

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.4
(341 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
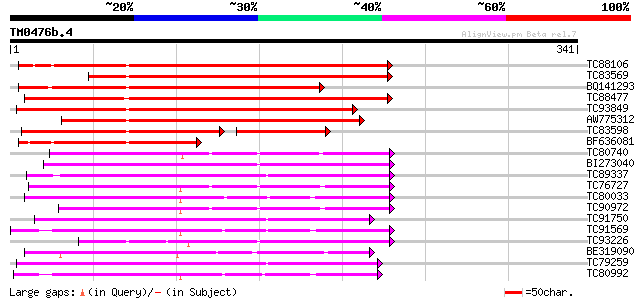
Score E
Sequences producing significant alignments: (bits) Value
TC88106 weakly similar to GP|6733842|emb|CAB69346.1 unnamed prot... 257 4e-69
TC83569 weakly similar to GP|6733842|emb|CAB69346.1 unnamed prot... 224 4e-59
BQ141293 weakly similar to GP|20466732|gb| unknown protein {Arab... 222 1e-58
TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase p... 198 2e-51
TC93849 similar to PIR|A86420|A86420 probable lipase/hydrolase ... 183 9e-47
AW775312 similar to GP|20466732|gb unknown protein {Arabidopsis ... 168 3e-42
TC83598 similar to PIR|H86419|H86419 probable lipase/hydrolase ... 128 3e-36
BF636081 weakly similar to GP|6733842|emb| unnamed protein produ... 144 5e-35
TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif lipase/h... 128 3e-30
BI273040 similar to GP|6721157|gb| putative GDSL-motif lipase/ac... 127 6e-30
TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif... 123 9e-29
TC76727 similar to GP|10638955|emb|CAB81548. putative proline-ri... 122 3e-28
TC80033 weakly similar to GP|21593518|gb|AAM65485.1 putative GDS... 120 6e-28
TC90972 similar to GP|10638955|emb|CAB81548. putative proline-ri... 120 6e-28
TC91750 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif... 118 3e-27
TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protei... 116 1e-26
TC93226 weakly similar to PIR|B96737|B96737 hypothetical protein... 111 4e-25
BE319090 weakly similar to GP|15054386|gb family II lipase EXL3 ... 110 6e-25
TC79259 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif... 109 2e-24
TC80992 weakly similar to GP|21593518|gb|AAM65485.1 putative GDS... 106 1e-23
>TC88106 weakly similar to GP|6733842|emb|CAB69346.1 unnamed protein product
{unidentified}, partial (28%)
Length = 788
Score = 257 bits (657), Expect = 4e-69
Identities = 133/226 (58%), Positives = 173/226 (75%), Gaps = 1/226 (0%)
Frame = +2
Query: 6 KTWLILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPT-SAKANFLSYG 64
K WL+ ++ ++ A+ +Q S V G+SQVPC+F+FGDSLSD GN+NNLP S ++N+ YG
Sbjct: 53 KPWLVFPIL-LLSANYLQQS-VNGKSQVPCLFIFGDSLSDGGNNNNLPANSPRSNYNPYG 226
Query: 65 IDFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKES 124
IDFP PT R++NGR ID I QLLGF+ FIPPFAN+NGSDILKGVNYASG AGIR E+
Sbjct: 227 IDFPMG-PTGRFTNGRTTIDIITQLLGFEKFIPPFANINGSDILKGVNYASGGAGIRMET 403
Query: 125 GSQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPN 184
S G+ ++LGLQL +HR IVS IA +LGG+DKA + L++CLYYV+I +ND+ NYFLP
Sbjct: 404 YSAKGYAISLGLQLRNHRVIVSQIASQLGGIDKAQEYLNKCLYYVHIGSNDYINNYFLPQ 583
Query: 185 VFNTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGC 230
++ +S +Y+P+QYA+ LI +LS LQ L+ GARK VL GL LGC
Sbjct: 584 LYLSSNVYSPEQYAENLIEELSLNLQALHEIGARKYVLPGLGLLGC 721
>TC83569 weakly similar to GP|6733842|emb|CAB69346.1 unnamed protein product
{unidentified}, partial (39%)
Length = 1185
Score = 224 bits (571), Expect = 4e-59
Identities = 114/183 (62%), Positives = 141/183 (76%)
Frame = +1
Query: 48 NDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDI 107
N+NNLPTSAK+N+ YGIDFP PT R++NGR ID I QLLGF+ FIPPFAN+NGSDI
Sbjct: 1 NNNNLPTSAKSNYKPYGIDFPMG-PTGRFTNGRTSIDIITQLLGFEKFIPPFANINGSDI 177
Query: 108 LKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLY 167
LKGVNYASG+AGIR E+ G ++LGLQL +H+ IVS IA +L G+DKA + LS+CLY
Sbjct: 178 LKGVNYASGAAGIRIETSITTGFVISLGLQLENHKVIVSRIASRLEGIDKAQEYLSKCLY 357
Query: 168 YVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDR 227
YVNI +ND+ NYF P + TS++Y+P+QYA+ LI +LS L TL+ GARK VLVGL
Sbjct: 358 YVNIGSNDYINNYFRPQFYPTSQIYSPEQYAEALIQELSLNLLTLHDIGARKYVLVGLGL 537
Query: 228 LGC 230
LGC
Sbjct: 538 LGC 546
>BQ141293 weakly similar to GP|20466732|gb| unknown protein {Arabidopsis
thaliana}, partial (24%)
Length = 583
Score = 222 bits (566), Expect = 1e-58
Identities = 113/184 (61%), Positives = 140/184 (75%)
Frame = +2
Query: 6 KTWLILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGI 65
KTWL++ +V + Q V G+SQVPC+F+FGDSLSDSGN+NNLPTSAK+N+ YGI
Sbjct: 41 KTWLVVLIVFLSANYFKQ--CVNGKSQVPCVFIFGDSLSDSGNNNNLPTSAKSNYKPYGI 214
Query: 66 DFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESG 125
DFP PT R++NGR ID I QLLGF+ FIPPFAN++GSDILKGVNYASG AGIR E+
Sbjct: 215 DFPMG-PTGRFTNGRTAIDIITQLLGFENFIPPFANISGSDILKGVNYASGGAGIRMETY 391
Query: 126 SQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNV 185
S G+ ++LGLQL +HR IVS IA +LGG+DKA Q L++CLYYVNI +N + N F P
Sbjct: 392 SAKGYAISLGLQLRNHRVIVSQIASRLGGIDKAQQYLNKCLYYVNIGSNXYINNXFXPXF 571
Query: 186 FNTS 189
+ TS
Sbjct: 572 YPTS 583
>TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase putative
{Arabidopsis thaliana}, partial (92%)
Length = 1415
Score = 198 bits (504), Expect = 2e-51
Identities = 98/221 (44%), Positives = 149/221 (67%)
Frame = +2
Query: 10 ILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPA 69
++++ ++V + S V + QVP F+FGDSL D+GN+N L + A+A++L YGIDF
Sbjct: 107 VINMFALLVVVLGLWSGVGADPQVPWYFIFGDSLVDNGNNNGLQSLARADYLPYGIDFGG 286
Query: 70 TFPTERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLG 129
PT R+SNG+ +D IA+LLGF +IPP+A+ + ILKGVNYAS +AGIR+E+G QLG
Sbjct: 287 --PTGRFSNGKTTVDAIAELLGFDDYIPPYASASDDAILKGVNYASAAAGIREETGRQLG 460
Query: 130 HNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTS 189
++ Q+ ++++ VS + + LG D+A LS+C+Y + + +ND+ NYF+P +NT
Sbjct: 461 ARLSFSAQVQNYQSTVSQVVNILGTEDQAASHLSKCIYSIGLGSNDYLNNYFMPQFYNTH 640
Query: 190 RMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGC 230
YTP +YA LI + L+TLY+ ARK VL G+ ++GC
Sbjct: 641 DQYTPDEYADDLIQSYTEQLRTLYNN*ARKMVLFGIGQIGC 763
>TC93849 similar to PIR|A86420|A86420 probable lipase/hydrolase
118270-120144 [imported] - Arabidopsis thaliana, partial
(48%)
Length = 673
Score = 183 bits (464), Expect = 9e-47
Identities = 92/207 (44%), Positives = 140/207 (67%), Gaps = 2/207 (0%)
Frame = +1
Query: 5 SKTWLILSLVTV-IVASIMQHSTVLGESQ-VPCIFVFGDSLSDSGNDNNLPTSAKANFLS 62
S +L LS++ + IV ++ + +G +Q VPC F+FGDSL D+GN+N L + AKAN+L
Sbjct: 52 SMAFLDLSMMNIGIVVLVLSLWSGIGVAQQVPCYFIFGDSLVDNGNNNQLTSIAKANYLP 231
Query: 63 YGIDFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRK 122
YGIDFP PT R+SNG+ +D IA+ LGF +IP +A+ G IL+GVNYAS +AGIR+
Sbjct: 232 YGIDFPGG-PTGRFSNGKTTVDVIAEQLGFNGYIPSYASARGRQILRGVNYASAAAGIRE 408
Query: 123 ESGSQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFL 182
E+G QLG ++ Q+ +++ VS + + G + A LS+C+Y + + +ND+ NYF+
Sbjct: 409 ETGQQLGQRISFRGQVQNYQRTVSQLVNYFGDENTAANYLSKCIYTIGLGSNDYLNNYFM 588
Query: 183 PNVFNTSRMYTPKQYAKVLIHQLSHYL 209
P +++TSR +TP+QYA VL+ + L
Sbjct: 589 PTIYSTSRQFTPQQYANVLLQAYAQQL 669
>AW775312 similar to GP|20466732|gb unknown protein {Arabidopsis thaliana},
partial (46%)
Length = 655
Score = 168 bits (425), Expect = 3e-42
Identities = 83/183 (45%), Positives = 120/183 (65%), Gaps = 1/183 (0%)
Frame = +3
Query: 32 QVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQLLG 91
QVPC F+FGDSL D+GN+N + T A+AN+ YGIDFP PT R++NGR +D +AQLLG
Sbjct: 105 QVPCFFIFGDSLVDNGNNNGILTLARANYRPYGIDFPQG-PTGRFTNGRTFVDALAQLLG 281
Query: 92 FQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSCIAHK 151
F+ +IPP + G D+L+GVNYASG+AGIR+E+GS LG + ++ Q+ + V +
Sbjct: 282 FRAYIPPNSRARGLDVLRGVNYASGAAGIREETGSNLGAHTSMTEQVTNFGNTVQEMRRL 461
Query: 152 L-GGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQLSHYLQ 210
G D LS+C+YY + +ND+ NYF+ + ++TS YTPK +A L+ + L
Sbjct: 462 FRGDNDALNSYLSKCIYYSGLGSNDYLNNYFMTDFYSTSTQYTPKAFASALLQDYARQLS 641
Query: 211 TLY 213
L+
Sbjct: 642 QLH 650
>TC83598 similar to PIR|H86419|H86419 probable lipase/hydrolase
114382-116051 [imported] - Arabidopsis thaliana, partial
(42%)
Length = 686
Score = 128 bits (321), Expect(2) = 3e-36
Identities = 63/122 (51%), Positives = 86/122 (69%)
Frame = +3
Query: 8 WLILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDF 67
W++ V ++V + S V + QVPC F+FGDSL D GN+NNL + AKAN+L YGIDF
Sbjct: 99 WIMNLCVMMVVVLGLWSSKVEADPQVPCYFIFGDSLVDDGNNNNLNSLAKANYLPYGIDF 278
Query: 68 PATFPTERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQ 127
PT R+SNG+ +D IA+LLGF+ +I P++ +IL+GVNYAS +AGIR+E+G Q
Sbjct: 279 NGG-PTGRFSNGKTTVDVIAELLGFEGYISPYSTARDQEILQGVNYASAAAGIREETGQQ 455
Query: 128 LG 129
LG
Sbjct: 456 LG 461
Score = 41.6 bits (96), Expect(2) = 3e-36
Identities = 18/57 (31%), Positives = 35/57 (60%)
Frame = +1
Query: 137 QLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYT 193
Q+ +++ VS + + LG D A+ L++C+Y + + +ND+ NYF+P + +YT
Sbjct: 484 QVQNYQKTVSQVVNLLGDEDTASNYLNKCIYSIGLGSNDYLNNYFMPCLSQW*TVYT 654
>BF636081 weakly similar to GP|6733842|emb| unnamed protein product
{unidentified}, partial (22%)
Length = 348
Score = 144 bits (363), Expect = 5e-35
Identities = 73/110 (66%), Positives = 88/110 (79%)
Frame = +1
Query: 6 KTWLILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGI 65
K WL++ L+ A+ +Q V G+SQVPC+F+FGDSLSDSGN+NNLPTSAK+N+ YGI
Sbjct: 28 KKWLVV-LIVFFSANYLQQC-VNGKSQVPCLFIFGDSLSDSGNNNNLPTSAKSNYNPYGI 201
Query: 66 DFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYAS 115
DFP PT R++NGR ID I QLLGF+ FIPPFAN+NGSDILKGVNYAS
Sbjct: 202 DFPMG-PTGRFTNGRTSIDIITQLLGFENFIPPFANINGSDILKGVNYAS 348
>TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif
lipase/hydrolase-like protein {Arabidopsis thaliana},
partial (94%)
Length = 1167
Score = 128 bits (322), Expect = 3e-30
Identities = 73/209 (34%), Positives = 121/209 (56%), Gaps = 2/209 (0%)
Frame = +3
Query: 25 STVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPID 84
+ V G+ VP +F+FGDS+ D+ N+NNL T K+NF YG DF PT R+ NG+ D
Sbjct: 159 NVVKGQPLVPALFIFGDSVVDARNNNNLYTIVKSNFPPYGRDFNNQMPTGRFCNGKLAAD 338
Query: 85 KIAQLLGFQTFIPPFANL--NGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHR 142
A+ LGF T+ P + NL ++L G N+ASG++G + ++L H ++L QL H++
Sbjct: 339 FTAENLGFTTYPPAYLNLQEKRKNLLNGANFASGASGY-FDPTAKLYHAISLEQQLEHYK 515
Query: 143 AIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLI 202
+ + + G A+ +S +Y V ++DF QNY++ + +++T Q++ +L+
Sbjct: 516 ECQNILV-GVAGKSNASSIISGAIYLVRAGSSDFVQNYYINPLL--YKVFTADQFSDILM 686
Query: 203 HQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
+ ++Q LY GARK + L LGC+
Sbjct: 687 QHYTIFIQNLYALGARKIGVTTLPPLGCL 773
>BI273040 similar to GP|6721157|gb| putative GDSL-motif lipase/acylhydrolase
{Arabidopsis thaliana}, partial (60%)
Length = 692
Score = 127 bits (319), Expect = 6e-30
Identities = 74/212 (34%), Positives = 115/212 (53%), Gaps = 1/212 (0%)
Frame = +2
Query: 21 IMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGR 80
++ +TV+ + + FVFGDSL D+GN+N L T+A+A+ YGID+P T R+SNG
Sbjct: 5 LVNFNTVVPQVEARAFFVFGDSLVDNGNNNYLATTARADSYPYGIDYPTHRATGRFSNGL 184
Query: 81 NPIDKIAQLLGFQTFIPPFA-NLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLL 139
N D I++ +G Q +P + LNG +L G N+AS GI ++G Q + + + QL
Sbjct: 185 NMPDLISERIGSQPTLPYLSPELNGEALLVGANFASAGIGILNDTGIQFFNIIRITRQLQ 364
Query: 140 HHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAK 199
+ ++ L G ++ + +++ LY + + NDF NYFL SR + Y
Sbjct: 365 YFEQYQQRVS-ALIGEEETVRLVNEALYLMTLGGNDFVNNYFLVPFSARSRQFXLPDYVV 541
Query: 200 VLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
LI + L LY GAR+ ++ G LGCV
Sbjct: 542 YLISEYRKILARLYELGARRVLVTGTXPLGCV 637
>TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(65%)
Length = 881
Score = 123 bits (309), Expect = 9e-29
Identities = 80/224 (35%), Positives = 121/224 (53%), Gaps = 3/224 (1%)
Frame = +1
Query: 11 LSLVTVIVASIMQHSTVLGESQVPCIF-VFGDSLSDSGNDNNLPTSAKANFLSYGIDF-P 68
++LV ++V I H +P F VFGDSL D+GN+N L T+A+A+ YGID+ P
Sbjct: 55 VALVILVVGGIFVHEI----EAIPRTFLVFGDSLVDNGNNNYLATTARADAPPYGIDYQP 222
Query: 69 ATFPTERYSNGRNPIDKIAQLLGFQTFIPPFA-NLNGSDILKGVNYASGSAGIRKESGSQ 127
+ PT R+SNG N D I+Q LG + +P + L G +L G N+AS GI ++G Q
Sbjct: 223 SHRPTGRFSNGYNIPDIISQKLGAEPTLPYLSPELRGEKLLVGANFASAGIGILNDTGIQ 402
Query: 128 LGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFN 187
+ + + Q + + S ++ +G +A ++Q L + + NDF NY+L
Sbjct: 403 FINIIRMYRQYEYFQEYQSRLSALIGA-SQAKSRVNQALVLITVGGNDFVNNYYLVPYSA 579
Query: 188 TSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
SR Y +Y K LI + LQ LY GAR+ ++ G +GCV
Sbjct: 580 RSRQYPLPEYVKYLISEYQKLLQKLYDLGARRVLVTGTGPMGCV 711
>TC76727 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (92%)
Length = 1919
Score = 122 bits (305), Expect = 3e-28
Identities = 74/222 (33%), Positives = 120/222 (53%), Gaps = 2/222 (0%)
Frame = -2
Query: 12 SLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATF 71
+LV +IV+ + + ++ VP I FGDS D GN++ LPT KAN+ YG DF
Sbjct: 1156 TLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQ 977
Query: 72 PTERYSNGRNPIDKIAQLLGFQTFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQLG 129
PT R+ NG+ D A+ LGF +F P + + +G ++L G N+AS ++G E + L
Sbjct: 976 PTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGY-DEKAATLN 800
Query: 130 HNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTS 189
H + L QL + + +A ++ G KA + LY ++ ++DF QNY+ N
Sbjct: 799 HAIPLSQQLEYFKEYQGKLA-QVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWIN-- 629
Query: 190 RMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
+ T QY+ L+ +++++ +Y GARK + L LGC+
Sbjct: 628 QAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCL 503
>TC80033 weakly similar to GP|21593518|gb|AAM65485.1 putative GDSL-motif
lipase/hydrolase {Arabidopsis thaliana}, partial (89%)
Length = 1164
Score = 120 bits (302), Expect = 6e-28
Identities = 75/224 (33%), Positives = 124/224 (54%), Gaps = 2/224 (0%)
Frame = +1
Query: 10 ILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPA 69
+L+L ++ + ++ ++VP I VFGDS D+GN+N +PT A++NF YG DF
Sbjct: 58 LLALCSLHILCLLLFHLNKVSAKVPAIIVFGDSSVDAGNNNFIPTVARSNFQPYGRDFQG 237
Query: 70 TFPTERYSNGRNPIDKIAQLLGFQTFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQ 127
T R+SNGR P D IA+ G + +P + + N SD GV++AS + G +
Sbjct: 238 GKATGRFSNGRIPTDFIAESFGIKESVPAYLDPKYNISDFATGVSFASAATGYDNATSDV 417
Query: 128 LGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFN 187
L + L QL +++ ++ LG KA + +S+ ++ +++ TNDF +NY+ +
Sbjct: 418 LS-VIPLWKQLEYYKDYQKNLSSYLGEA-KAKETISESVHLMSMGTNDFLENYY--TMPG 585
Query: 188 TSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
+ YTP+QY L ++++ LY GARK L GL +GC+
Sbjct: 586 RASQYTPQQYQTFLAGIAENFIRNLYALGARKISLGGLPPMGCL 717
>TC90972 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (88%)
Length = 1258
Score = 120 bits (302), Expect = 6e-28
Identities = 72/204 (35%), Positives = 115/204 (56%), Gaps = 2/204 (0%)
Frame = +3
Query: 30 ESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQL 89
++ VP I FGDS D GN++ L T KAN+ YG DF PT R+ NG+ D A+
Sbjct: 93 DTVVPAIVTFGDSAVDVGNNDYLFTLFKANYPPYGRDFVXHKPTGRFCNGKLATDITAET 272
Query: 90 LGFQTFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSC 147
LGF+++ P + + G ++L G N+AS ++G E + L H + L QL +++ S
Sbjct: 273 LGFKSYAPAYLSPQATGKNLLIGANFASAASGY-DEKAAILNHAIPLSQQLKYYKEYQSK 449
Query: 148 IAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQLSH 207
++ K+ G KA + LY ++ ++DF QNY++ + N ++ TP QY+ L+ S
Sbjct: 450 LS-KIAGSKKAASIIKGALYLLSGGSSDFIQNYYVNPLIN--KVVTPDQYSAYLVDTYSS 620
Query: 208 YLQTLYHFGARKSVLVGLDRLGCV 231
+++ LY GARK + L LGC+
Sbjct: 621 FVKDLYKLGARKIGVTSLPPLGCL 692
>TC91750 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(50%)
Length = 686
Score = 118 bits (296), Expect = 3e-27
Identities = 71/205 (34%), Positives = 108/205 (52%), Gaps = 1/205 (0%)
Frame = +3
Query: 16 VIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTER 75
VI+ + S G FVFGDSL D+GN+N L T+A+A+ YGID+P PT R
Sbjct: 57 VIILMVALTSCFKGTVAQRAFFVFGDSLVDNGNNNYLATTARADAPPYGIDYPTRRPTGR 236
Query: 76 YSNGRNPIDKIAQLLGFQTFIPPFA-NLNGSDILKGVNYASGSAGIRKESGSQLGHNVNL 134
+SNG N D I+Q LG + +P + LNG +L G N+AS GI ++G Q + + +
Sbjct: 237 FSNGYNIPDFISQALGAEPTLPYLSPELNGEALLVGANFASAGIGILNDTGIQFINIIRI 416
Query: 135 GLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTP 194
QL + + ++ +G ++ ++ L + + NDF NY+L SR Y
Sbjct: 417 FRQLEYFQQYQQRVSGLIGP-EQTQSLVNGALVLITLGGNDFVNNYYLVPFSARSRQYNL 593
Query: 195 KQYAKVLIHQLSHYLQTLYHFGARK 219
Y + +I + L+ LYH GAR+
Sbjct: 594 PDYVRYIISEYKKILRRLYHLGARR 668
>TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protein
{Arabidopsis thaliana}, partial (60%)
Length = 1398
Score = 116 bits (290), Expect = 1e-26
Identities = 79/234 (33%), Positives = 121/234 (50%), Gaps = 3/234 (1%)
Frame = +1
Query: 1 MGCESKTWLILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANF 60
MG WLIL ++V T E+ VP + VFGDS DSGN+N + T K+NF
Sbjct: 52 MGNICIVWLILITQMIMV-------TCNNENYVPAVIVFGDSSVDSGNNNMISTFLKSNF 210
Query: 61 LSYGIDFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFAN--LNGSDILKGVNYASGSA 118
YG D PT R+SNGR P D I++ G ++ IP + + D + GV +AS
Sbjct: 211 RPYGRDIDGGRPTGRFSNGRIPPDFISEAFGIKSLIPAYLDPAYTIDDFVTGVCFASAGT 390
Query: 119 GIRKESGSQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQ 178
G + + L + + L ++ ++ + +G +K+ + +S+ LY +++ TNDF
Sbjct: 391 GYDNATSAIL-NVIPLWKEVEFYKEYQDKLKAHIGE-EKSIEIISEALYIISLGTNDFLG 564
Query: 179 NYFLPNVFNTSRM-YTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
NY+ F T R YT QY LI ++++ LY GARK + GL +GC+
Sbjct: 565 NYY---GFTTLRFRYTISQYQDYLIGIAENFIRQLYSLGARKLAITGLIPMGCL 717
>TC93226 weakly similar to PIR|B96737|B96737 hypothetical protein F3I17.10
[imported] - Arabidopsis thaliana, partial (60%)
Length = 847
Score = 111 bits (278), Expect = 4e-25
Identities = 69/192 (35%), Positives = 104/192 (53%), Gaps = 2/192 (1%)
Frame = +2
Query: 42 SLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFAN 101
SL + GN+N L T AK+NF YGID+ PT R+SNG++ ID I +LG + PPF +
Sbjct: 2 SLVEVGNNNFLSTFAKSNFYPYGIDYNGR-PTGRFSNGKSLIDFIGDMLGVPS-PPPFLD 175
Query: 102 LNGSD--ILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKAT 159
++ +L GVNYASGS GI +SG G ++ QL + ++ K+ +
Sbjct: 176 PTSTENKLLNGVNYASGSGGILDDSGRHYGDRHSMSRQLQNFERTLNQYK-KMMNETALS 352
Query: 160 QCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARK 219
Q L++ + V +ND+ NY P + TSR Y+ Q+ +L++ + LY RK
Sbjct: 353 QFLAKSIVIVVTGSNDYINNYLRPEYYGTSRNYSVPQFGNLLLNTFGRQILALYSLRLRK 532
Query: 220 SVLVGLDRLGCV 231
L G+ LGC+
Sbjct: 533 FFLAGVGPLGCI 568
>BE319090 weakly similar to GP|15054386|gb family II lipase EXL3 {Arabidopsis
thaliana}, partial (42%)
Length = 664
Score = 110 bits (276), Expect = 6e-25
Identities = 68/216 (31%), Positives = 115/216 (52%), Gaps = 6/216 (2%)
Frame = +1
Query: 10 ILSLVTVIVASIMQHSTVLG---ESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGID 66
I+ +T++ A + +L + P +FVFGDS+ D+GN+NN PT + F YG D
Sbjct: 31 IILFLTMLFAIFSKTKAILKLPPNASFPAVFVFGDSIMDTGNNNNRPTPTQCKFPPYGKD 210
Query: 67 FPATFPTERYSNGRNPIDKIAQLLGFQTFIPPF--ANLNGSDILKGVNYASGSAGIRKES 124
F PT R+SNG+ P D I + LG + ++P + NL S+++ GVN+ASG AG +
Sbjct: 211 FQGGIPTGRFSNGKVPADLIVEELGIKEYLPAYLDPNLQPSELVTGVNFASGGAGYDPLT 390
Query: 125 GSQLGHNVNLGLQL-LHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLP 183
S++ +++ Q+ L IV + G D+ L+ +Y+V + +ND YFL
Sbjct: 391 -SKIEAAISMSAQIELFKEYIVK--LKGIVGEDRTNFILANSIYFVLVGSNDISNTYFLF 561
Query: 184 NVFNTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARK 219
+ + Y Y+ +L+ ++ + +Y GAR+
Sbjct: 562 HARQVN--YDFPSYSDLLVDSAYNFYKEMYQLGARR 663
>TC79259 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(52%)
Length = 712
Score = 109 bits (272), Expect = 2e-24
Identities = 73/222 (32%), Positives = 116/222 (51%), Gaps = 2/222 (0%)
Frame = +3
Query: 5 SKTWLILSLVTVIVASIMQHSTVLGESQVPCIF-VFGDSLSDSGNDNNLPTSAKANFLSY 63
S+ + I ++++A + S V P F VFGDSL DSGN++ L T+A+A+ Y
Sbjct: 45 SQLFCIAMATSLVIAFCVMISFVGCAYAQPRAFGVFGDSLVDSGNNDFLATTARADNYPY 224
Query: 64 GIDFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFAN-LNGSDILKGVNYASGSAGIRK 122
GID+P+ PT R+SNG N D I+ LG + +P + L G +L G N+AS GI
Sbjct: 225 GIDYPSHRPTGRFSNGYNIPDLISLELGLEPTLPYLSPLLVGEKLLIGANFASAGIGILN 404
Query: 123 ESGSQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFL 182
++G Q H + + QL ++ +G + A +++ L + + NDF NY+L
Sbjct: 405 DTGFQFIHIIRIYKQLRLFELYQKRVSAHIGS-EGARNLVNRALVLITLGGNDFVNNYYL 581
Query: 183 PNVFNTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVG 224
SR ++ Y + LI + L+ LY GAR+ ++ G
Sbjct: 582 VPFSARSRQFSLPDYVRYLISEYRKVLRRLYDLGARRVLVTG 707
>TC80992 weakly similar to GP|21593518|gb|AAM65485.1 putative GDSL-motif
lipase/hydrolase {Arabidopsis thaliana}, partial (53%)
Length = 812
Score = 106 bits (264), Expect = 1e-23
Identities = 72/225 (32%), Positives = 113/225 (50%), Gaps = 3/225 (1%)
Frame = +1
Query: 3 CESKTWLILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLS 62
C WLIL ++V T ++ VP + VFGDS DSGN+N + T K+NF
Sbjct: 64 CICIAWLILITQIIMV-------TCKTKNHVPAVIVFGDSSVDSGNNNRIATLLKSNFKP 222
Query: 63 YGIDFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFAN--LNGSDILKGVNYASGSAGI 120
YG DF PT R+ NGR P D IA+ G + IP + + D + GV +AS G
Sbjct: 223 YGRDFEGGRPTGRFCNGRTPPDFIAEAFGVKRNIPAYLDPAYTIDDFVTGVCFASAGTGY 402
Query: 121 RKESGSQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNY 180
+ L + + L ++ + + + G KA + +S+ LY +++ TNDF +NY
Sbjct: 403 DNATSDVL-NVIPLWKEIEFFKEYQEKLRVHV-GKKKANEIISEALYLISLGTNDFLENY 576
Query: 181 FLPNVFNTSRM-YTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVG 224
+ +F T ++ +T QY L+ +++ L+ GARK + G
Sbjct: 577 Y---IFPTRQLHFTVSQYQDFLVDIAEDFVRKLHSLGARKLSITG 702
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.336 0.145 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,590,804
Number of Sequences: 36976
Number of extensions: 149195
Number of successful extensions: 1254
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 1191
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1197
length of query: 341
length of database: 9,014,727
effective HSP length: 97
effective length of query: 244
effective length of database: 5,428,055
effective search space: 1324445420
effective search space used: 1324445420
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0476b.4


