL Y+ I +N L+ GP+ Q + +++ L
Sbjct: 341 SKLKFLDGIYLINLLKISGPFPDFLFKLPNLKYIYIENNTLS---GPIPQNIGSMNQLEA 511
Query: 59 LDLHNNQLQGPIPIFP---VNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGN 115
L N+ GPIP + + N + IP + + +++ +LSL N+ GN
Sbjct: 512 FSLQENKFTGPIPSSISALTKLTQLKLGNNFLTGTIPVSLKN-LTNLTYLSLQGNQLSGN 688
Query: 116 IPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASC 175
IPD + L +L LS N FSG IP + ++ L L L N+L G IPD
Sbjct: 689 IPDIFTSLKNLIILQLSHNKFSGNIPLSISSLYP--TLRYLELGHNSLSGKIPDFLGKFK 862
Query: 176 FLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRF 235
L TL+L NQ G +PKS A + + LDL N + FP + N+ + L L N F
Sbjct: 863 ALDTLDLSKNQFKGTVPKSFANLTKIFNLDLSDNFLVDPFP--VMNVKGIESLDLSRNMF 1036
Score = 83.2 bits (204), Expect = 1e-16
Identities = 71/206 (34%), Positives = 96/206 (46%), Gaps = 6/206 (2%)
Frame = +2
Query: 7 LTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQ-KLNNVSSLHYLDLHNN 64
L L +N+ G +P+ + L L L + +N LT G + L N+++L YL L N
Sbjct: 503 LEAFSLQENKFTGPIPSSISALTKLTQLKLGNNFLT---GTIPVSLKNLTNLTYLSLQGN 673
Query: 65 QLQGPIP-IFPV--NVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLC 121
QL G IP IF N++ + S N+FS IP I + +L L N G IPD L
Sbjct: 674 QLSGNIPDIFTSLKNLIILQLSHNKFSGNIPLSISSLYPTLRYLELGHNSLSGKIPDFLG 853
Query: 122 NATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLN 181
L LDLS N F GT+P + K NL+ L DN L P M + +L+
Sbjct: 854 KFKALDTLDLSKNQFKGTVPKSFANLTKIFNLD---LSDNFLVDPFPVMNVKG--IESLD 1018
Query: 182 LRGNQLH-GPIPKSLAQCSTLEVLDL 206
L N H IPK +A + L L
Sbjct: 1019LSRNMFHLKEIPKWVATSPIIYSLKL 1096
Score = 68.2 bits (165), Expect = 4e-12
Identities = 52/169 (30%), Positives = 81/169 (47%)
Frame = +2
Query: 98 YMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLN 157
++S I SLS KF G+ +++L SG P L K NL+ +
Sbjct: 311 FLSGTISPSLSKLKF----------LDGIYLINLL--KISGPFPDFLF---KLPNLKYIY 445
Query: 158 LRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPC 217
+ +N L G IP + L +L+ N+ GPIP S++ + L L LG N +TG P
Sbjct: 446 IENNTLSGPIPQNIGSMNQLEAFSLQENKFTGPIPSSISALTKLTQLKLGNNFLTGTIPV 625
Query: 218 FLKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTL 266
LKN++ L L L+ N+ G++ + K L I+ ++ N FSG +
Sbjct: 626 SLKNLTNLTYLSLQGNQLSGNIPDIFTS---LKNLIILQLSHNKFSGNI 763
Score = 64.7 bits (156), Expect = 4e-11
Identities = 59/194 (30%), Positives = 91/194 (46%), Gaps = 5/194 (2%)
Frame = +2
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQKLNNVSSLHYL 59
L+N + LTYL L NQ+ G +P+ L NL L +S N + PL + +L YL
Sbjct: 629 LKNLTNLTYLSLQGNQLSGNIPDIFTSLKNLIILQLSHNKFSG-NIPLSISSLYPTLRYL 805
Query: 60 DLHNNQLQGPIPIFPVN---VVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNI 116
+L +N L G IP F + +D S+N+F +P+ + ++ L LSDN
Sbjct: 806 ELGHNSLSGKIPDFLGKFKALDTLDLSKNQFKGTVPKSFAN-LTKIFNLDLSDNFLVDPF 982
Query: 117 PDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCF 176
P + N G++ LDLS N F + +A + L L +K + D P F
Sbjct: 983 P--VMNVKGIESLDLSRNMFH--LKEIPKWVATSPIIYSLKLAHCGIKMKLDDWKPLETF 1150
Query: 177 L-STLNLRGNQLHG 189
++L GN++ G
Sbjct: 1151FYDYIDLSGNEISG 1192
>BG648572 weakly similar to GP|9802748|gb Unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 697
Score = 100 bits (250), Expect = 5e-22
Identities = 65/188 (34%), Positives = 95/188 (49%), Gaps = 3/188 (1%)
Frame = +2
Query: 55 SLHYLDLHNNQLQGPIP---IFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNK 111
SL LD +N G +P F + ++ N+F I D+G ++ L L DN
Sbjct: 2 SLVQLDFTSNNFNGTLPPNLCFGKKLAKLNMGENQFIGRITSDVGS-CTTLTRLKLEDNY 178
Query: 112 FHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMF 171
F G +PD N + + L + NN +GTIPS L NL +L+L N+L G +P
Sbjct: 179 FTGPLPDFETNPS-ISYLSIGNNNINGTIPSSLSNCT---NLSLLDLSMNSLTGFVPLEL 346
Query: 172 PASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILR 231
L +L L N L GP+P L++C+ + V D+G N + G FP L++ + L L LR
Sbjct: 347 GNLLNLQSLKLSYNNLEGPLPHQLSKCTKMSVFDVGFNFLNGSFPSSLRSWTALTSLTLR 526
Query: 232 NNRFQGSL 239
NRF G +
Sbjct: 527 ENRFSGGI 550
Score = 97.4 bits (241), Expect = 6e-21
Identities = 70/231 (30%), Positives = 104/231 (44%), Gaps = 1/231 (0%)
Frame = +2
Query: 6 TLTYLDLSKNQIHGVLP-NWLWELNLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNN 64
+L LD + N +G LP N + L LN+ N + + + ++L L L +N
Sbjct: 2 SLVQLDFTSNNFNGTLPPNLCFGKKLAKLNMGENQF--IGRITSDVGSCTTLTRLKLEDN 175
Query: 65 QLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNAT 124
GP+P F N S +LS+ +N +G IP SL N T
Sbjct: 176 YFTGPLPDFETN-----------------------PSISYLSIGNNNINGTIPSSLSNCT 286
Query: 125 GLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRG 184
L +LDLS+N+ +G +P + + NL+ L L NNL+G +P +S ++
Sbjct: 287 NLSLLDLSMNSLTGFVP---LELGNLLNLQSLKLSYNNLEGPLPHQLSKCTKMSVFDVGF 457
Query: 185 NQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRF 235
N L+G P SL + L L L +N +GG P FL L L L N+F
Sbjct: 458 NFLNGSFPSSLRSWTALTSLTLRENRFSGGIPDFLSAFENLNELKLDGNKF 610
>CB892620 weakly similar to GP|22655012|g At1g73080/F3N23_28 {Arabidopsis
thaliana}, partial (9%)
Length = 704
Score = 99.8 bits (247), Expect = 1e-21
Identities = 70/215 (32%), Positives = 108/215 (49%), Gaps = 5/215 (2%)
Frame = +1
Query: 7 LTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNNQ 65
L L++ NQ+ G +P+ L L L ++ N T G L + +L Y+D+ N
Sbjct: 64 LLELNMGINQLQGGIPSDLGRCATLRRLFLNQNNFT---GSLPDFASNLNLKYMDISKNN 234
Query: 66 LQGPIPIFP---VNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCN 122
+ GPIP N+ Y++ SRN+F+ +IP ++G+ ++ I L LS N G +P L N
Sbjct: 235 ISGPIPSSLGNCTNLTYINLSRNKFARLIPSELGNLLNLVI-LELSHNNLEGPLPHQLSN 411
Query: 123 ATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNL 182
+ + D+ N +G++PS L + N+ L LR+N G IP+ L L L
Sbjct: 412 CSHMDRFDIGFNFLNGSLPSNLRSWT---NITTLILRENYFTGGIPEFLAKFRNLRELQL 582
Query: 183 RGNQLHGPIPKSLAQCSTLEV-LDLGKNHITGGFP 216
GN L G IP+S+ L L+L N + GG P
Sbjct: 583 GGNLLGGKIPRSIVTLRNLFYGLNLSANGLIGGIP 687
Score = 94.4 bits (233), Expect = 5e-20
Identities = 63/184 (34%), Positives = 91/184 (49%), Gaps = 3/184 (1%)
Frame = +1
Query: 59 LDLHNNQLQGPIP---IFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGN 115
LD NN+ G IP F +++ ++ N+ IP D+G ++ L L+ N F G+
Sbjct: 1 LDCMNNKFNGNIPPNLCFGKHLLELNMGINQLQGGIPSDLGR-CATLRRLFLNQNNFTGS 177
Query: 116 IPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASC 175
+PD N L+ +D+S NN SG IPS L NL +NL N IP
Sbjct: 178 LPDFASNLN-LKYMDISKNNISGPIPSSLGNCT---NLTYINLSRNKFARLIPSELGNLL 345
Query: 176 FLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRF 235
L L L N L GP+P L+ CS ++ D+G N + G P L++ + + LILR N F
Sbjct: 346 NLVILELSHNNLEGPLPHQLSNCSHMDRFDIGFNFLNGSLPSNLRSWTNITTLILRENYF 525
Query: 236 QGSL 239
G +
Sbjct: 526 TGGI 537
>BF648743 weakly similar to PIR|H84632|H846 probable receptor-like protein
kinase [imported] - Arabidopsis thaliana, partial (9%)
Length = 648
Score = 97.4 bits (241), Expect = 6e-21
Identities = 73/211 (34%), Positives = 108/211 (50%), Gaps = 5/211 (2%)
Frame = +1
Query: 29 NLHYLNISSNLLTDLEGPLQ-KLNNVSSLHYLDLHNNQLQGPIPI---FPVNVVYVDYSR 84
NL YL + N + GP+ ++ +++L +L + + L G IP F N+ Y+D S+
Sbjct: 10 NLSYLILGGNNWSG--GPIPPEIGKLNNLLHLAIQKSNLVGSIPQEIGFLTNLAYIDLSK 183
Query: 85 NRFSSVIPQDIGDYMSSAIFLSLSDN-KFHGNIPDSLCNATGLQVLDLSINNFSGTIPSC 143
N S IP+ IG+ +S L LS+N K G IP SL N + L VL SG+IP
Sbjct: 184 NSLSGGIPETIGN-LSKLDTLVLSNNTKMSGPIPHSLWNMSSLTVLYFDNIGLSGSIPDS 360
Query: 144 LMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEV 203
+ + NL+ L L N+L G+IP L L L N L GPIP S+ L+V
Sbjct: 361 IQNLV---NLKELALDINHLSGSIPSTIGDLKNLIKLYLGSNNLSGPIPASIGNLINLQV 531
Query: 204 LDLGKNHITGGFPCFLKNISTLRVLILRNNR 234
L + +N++TG P + N+ L V + N+
Sbjct: 532 LSVQENNLTGTIPASIGNLKWLTVFEVXTNK 624
Score = 70.9 bits (172), Expect = 6e-13
Identities = 65/218 (29%), Positives = 102/218 (45%), Gaps = 28/218 (12%)
Frame = +1
Query: 3 NHSTLTYLDLSKNQIHG-VLPNWLWELN--LHYLNISSNLLTDLEGPLQKLNNVSSLHYL 59
N + L+YL L N G +P + +LN LH SNL+ + Q++ +++L Y+
Sbjct: 1 NLTNLSYLILGGNNWSGGPIPPEIGKLNNLLHLAIQKSNLVGSIP---QEIGFLTNLAYI 171
Query: 60 DLHNNQLQGPIPIFPVNVVYVDY----SRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGN 115
DL N L G IP N+ +D + + S IP + + MSS L + G+
Sbjct: 172 DLSKNSLSGGIPETIGNLSKLDTLVLSNNTKMSGPIPHSLWN-MSSLTVLYFDNIGLSGS 348
Query: 116 IPDSLCNATGLQVLDLSINNFSGTIPSCL---------------------MTMAKPENLE 154
IPDS+ N L+ L L IN+ SG+IPS + ++ NL+
Sbjct: 349 IPDSIQNLVNLKELALDINHLSGSIPSTIGDLKNLIKLYLGSNNLSGPIPASIGNLINLQ 528
Query: 155 VLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIP 192
VL++++NNL GTIP +L+ + N+ IP
Sbjct: 529 VLSVQENNLTGTIPASIGNLKWLTVFEVXTNKPSWRIP 642
>TC86633 weakly similar to SP|Q00874|D100_ARATH DNA-damage-repair/toleration
protein DRT100 precursor. [Mouse-ear cress] {Arabidopsis
thaliana}, partial (93%)
Length = 1407
Score = 96.7 bits (239), Expect = 1e-20
Identities = 74/237 (31%), Positives = 120/237 (50%), Gaps = 7/237 (2%)
Frame = +3
Query: 17 IHGVLPNWLWELN-LHYLNISSNLLT-DLEGPLQKLNNVSSLHYLDLHNNQLQGPIPIFP 74
I G +P+ + L+ L L+++ N ++ ++ G + KL +++ L+ D N + G IP+
Sbjct: 372 ISGEIPSCITSLSSLRILDLTGNKISGNIPGNIGKLQHLTVLNLAD---NAISGEIPMSI 542
Query: 75 VNV---VYVDYSRNRFSSVIPQDIGDY--MSSAIFLSLSDNKFHGNIPDSLCNATGLQVL 129
V + +++D + N+ S +P DIG +S A+F S N+ G+IPDS+ L L
Sbjct: 543 VRISGLMHLDLAGNQISGELPSDIGKLRRLSRALF---SRNQLTGSIPDSVLKMNRLADL 713
Query: 130 DLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHG 189
DLS+N +G+IP+ + K L L L N++ G IP ++ + LNL N G
Sbjct: 714 DLSMNRITGSIPA---RIGKMRVLSTLKLDGNSMTGQIPSTLLSNTGMGILNLSRNGFEG 884
Query: 190 PIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVND 246
IP S VLDL N +TG P L + + L + NN G++ G D
Sbjct: 885 TIPDVFGSKSYFMVLDLSFNKLTGRIPGSLSSAKFMGHLDISNNHLCGTIPIGSPFD 1055
Score = 75.1 bits (183), Expect = 3e-14
Identities = 63/191 (32%), Positives = 86/191 (44%), Gaps = 3/191 (1%)
Frame = +3
Query: 5 STLTYLDLSKNQIHGVLPNWLWELNLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNN 64
S L +LDL+ NQI G LP+ + G L++L+ N
Sbjct: 552 SGLMHLDLAGNQISGELPSDI-------------------GKLRRLSRAL------FSRN 656
Query: 65 QLQGPIP--IFPVN-VVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLC 121
QL G IP + +N + +D S NR + IP IG M L L N G IP +L
Sbjct: 657 QLTGSIPDSVLKMNRLADLDLSMNRITGSIPARIGK-MRVLSTLKLDGNSMTGQIPSTLL 833
Query: 122 NATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLN 181
+ TG+ +L+LS N F GTIP VL+L N L G IP ++ F+ L+
Sbjct: 834 SNTGMGILNLSRNGFEGTIPD---VFGSKSYFMVLDLSFNKLTGRIPGSLSSAKFMGHLD 1004
Query: 182 LRGNQLHGPIP 192
+ N L G IP
Sbjct: 1005ISNNHLCGTIP 1037
Score = 72.8 bits (177), Expect = 2e-13
Identities = 47/160 (29%), Positives = 75/160 (46%)
Frame = +3
Query: 114 GNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPA 173
G IP + + + L++LDL+ N SG IP + K ++L VLNL DN + G IP
Sbjct: 378 GEIPSCITSLSSLRILDLTGNKISGNIPG---NIGKLQHLTVLNLADNAISGEIPMSIVR 548
Query: 174 SCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNN 233
L L+L GNQ+ G +P + + L +N +TG P + ++ L L L N
Sbjct: 549 ISGLMHLDLAGNQISGELPSDIGKLRRLSRALFSRNQLTGSIPDSVLKMNRLADLDLSMN 728
Query: 234 RFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKYFKN 273
R GS+ ++L + + N+ +G + N
Sbjct: 729 RITGSI---PARIGKMRVLSTLKLDGNSMTGQIPSTLLSN 839
>BQ147768 weakly similar to PIR|H84632|H84 probable receptor-like protein
kinase [imported] - Arabidopsis thaliana, partial (12%)
Length = 712
Score = 96.3 bits (238), Expect = 1e-20
Identities = 73/228 (32%), Positives = 99/228 (43%), Gaps = 3/228 (1%)
Frame = +3
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWELNLHYLNISSNLLTDLEGPLQKLNNVSSLHYLD 60
L N S LT+LD S N + G +PN L N L YLD
Sbjct: 6 LGNLSKLTHLDFSYNSLEGEIPN-------------------------SLGNHRQLKYLD 110
Query: 61 LHNNQLQGPIPIFPVNVVYV---DYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIP 117
+ NN L G IP + Y+ + S NR S IP +G+ + L + N G IP
Sbjct: 111 ISNNNLNGSIPHELGFIKYLGSLNLSTNRISGDIPPSLGNLVKLT-HLVIYGNSLVGKIP 287
Query: 118 DSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFL 177
S+ N L+ L++S N G+IP L + +NL L L N +KG IP L
Sbjct: 288 PSIGNLRSLESLEISDNYIQGSIPPRLGLL---KNLTTLRLSHNRIKGEIPPSLGNLKQL 458
Query: 178 STLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTL 225
L++ N + G +P L L LDL N + G P LKN++ L
Sbjct: 459 EELDISNNNIQGFLPFELGLLKNLTTLDLSHNRLNGNLPISLKNLTQL 602
Score = 67.4 bits (163), Expect = 6e-12
Identities = 57/175 (32%), Positives = 80/175 (45%), Gaps = 27/175 (15%)
Frame = +3
Query: 119 SLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLS 178
SL N + L LD S N+ G IP+ L L+ L++ +NNL G+IP +L
Sbjct: 3 SLGNLSKLTHLDFSYNSLEGEIPNSL---GNHRQLKYLDISNNNLNGSIPHELGFIKYLG 173
Query: 179 TLNLR------------------------GNQLHGPIPKSLAQCSTLEVLDLGKNHITGG 214
+LNL GN L G IP S+ +LE L++ N+I G
Sbjct: 174 SLNLSTNRISGDIPPSLGNLVKLTHLVIYGNSLVGKIPPSIGNLRSLESLEISDNYIQGS 353
Query: 215 FP---CFLKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTL 266
P LKN++TLR L +NR +G + N K L+ +DI+ NN G L
Sbjct: 354 IPPRLGLLKNLTTLR---LSHNRIKGEIPPSLGN---LKQLEELDISNNNIQGFL 500
Score = 37.0 bits (84), Expect = 0.009
Identities = 30/78 (38%), Positives = 45/78 (57%), Gaps = 2/78 (2%)
Frame = +3
Query: 7 LTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQ-KLNNVSSLHYLDLHNN 64
LT L LS N+I G +P L L L L+IS+N +++G L +L + +L LDL +N
Sbjct: 384 LTTLRLSHNRIKGEIPPSLGNLKQLEELDISNN---NIQGFLPFELGLLKNLTTLDLSHN 554
Query: 65 QLQGPIPIFPVNVVYVDY 82
+L G +PI N+ + Y
Sbjct: 555 RLNGNLPISLKNLTQLIY 608
>TC90608 weakly similar to GP|10716599|gb|AAG21897.1 putative disease
resistance protein (3' partial) {Oryza sativa}, partial
(6%)
Length = 1494
Score = 94.7 bits (234), Expect = 4e-20
Identities = 95/296 (32%), Positives = 138/296 (46%), Gaps = 31/296 (10%)
Frame = +1
Query: 1 LRNHSTLTYLDL----------SKNQIHGVLPNWLWELNLHYLNISSNLLTDLEGPLQKL 50
L N ++LT+L L S ++ G LP L EL+L ++S + + L K
Sbjct: 274 LSNLTSLTHLHLMSISNLDKFNSWLKMVGKLPK-LRELSLRNCDLSDHFIHSLSQ--SKF 444
Query: 51 NNVSSLHYLDLHNNQLQGPIPIFPV------NVVYVDYSRNRFSSVIPQDIGDYMSSAIF 104
N +SL LDL N + IFP+ N+V +D S N + D G M+S
Sbjct: 445 NFSNSLSILDLSVNNFVSSM-IFPLLSNISSNLVELDLSFNHLEAPPSIDYGIVMNSLER 621
Query: 105 LSLSDNKFHGNIPDSLCNATGLQVLDLSI-NNFSGTIPSCLMTMAKP---ENLEVLNLRD 160
L LS N+ G + S N L LDLS NN + + L ++ +L+VL++
Sbjct: 622 LGLSGNRLKGGVFKSFMNVCTLSSLDLSRQNNLTEDLQIILQNLSSGCVRNSLQVLDISY 801
Query: 161 NNLKGTIPDMFPASCF--LSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFP-C 217
N + GT+PD+ S F L TL+L NQL G IP+ + LE D+ N + GG P
Sbjct: 802 NEIAGTLPDL---SIFTSLKTLDLSSNQLSGKIPEGSSLPFQLEYFDIRSNSLEGGIPKS 972
Query: 218 FLKNISTLRVLILRNNRFQGSL--------GCGQVNDEPWKILQIMDIAFNNFSGT 265
F N L+ L L NRF G L C + + L+ +D++FN +GT
Sbjct: 973 FWMNACKLKSLTLSKNRFSGELQVIIDHLPKCARYS------LRELDLSFNQINGT 1122
Score = 43.1 bits (100), Expect = 1e-04
Identities = 46/172 (26%), Positives = 76/172 (43%), Gaps = 4/172 (2%)
Frame = +1
Query: 99 MSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNL 158
+S +L+LS N G IP L + + LQ LDLS N G+IPS L + + L + +
Sbjct: 10 LSHLKYLNLSWNHLDGLIPHQLGDLSNLQFLDLSYNFLEGSIPSQLGKLVNLQELYLGSA 189
Query: 159 RDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCF 218
+ TI ++ + ST + G L +L + L ++ + +
Sbjct: 190 YYDIANLTIDNIINLTIDNSTDHSGGQWL-----SNLTSLTHLHLMSISNLDKFNSWLKM 354
Query: 219 LKNISTLRVLILRN----NRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTL 266
+ + LR L LRN + F SL + N L I+D++ NNF ++
Sbjct: 355 VGKLPKLRELSLRNCDLSDHFIHSLSQSKFNFS--NSLSILDLSVNNFVSSM 504
Score = 40.0 bits (92), Expect = 0.001
Identities = 30/109 (27%), Positives = 45/109 (40%)
Frame = +1
Query: 171 FPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLIL 230
F + L LNL N L G IP L S L+ LDL N + G P L + L+ L
Sbjct: 1 FESLSHLKYLNLSWNHLDGLIPHQLGDLSNLQFLDLSYNFLEGSIPSQLGKLVNLQELY- 177
Query: 231 RNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKYFKNWETMMH 279
LG + I I+++ +N + G++ N ++ H
Sbjct: 178 --------LGSAYYDIANLTIDNIINLTIDNSTDHSGGQWLSNLTSLTH 300
>TC86726 similar to GP|14626935|gb|AAK70805.1 leucine-rich repeat resistance
protein-like protein {Gossypium hirsutum}, partial (92%)
Length = 1507
Score = 94.4 bits (233), Expect = 5e-20
Identities = 70/198 (35%), Positives = 103/198 (51%), Gaps = 8/198 (4%)
Frame = +2
Query: 50 LNNVSSLHYLDLHNNQLQGPIP-----IFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIF 104
+ ++ L LDLHNN+L GPIP + + ++ + + N+ IP +IG+ + S
Sbjct: 479 VTSLLDLTRLDLHNNKLTGPIPPQIGRLKRLKILNLRW--NKLQDAIPPEIGE-LKSLTH 649
Query: 105 LSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLK 164
L LS N F G IP L + L+ L L N +G IP L T+ +NL L+ +N+L
Sbjct: 650 LYLSFNSFKGEIPRELADLPDLRYLYLHENRLTGRIPPELGTL---QNLRHLDAGNNHLV 820
Query: 165 GTIPDMFP-ASCF--LSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKN 221
GTI ++ CF L L L N G IP LA S+LE+L L N ++G P + +
Sbjct: 821 GTIRELIRIEGCFPSLRNLYLNNNYFTGGIPAQLANLSSLEILYLSYNKMSGVIPSSVAH 1000
Query: 222 ISTLRVLILRNNRFQGSL 239
I L L L +N+F G +
Sbjct: 1001IPKLTYLYLDHNQFSGRI 1054
Score = 60.5 bits (145), Expect = 8e-10
Identities = 48/153 (31%), Positives = 68/153 (44%)
Frame = +2
Query: 87 FSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMT 146
+S V +GDY L + G P ++ + L LDL N +G IP
Sbjct: 383 WSGVTCSTVGDYRV-VTELEVYAVSIVGPFPTAVTSLLDLTRLDLHNNKLTGPIPP---Q 550
Query: 147 MAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDL 206
+ + + L++LNLR N L+ IP L+ L L N G IP+ LA L L L
Sbjct: 551 IGRLKRLKILNLRWNKLQDAIPPEIGELKSLTHLYLSFNSFKGEIPRELADLPDLRYLYL 730
Query: 207 GKNHITGGFPCFLKNISTLRVLILRNNRFQGSL 239
+N +TG P L + LR L NN G++
Sbjct: 731 HENRLTGRIPPELGTLQNLRHLDAGNNHLVGTI 829
>AW690301 weakly similar to GP|18175662|gb putative receptor protein kinase
{Arabidopsis thaliana}, partial (18%)
Length = 673
Score = 91.7 bits (226), Expect = 3e-19
Identities = 61/163 (37%), Positives = 81/163 (49%), Gaps = 1/163 (0%)
Frame = +1
Query: 110 NKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPD 169
NK G IP+SL LQ LDLS NN +G+IP L + NL L L N+L+G IP
Sbjct: 19 NKLTGKIPNSLSECQNLQALDLSYNNLTGSIPKQLFVL---RNLTQLMLISNDLEGLIPP 189
Query: 170 MFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLI 229
L L L N+L G IP +A L LDL NH+ G P +S L VL
Sbjct: 190 DIGNCTSLYRLRLNQNRLVGTIPSEIANLKNLNFLDLHYNHLVGEIPSQFSGLSKLGVLD 369
Query: 230 LRNNRFQGSL-GCGQVNDEPWKILQIMDIAFNNFSGTLKGKYF 271
L +N+ G+L +++ L ++++FN FSG L F
Sbjct: 370 LSHNKLSGNLDAISNLHN-----LVSLNVSFNEFSGELPNSPF 483
Score = 64.7 bits (156), Expect = 4e-11
Identities = 51/148 (34%), Positives = 72/148 (48%), Gaps = 3/148 (2%)
Frame = +1
Query: 50 LNNVSSLHYLDLHNNQLQGPIP--IFPV-NVVYVDYSRNRFSSVIPQDIGDYMSSAIFLS 106
L+ +L LDL N L G IP +F + N+ + N +IP DIG+ +S L
Sbjct: 49 LSECQNLQALDLSYNNLTGSIPKQLFVLRNLTQLMLISNDLEGLIPPDIGN-CTSLYRLR 225
Query: 107 LSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGT 166
L+ N+ G IP + N L LDL N+ G IPS ++K L VL+L N L G
Sbjct: 226 LNQNRLVGTIPSEIANLKNLNFLDLHYNHLVGEIPSQFSGLSK---LGVLDLSHNKLSGN 396
Query: 167 IPDMFPASCFLSTLNLRGNQLHGPIPKS 194
+ D L +LN+ N+ G +P S
Sbjct: 397 L-DAISNLHNLVSLNVSFNEFSGELPNS 477
>BG447915 similar to GP|7268809|emb leucine rich repeat-like protein
{Arabidopsis thaliana}, partial (4%)
Length = 669
Score = 90.5 bits (223), Expect = 7e-19
Identities = 81/231 (35%), Positives = 109/231 (47%), Gaps = 9/231 (3%)
Frame = +3
Query: 45 GPLQKLNNVSSLHYLDLHNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIF 104
G L L+ +SL LDL +NQL G IP + RN S G +S
Sbjct: 18 GTLPDLSAFTSLKTLDLSSNQLTGEIPGLQTIL------RNLSS-------GCVRNSLQV 158
Query: 105 LSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSC------LMTMAKPENLEVLNL 158
L LSDN+ G +PD L T L+ L LS N +G IP L + +L+VL+L
Sbjct: 159 LDLSDNRITGTLPD-LSAFTSLKTLYLSSNQLTGEIPGLQTILRNLSSGCVRNSLQVLDL 335
Query: 159 RDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFP-C 217
DN + GT+PD+ A L TL+L NQL G IP + LE L + N + G P
Sbjct: 336 SDNRITGTLPDL-SAFTSLKTLDLSSNQLSGEIPGGSSLPYQLEHLSITSNTLEGVIPKS 512
Query: 218 FLKNISTLRVLILRNNRFQGSLG--CGQVNDEPWKILQIMDIAFNNFSGTL 266
F N L+ L + NN F G L ++ LQ +D++ N +GTL
Sbjct: 513 FWMNACKLKSLKMSNNSFSGELQVIIHHLSXCAXYSLQELDLSXNKINGTL 665
Score = 82.0 bits (201), Expect = 2e-16
Identities = 79/228 (34%), Positives = 106/228 (45%), Gaps = 16/228 (7%)
Frame = +3
Query: 5 STLTYLDLSKNQIHGVLPNWLWEL----------NLHYLNISSNLLTDLEGPLQKLNNVS 54
++L LDLS NQ+ G +P L +L L++S N +T G L L+ +
Sbjct: 45 TSLKTLDLSSNQLTGEIPGLQTILRNLSSGCVRNSLQVLDLSDNRIT---GTLPDLSAFT 215
Query: 55 SLHYLDLHNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHG 114
SL L L +NQL G IP + RN S G +S L LSDN+ G
Sbjct: 216 SLKTLYLSSNQLTGEIPGLQTIL------RNLSS-------GCVRNSLQVLDLSDNRITG 356
Query: 115 NIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMF-PA 173
+PD L T L+ LDLS N SG IP + P LE L++ N L+G IP F
Sbjct: 357 TLPD-LSAFTSLKTLDLSSNQLSGEIPG---GSSLPYQLEHLSITSNTLEGVIPKSFWMN 524
Query: 174 SCFLSTLNLRGNQLHGP---IPKSLAQCS--TLEVLDLGKNHITGGFP 216
+C L +L + N G I L+ C+ +L+ LDL N I G P
Sbjct: 525 ACKLKSLKMSNNSFSGELQVIIHHLSXCAXYSLQELDLSXNKINGTLP 668
Score = 73.2 bits (178), Expect = 1e-13
Identities = 61/179 (34%), Positives = 88/179 (49%), Gaps = 15/179 (8%)
Frame = +3
Query: 110 NKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSC------LMTMAKPENLEVLNLRDNNL 163
N G +PD L T L+ LDLS N +G IP L + +L+VL+L DN +
Sbjct: 6 NGIIGTLPD-LSAFTSLKTLDLSSNQLTGEIPGLQTILRNLSSGCVRNSLQVLDLSDNRI 182
Query: 164 KGTIPDMFPASCFLSTLNLRGNQLHGPIP---------KSLAQCSTLEVLDLGKNHITGG 214
GT+PD+ A L TL L NQL G IP S ++L+VLDL N ITG
Sbjct: 183 TGTLPDL-SAFTSLKTLYLSSNQLTGEIPGLQTILRNLSSGCVRNSLQVLDLSDNRITGT 359
Query: 215 FPCFLKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKYFKN 273
P L ++L+ L L +N+ G + G + P++ L+ + I N G + ++ N
Sbjct: 360 LP-DLSAFTSLKTLDLSSNQLSGEIPGG--SSLPYQ-LEHLSITSNTLEGVIPKSFWMN 524
>BE320710 weakly similar to GP|16924044|gb| Putative protein kinase {Oryza
sativa}, partial (12%)
Length = 476
Score = 90.5 bits (223), Expect = 7e-19
Identities = 57/144 (39%), Positives = 80/144 (54%), Gaps = 3/144 (2%)
Frame = +3
Query: 56 LHYLDLHNNQLQGPIPIFPVNV---VYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKF 112
L L L NN L G +P+ ++ ++ + N S IP+ +G +S + L+LS NKF
Sbjct: 6 LFKLSLSNNHLSGEVPVQIASLHQLTALELAINNLSGFIPKKLG-MLSMLLQLNLSQNKF 182
Query: 113 HGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFP 172
GNIP ++ LDLS N+ +GTIP+ L + +LE LNL NNL GTIP F
Sbjct: 183 EGNIPVEFGQLNVIENLDLSGNSMNGTIPAMLGQL---NHLETLNLSHNNLSGTIPSSFV 353
Query: 173 ASCFLSTLNLRGNQLHGPIPKSLA 196
L+T+++ NQL GPIP A
Sbjct: 354 DMLSLTTVDISYNQLEGPIPNVTA 425
Score = 66.2 bits (160), Expect = 1e-11
Identities = 45/138 (32%), Positives = 63/138 (45%)
Frame = +3
Query: 129 LDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLH 188
L LS N+ SG +P + +A L L L NNL G IP L LNL N+
Sbjct: 15 LSLSNNHLSGEVP---VQIASLHQLTALELAINNLSGFIPKKLGMLSMLLQLNLSQNKFE 185
Query: 189 GPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVNDEP 248
G IP Q + +E LDL N + G P L ++ L L L +N G++ V+
Sbjct: 186 GNIPVEFGQLNVIENLDLSGNSMNGTIPAMLGQLNHLETLNLSHNNLSGTIPSSFVD--- 356
Query: 249 WKILQIMDIAFNNFSGTL 266
L +DI++N G +
Sbjct: 357 MLSLTTVDISYNQLEGPI 410
Score = 41.2 bits (95), Expect = 5e-04
Identities = 27/64 (42%), Positives = 39/64 (60%), Gaps = 2/64 (3%)
Frame = +3
Query: 10 LDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPL-QKLNNVSSLHYLDLHNNQLQ 67
LDLS N ++G +P L +LN L LN+S N +L G + ++ SL +D+ NQL+
Sbjct: 231 LDLSGNSMNGTIPAMLGQLNHLETLNLSHN---NLSGTIPSSFVDMLSLTTVDISYNQLE 401
Query: 68 GPIP 71
GPIP
Sbjct: 402 GPIP 413
>BE205412 weakly similar to GP|14495542|gb receptor-like protein kinase
INRPK1 {Ipomoea nil}, partial (7%)
Length = 627
Score = 90.1 bits (222), Expect = 9e-19
Identities = 61/189 (32%), Positives = 106/189 (55%), Gaps = 4/189 (2%)
Frame = +2
Query: 55 SLHYLDLHNNQLQGPI-PIFP--VNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNK 111
+L ++ L N+L G + P + +++ ++ S N+ S IP D+ +S FLSL N+
Sbjct: 32 NLSFISLSRNRLIGYLSPDWGKCISLTEMEMSGNKLSGKIPIDLNK-LSKLQFLSLHSNE 208
Query: 112 FHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMF 171
F GNIP + N + L +L+LS N+ SG IP + +A+ L +++L DNN G+IP+
Sbjct: 209 FTGNIPHEIGNISLLFMLNLSRNHLSGEIPKSIGRLAQ---LNIVDLSDNNFSGSIPNEL 379
Query: 172 PASCFLSTLNLRGNQLHGPIPKSLAQCSTLE-VLDLGKNHITGGFPCFLKNISTLRVLIL 230
L ++NL N L G IP L +L+ +LDL N+++ P L+ +++L + +
Sbjct: 380 GNCNRLLSMNLSHNDLSGMIPYELGNLYSLQSLLDLSSNNLSREIPQNLQKLASLEIFNV 559
Query: 231 RNNRFQGSL 239
+N G++
Sbjct: 560 SHNNLSGTI 586
Score = 89.4 bits (220), Expect = 2e-18
Identities = 65/194 (33%), Positives = 103/194 (52%), Gaps = 26/194 (13%)
Frame = +2
Query: 4 HSTLTYLDLSKNQIHGVL-PNWLWELNLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLH 62
H L+++ LS+N++ G L P+W ++L + +S N L+ + P+ LN +S L +L LH
Sbjct: 26 HPNLSFISLSRNRLIGYLSPDWGKCISLTEMEMSGNKLSG-KIPID-LNKLSKLQFLSLH 199
Query: 63 NNQLQGPIP--IFPVNVVYV-DYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDS 119
+N+ G IP I ++++++ + SRN S IP+ IG ++ + LSDN F G+IP+
Sbjct: 200 SNEFTGNIPHEIGNISLLFMLNLSRNHLSGEIPKSIGR-LAQLNIVDLSDNNFSGSIPNE 376
Query: 120 LCNATGLQVLDLSINNFSGTIPSCL----------------------MTMAKPENLEVLN 157
L N L ++LS N+ SG IP L + K +LE+ N
Sbjct: 377 LGNCNRLLSMNLSHNDLSGMIPYELGNLYSLQSLLDLSSNNLSREIPQNLQKLASLEIFN 556
Query: 158 LRDNNLKGTIPDMF 171
+ NNL GTIP F
Sbjct: 557 VSHNNLSGTIPQSF 598
Score = 73.6 bits (179), Expect = 9e-14
Identities = 50/160 (31%), Positives = 77/160 (47%)
Frame = +2
Query: 104 FLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNL 163
F+SLS N+ G + L +++S N SG IP L ++K L+ L+L N
Sbjct: 41 FISLSRNRLIGYLSPDWGKCISLTEMEMSGNKLSGKIPIDLNKLSK---LQFLSLHSNEF 211
Query: 164 KGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNIS 223
G IP L LNL N L G IPKS+ + + L ++DL N+ +G P L N +
Sbjct: 212 TGNIPHEIGNISLLFMLNLSRNHLSGEIPKSIGRLAQLNIVDLSDNNFSGSIPNELGNCN 391
Query: 224 TLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFS 263
L + L +N G + N + + ++D++ NN S
Sbjct: 392 RLLSMNLSHNDLSGMIPYELGN--LYSLQSLLDLSSNNLS 505
Score = 58.5 bits (140), Expect = 3e-09
Identities = 39/117 (33%), Positives = 59/117 (50%), Gaps = 2/117 (1%)
Frame = +2
Query: 152 NLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHI 211
NL ++L N L G + + L+ + + GN+L G IP L + S L+ L L N
Sbjct: 32 NLSFISLSRNRLIGYLSPDWGKCISLTEMEMSGNKLSGKIPIDLNKLSKLQFLSLHSNEF 211
Query: 212 TGGFPCFLKNISTLRVLILRNNRFQGSL--GCGQVNDEPWKILQIMDIAFNNFSGTL 266
TG P + NIS L +L L N G + G++ L I+D++ NNFSG++
Sbjct: 212 TGNIPHEIGNISLLFMLNLSRNHLSGEIPKSIGRLAQ-----LNIVDLSDNNFSGSI 367
>TC90857 weakly similar to GP|16930691|gb|AAL32011.1 AT4g26540/M3E9_30
{Arabidopsis thaliana}, partial (22%)
Length = 829
Score = 89.4 bits (220), Expect = 2e-18
Identities = 80/262 (30%), Positives = 117/262 (44%), Gaps = 10/262 (3%)
Frame = +3
Query: 15 NQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPL-QKLNNVSSLHYLDLHNNQLQGPIPI 72
N + G +P L L NL L + N +L G + ++ N L +D N + G IP
Sbjct: 3 NSLTGSIPTKLGNLKNLKNLLLWQN---NLVGTIPSEIGNCYQLSVIDASMNSITGSIPK 173
Query: 73 FPVNVVYVD---YSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVL 129
N+ + S N+ S IP ++G+ + + +N G IP L N L +L
Sbjct: 174 TFGNLTLLQELQLSVNQISGEIPAELGN-CQQLTHVEIDNNLITGTIPSELGNLGNLTLL 350
Query: 130 DLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHG 189
L N G IPS T++ +NLE ++L N L G IP L+ L L N L G
Sbjct: 351 FLWHNKLQGNIPS---TLSNCQNLEAIDLSQNLLTGPIPKGIFQLQNLNKLLLLSNNLSG 521
Query: 190 PIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSL-----GCGQV 244
IP + CS+L N+ITG P + N+ L L L +NR +G + GC
Sbjct: 522 KIPSQIGNCSSLIRFRANNNNITGFIPSQIGNLKNLNFLDLGSNRIEGIIPEKISGC--- 692
Query: 245 NDEPWKILQIMDIAFNNFSGTL 266
+ L +D+ N +GTL
Sbjct: 693 -----RNLTFLDLHSNYIAGTL 743
Score = 76.6 bits (187), Expect = 1e-14
Identities = 53/163 (32%), Positives = 79/163 (47%)
Frame = +3
Query: 110 NKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPD 169
N G+IP L N L+ L L NN GTIPS + L V++ N++ G+IP
Sbjct: 3 NSLTGSIPTKLGNLKNLKNLLLWQNNLVGTIPS---EIGNCYQLSVIDASMNSITGSIPK 173
Query: 170 MFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLI 229
F L L L NQ+ G IP L C L +++ N ITG P L N+ L +L
Sbjct: 174 TFGNLTLLQELQLSVNQISGEIPAELGNCQQLTHVEIDNNLITGTIPSELGNLGNLTLLF 353
Query: 230 LRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKYFK 272
L +N+ QG++ N + L+ +D++ N +G + F+
Sbjct: 354 LWHNKLQGNIPSTLSNCQN---LEAIDLSQNLLTGPIPKGIFQ 473
Score = 71.2 bits (173), Expect = 4e-13
Identities = 55/172 (31%), Positives = 87/172 (49%), Gaps = 5/172 (2%)
Frame = +3
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQK-LNNVSSLHY 58
L N LT L L N++ G +P+ L NL +++S NLLT GP+ K + + +L+
Sbjct: 321 LGNLGNLTLLFLWHNKLQGNIPSTLSNCQNLEAIDLSQNLLT---GPIPKGIFQLQNLNK 491
Query: 59 LDLHNNQLQGPIPIFPVN---VVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGN 115
L L +N L G IP N ++ + N + IP IG+ + + FL L N+ G
Sbjct: 492 LLLLSNNLSGKIPSQIGNCSSLIRFRANNNNITGFIPSQIGN-LKNLNFLDLGSNRIEGI 668
Query: 116 IPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTI 167
IP+ + L LDL N +GT+P L + +L+ L+ DN ++G +
Sbjct: 669 IPEKISGCRNLTFLDLHSNYIAGTLPDSLSELV---SLQFLDFSDNMIEGAL 815
>AW299082 weakly similar to GP|10177183|d receptor protein kinase-like
protein {Arabidopsis thaliana}, partial (16%)
Length = 565
Score = 88.6 bits (218), Expect = 3e-18
Identities = 59/156 (37%), Positives = 79/156 (49%)
Frame = +2
Query: 84 RNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSC 143
+N IP +IG+ SS + LS N G IP SL + L+ +S NN SG+IP+
Sbjct: 5 QNGLVGAIPNEIGN-CSSLRNIDLSLNSLSGTIPLSLGSLLELEEFMISDNNVSGSIPA- 178
Query: 144 LMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEV 203
T++ ENL+ L + N L G IP L NQL G IP SL CS L+
Sbjct: 179 --TLSNAENLQQLQVDTNQLSGLIPPEIGKLSNLLVFFAWQNQLEGSIPSSLGNCSKLQA 352
Query: 204 LDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSL 239
LDL +N +TG P L + L L+L +N GS+
Sbjct: 353 LDLSRNSLTGSIPSRLFQLQNLTKLLLISNDISGSI 460
Score = 82.4 bits (202), Expect = 2e-16
Identities = 64/191 (33%), Positives = 84/191 (43%)
Frame = +2
Query: 49 KLNNVSSLHYLDLHNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLS 108
++ N SSL +DL N L G IP+ +G + F+ +S
Sbjct: 35 EIGNCSSLRNIDLSLNSLSGTIPL---------------------SLGSLLELEEFM-IS 148
Query: 109 DNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIP 168
DN G+IP +L NA LQ L + N SG IP + K NL V N L+G+IP
Sbjct: 149 DNNVSGSIPATLSNAENLQQLQVDTNQLSGLIPP---EIGKLSNLLVFFAWQNQLEGSIP 319
Query: 169 DMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVL 228
L L+L N L G IP L Q L L L N I+G P + + +L L
Sbjct: 320 SSLGNCSKLQALDLSRNSLTGSIPSRLFQLQNLTKLLLISNDISGSIPSEIGSCKSLIRL 499
Query: 229 ILRNNRFQGSL 239
L NNR GS+
Sbjct: 500 RLGNNRITGSI 532
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.139 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,695,015
Number of Sequences: 36976
Number of extensions: 163286
Number of successful extensions: 1930
Number of sequences better than 10.0: 246
Number of HSP's better than 10.0 without gapping: 1093
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1480
length of query: 299
length of database: 9,014,727
effective HSP length: 96
effective length of query: 203
effective length of database: 5,465,031
effective search space: 1109401293
effective search space used: 1109401293
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0490.10


