

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.3
(862 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
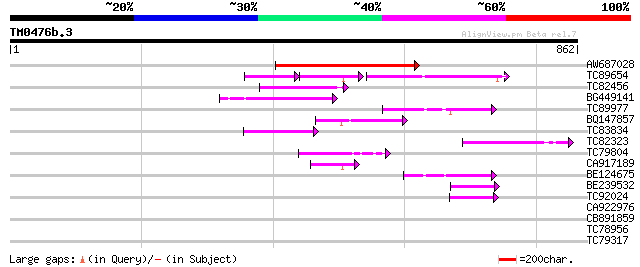
Score E
Sequences producing significant alignments: (bits) Value
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 199 3e-51
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 100 5e-48
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 115 9e-26
BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidop... 82 7e-16
TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 77 4e-14
BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {A... 73 5e-13
TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabid... 65 9e-11
TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea ma... 59 8e-09
TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like... 55 2e-07
CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.3... 52 1e-06
BE124675 weakly similar to GP|13365573|db hypothetical protein~s... 52 1e-06
BE239532 50 5e-06
TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10... 46 5e-05
CA922976 weakly similar to GP|9502168|gb|A contains simlarity to... 42 0.001
CB891859 similar to GP|6175165|gb| Mutator-like transposase {Ara... 35 0.16
TC78956 similar to PIR|G86434|G86434 protein F17F8.23 [imported]... 31 2.3
TC79317 similar to GP|7673359|gb|AAF66825.1| poly(A)-binding pro... 30 4.0
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 199 bits (507), Expect = 3e-51
Identities = 100/219 (45%), Positives = 136/219 (61%), Gaps = 1/219 (0%)
Frame = +1
Query: 405 DALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKK 464
D LAFD TYKKNKY PL IFS NH +QT + G A++ +E E+Y W+L + LE M+ K
Sbjct: 1 DVLAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECMENK 180
Query: 465 SPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIPDFLPKFKTCMFGDFQI 524
P V+TDGD +MR AI++VFP+ASHRLCAWHL NA +NIK FL F+ M+ +F
Sbjct: 181 FPKAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQENIKKTPFLEGFRKAMYSNFTP 360
Query: 525 GDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQLGK 584
F+ W L+++ L N W+I YA + WA AY R KFF RTTS+CE NA +
Sbjct: 361 EQFEDFWSELIQKNELEGNAWVIKTYANKSLWATAYLRDKFFGRIRTTSQCEAINAIVKT 540
Query: 585 FVK-RRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDP 622
+ + +R N + + E R+ E+ ADF+S + +P
Sbjct: 541 YSRSQRQNF*IYA*FLNRF*EGYRNNELVADFKSKFTEP 657
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 100 bits (248), Expect(3) = 5e-48
Identities = 62/223 (27%), Positives = 100/223 (44%), Gaps = 6/223 (2%)
Frame = +1
Query: 543 NPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHC 602
N W MY R W Y RG FF E N V L+ ++ +
Sbjct: 697 NEWPQSMYNARQHWVPVYLRGSFFGEIPLNDGNECLNFFFDGHVNASTTLQLLVRQYEKA 876
Query: 603 LEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIIT 662
+ RE++ADF + +P L+ P +E A+ + TR++F +F+ +V T
Sbjct: 877 VSTWHERELKADFETSNSNPVLRTP---SPMEKQAASLYTRKMFMKFQEELVGTMANPAT 1047
Query: 663 NCEDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNI 722
ED+ ++ + V + E+ V++ ++ + CSC E G+ C HI+AV N+
Sbjct: 1048KIEDSGTITTYRVAKFGENQTSHTVAFNSSEMKASCSCQMFEYSGIVCRHILAVFRAKNV 1227
Query: 723 HNIPKSLVLERWSQGAK------DHLCETPGSSPSLFRDSAVL 759
+P VL+RW+ AK +H E P SS R+SA +
Sbjct: 1228LTLPSHYVLKRWTMNAKTGVVLDEHTSELPNSS----RESATV 1344
Score = 68.2 bits (165), Expect(3) = 5e-48
Identities = 33/84 (39%), Positives = 43/84 (50%)
Frame = +2
Query: 357 GRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALAFDATYKKN 416
G LDYL + ++ + F D D ++FW D R NYS FGD + FD TYK N
Sbjct: 122 GHQVLDYLKRMRAENHAFFYAVQSDVDNAGGNIFWADETCRTNYSYFGDTVIFDTTYKTN 301
Query: 417 KYRRPLVIFSCVNHRNQTTVVGGA 440
+YR P F+ NH Q + G A
Sbjct: 302 QYRVPFASFTGFNHHGQPVLFGCA 373
Score = 63.2 bits (152), Expect(3) = 5e-48
Identities = 31/103 (30%), Positives = 51/103 (49%), Gaps = 5/103 (4%)
Frame = +3
Query: 441 VIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCN 500
+I NE E +Y+WL + L A+ + P ++ TD D ++ A+ +V P HR C W +
Sbjct: 375 LILNESEPSYIWLFKTWLRAVSGRPPVSITTDLDPVIQVAVAQVLPPTRHRFCKWSIFRE 554
Query: 501 ATDNI-----KIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEF 538
+ P F +FK C+ I +F+ W SL+E +
Sbjct: 555 NRSKLAHLYQSNPTFDNEFKKCVHESVTIDEFESCWRSLLERY 683
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 115 bits (287), Expect = 9e-26
Identities = 56/134 (41%), Positives = 77/134 (56%)
Frame = +2
Query: 381 DADKNLEHLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGA 440
D +N+++LFW D R Y FG+ + D TY +KY PLV F VNH QT ++G A
Sbjct: 791 DDKRNIQNLFWADARCRAAYEYFGEVITLDTTYLTSKYDLPLVPFVGVNHHGQTILLGCA 970
Query: 441 VIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCN 500
++ N +T WL + LE M +P +IT+ DKAM+NAI FP A HR C WH+M
Sbjct: 971 ILSNLDAKTLTWLFTRWLECMHGHAPNGIITEEDKAMKNAIEVAFPKARHRWCLWHIM-- 1144
Query: 501 ATDNIKIPDFLPKF 514
K+P+ L K+
Sbjct: 1145----KKVPEMLGKY 1174
>BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (33%)
Length = 690
Score = 82.4 bits (202), Expect = 7e-16
Identities = 46/182 (25%), Positives = 88/182 (48%), Gaps = 3/182 (1%)
Frame = +3
Query: 320 RRLQFTCCIGCGYNNTQYILKDLQNQVGKQRRAHVSDGRSALDYLA---SLGTKDPSMFV 376
R ++ C+ GY + KD++N + R+ + LD L ++ KDP+
Sbjct: 126 RLMELEKCVEPGY--LPFTEKDVRNLLQSFRKLDPEE--ETLDLLRMCRNIKDKDPNFKF 293
Query: 377 RHTEDADKNLEHLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTV 436
+T DA+ LE++ W Y +FGDA+ FD T++ + PL ++ +N+
Sbjct: 294 EYTLDANNRLENIAWSYASSIQLYDIFGDAVVFDTTHRLTAFDMPLGLWVGINNYGMPCF 473
Query: 437 VGGAVIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWH 496
G ++ +E ++ W ++ L M+ K+P T++TD + ++ A+ P H C W
Sbjct: 474 FGCVLLRDETVRSFSWAIKAFLGFMNGKAPQTILTDQNICLKEALSAEMPMTKHAFCIWM 653
Query: 497 LM 498
++
Sbjct: 654 IV 659
>TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (35%)
Length = 1523
Score = 76.6 bits (187), Expect = 4e-14
Identities = 47/177 (26%), Positives = 83/177 (46%), Gaps = 4/177 (2%)
Frame = +2
Query: 567 AGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKA 626
AG T R E N+ K++ ++ LKEF++ + L E ADF + + P LK+
Sbjct: 2 AGTSTAQRSESMNSFFDKYIHKKITLKEFVKQYRLILLNRYDEEEIADFDTLHKQPALKS 181
Query: 627 PPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTL----GMSMHTVKRSQESS 682
P + E S I T +F +F+ ++ A C+ + G ++ + + E
Sbjct: 182 PSPW---EKQMSTIYTHAIFKKFQVEVLGVA-----GCQSRIEVGDGTAVRYIVQDYEKD 337
Query: 683 REWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
E+LV++ + E C C E G C H ++VL ++P +++RW++ AK
Sbjct: 338 EEFLVTWKELSSEVSCFCRLFEYKGFLCRHALSVLQRCGCSSVPSHYIMKRWTKDAK 508
>BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (26%)
Length = 560
Score = 72.8 bits (177), Expect = 5e-13
Identities = 37/147 (25%), Positives = 70/147 (47%), Gaps = 6/147 (4%)
Frame = +1
Query: 465 SPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATD------NIKIPDFLPKFKTCM 518
+P T++TD + ++ AI P + H C WH++ +D + ++ F +
Sbjct: 1 APQTLLTDHNTWLKEAIAVEMPESKHAFCIWHILSKFSDWXYLLLGSQYDEWKADFHR-L 177
Query: 519 FGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGF 578
+ DF++ W +V+++ LH N II +Y+ R WA + R FFAG + + E
Sbjct: 178 YNLEMXEDFEKSWRQMVDKYGLHANKHIISLYSLRTFWALPFLRRYFFAGLTSXCQTESI 357
Query: 579 NAQLGKFVKRRYNLKEFLQHFHHCLEY 605
N + +F+ + FLQ +++
Sbjct: 358 NVFIQRFLSAQSQPXRFLQQVADIVDF 438
>TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 1032
Score = 65.5 bits (158), Expect = 9e-11
Identities = 32/114 (28%), Positives = 56/114 (49%)
Frame = +3
Query: 356 DGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALAFDATYKK 415
D ++ ++L + +P+ F + + +L + W D R F D + FD TY
Sbjct: 690 DTQAIYNFLCRMQLTNPNFFYLMDFNDEGHLRNALWVDAKSRAACGYFSDVIYFDNTYLV 869
Query: 416 NKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPTTV 469
NKY PLV +NH Q+ ++G ++ E E+Y WL ++ + + SP T+
Sbjct: 870 NKYEIPLVALVGINHHGQSVLLGCGLLAGEIIESYKWLFRTWIKCIPRCSPQTI 1031
>TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea mays},
partial (2%)
Length = 1165
Score = 58.9 bits (141), Expect = 8e-09
Identities = 45/173 (26%), Positives = 72/173 (41%), Gaps = 4/173 (2%)
Frame = +1
Query: 689 YYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHLCETPGS 748
Y + EF+CSC ES G+ C H+ V+ + ++ +IPK LV+ RW++ AK + S
Sbjct: 4 YNAASLEFQCSCRLFESRGIPCCHVFFVMKEEDVDHIPKCLVMTRWTKNAKGEFLNS-DS 180
Query: 749 SPSLFRDSAVLVRYVCMLEACRKMCTLACPLIDDFRQTW-EIVGGHTQHL---ENKNNPS 804
+ + + L R+ A C A FR EI+ ++ ++ + S
Sbjct: 181 NGEIDANMIELARFGAYCSAFTTFCKEASKKNGVFRDIMDEILNLQKKYCDVEDSTRSKS 360
Query: 805 VPEGVTGTPSANAGLTTHRRRGRPCGPSTSRLRAKRPLKCSTCKVIGHNRLTC 857
+ G P T + +G P + AK CS CK H C
Sbjct: 361 FTDDQVGDP------VTVKSKG---APKKKKNAAKSVRHCSRCKSTSHTARHC 492
>TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like
transposase {Arabidopsis thaliana}, partial (57%)
Length = 2129
Score = 54.7 bits (130), Expect = 2e-07
Identities = 40/144 (27%), Positives = 67/144 (45%), Gaps = 4/144 (2%)
Frame = +1
Query: 440 AVIGNEKEETYVWLLEQLLEAMDKKS---PTTV-ITDGDKAMRNAIRRVFPNASHRLCAW 495
AV+ E +E++ W L +L A++ + P + ++DG K + +AIRR FP +SH C
Sbjct: 859 AVVDVENDESWTWFLSELHNALEVNTECMPQIIFLSDGQKGIVDAIRRKFPRSSHAFCMR 1038
Query: 496 HLMCNATDNIKIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHT 555
HL N K + + + I F+ K +EE + + + W+ + +
Sbjct: 1039HLSENIGKEFKNSRLIHLLWSAAYAT-TINAFREKMAE-IEEVSPNASMWLQHFHPSQ-- 1206
Query: 556 WAAAYFRGKFFAGFRTTSRCEGFN 579
WA YF G + +S E FN
Sbjct: 1207WALVYFEGTRYG--HLSSNIEEFN 1272
>CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (12%)
Length = 312
Score = 52.0 bits (123), Expect = 1e-06
Identities = 24/79 (30%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Frame = +1
Query: 458 LEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDN-----IKIPDFLP 512
L AM + P ++ TD D +++AI +VFP+ HR C WH+ + ++ P+F
Sbjct: 1 LTAMSGRPPLSITTDHDSVIQSAIMQVFPDTRHRFCKWHIFKQCQEKLSHIFLQFPNFEA 180
Query: 513 KFKTCMFGDFQIGDFKRKW 531
+F C+ I +F+ W
Sbjct: 181 EFHKCVNLTDSIDEFESCW 237
>BE124675 weakly similar to GP|13365573|db hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (4%)
Length = 448
Score = 51.6 bits (122), Expect = 1e-06
Identities = 35/141 (24%), Positives = 66/141 (45%)
Frame = +2
Query: 599 FHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRILTRQLFFRFRPTIVSAAD 658
F +E R++E+EA P L + + AS T + F F+ + +
Sbjct: 2 FERIVEEQRYKEIEASDEMKGCLPKLMGNVV---VLKHASVAYTPRAFEVFQQRYEKSLN 172
Query: 659 MIITNCEDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLV 718
+I+ N G + +R++ V++ ++ CSCM+ E +G C+H + VL
Sbjct: 173 VIV-NQHKRDGYLFEYKVNTYGHARQYTVTFSSSDNTVVCSCMKFEHVGFLCSHALKVLD 349
Query: 719 DLNIHNIPKSLVLERWSQGAK 739
+ NI +P +L+RW++ A+
Sbjct: 350 NRNIKVVPSRYILKRWTKDAR 412
>BE239532
Length = 645
Score = 49.7 bits (117), Expect = 5e-06
Identities = 20/75 (26%), Positives = 43/75 (56%)
Frame = +3
Query: 670 MSMHTVKRSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSL 729
+S H V + + ++++ V++ ++ + CSC + +S+G+ C+H + + I IP
Sbjct: 156 LSNHEVMLNHKPNKKYAVTFDSSNMKINCSCRKFDSMGILCSHALRIYNIKGILRIPDQY 335
Query: 730 VLERWSQGAKDHLCE 744
L+RWS+ A+ + E
Sbjct: 336 FLKRWSKNARSVIYE 380
>TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (11%)
Length = 938
Score = 46.2 bits (108), Expect = 5e-05
Identities = 23/75 (30%), Positives = 38/75 (50%), Gaps = 1/75 (1%)
Frame = +3
Query: 669 GMSMHTV-KRSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPK 727
G S+H + E + + V + + CSC E GL C HI+AV N+ +P
Sbjct: 3 GRSLHIM*LNLGEDHKAYNVRFNVLEMKATCSCQMFEFSGLLCRHILAVFRVTNVLTLPS 182
Query: 728 SLVLERWSQGAKDHL 742
+L+RW++ AK ++
Sbjct: 183 HYILKRWTKNAKSNV 227
>CA922976 weakly similar to GP|9502168|gb|A contains simlarity to Arabidopsis
thaliana far-red impaired response protein
(GB:AAD51282.1), partial (11%)
Length = 827
Score = 41.6 bits (96), Expect = 0.001
Identities = 18/62 (29%), Positives = 30/62 (48%)
Frame = -3
Query: 673 HTVKRSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLE 732
+ ++ S++ E V + + CSC ES G+ C H + +LV N +P+ L
Sbjct: 777 YIIRHSKKLDGERHVLWLPEKEQILCSCKEFESSGILCRHALRILVTKNYFQLPEKYYLS 598
Query: 733 RW 734
RW
Sbjct: 597 RW 592
>CB891859 similar to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (27%)
Length = 625
Score = 34.7 bits (78), Expect = 0.16
Identities = 35/128 (27%), Positives = 56/128 (43%), Gaps = 4/128 (3%)
Frame = +1
Query: 457 LLEAMDKKSPT-TVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNAT---DNIKIPDFLP 512
LLE + P T+++D + + + + FP A H C HL + +N + + L
Sbjct: 1 LLEVNTENMPRLTILSDRQQGIVDGVEANFPTAFHGFCMRHLSDSFRKEFNNTMLVNLLW 180
Query: 513 KFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTT 572
+ C+ I +F+ K +EE + WI + R WA AYF G F G T
Sbjct: 181 EAANCL----TIIEFEGKVME-IEEISQDAAYWIRRVPPR--LWATAYFEGHRF-GHLTA 336
Query: 573 SRCEGFNA 580
+ E N+
Sbjct: 337 NIVEALNS 360
>TC78956 similar to PIR|G86434|G86434 protein F17F8.23 [imported] -
Arabidopsis thaliana, partial (47%)
Length = 1744
Score = 30.8 bits (68), Expect = 2.3
Identities = 18/63 (28%), Positives = 31/63 (48%)
Frame = +1
Query: 665 EDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHN 724
E+ + +H V + E+ R W+ Y T + RCS M+ + CA ++ +DL +
Sbjct: 31 ENLRQLCVHLV--ANETGRSWVWWDYVTDFHVRCS-MKEKKYSKDCAEVVMKSLDLPLDK 201
Query: 725 IPK 727
I K
Sbjct: 202 IKK 210
>TC79317 similar to GP|7673359|gb|AAF66825.1| poly(A)-binding protein
{Nicotiana tabacum}, partial (32%)
Length = 848
Score = 30.0 bits (66), Expect = 4.0
Identities = 18/54 (33%), Positives = 23/54 (42%)
Frame = -2
Query: 804 SVPEGVTGTPSANAGLTTHRRRGRPCGPSTSRLRAKRPLKCSTCKVIGHNRLTC 857
+V G P A G+ G PCG R RPL C C +IG+ +C
Sbjct: 214 AVARACMGVPPATPGM------GIPCGLRPVGYR*TRPLGCICCCIIGNECGSC 71
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.351 0.156 0.561
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,551,964
Number of Sequences: 36976
Number of extensions: 458744
Number of successful extensions: 4269
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 1753
Number of HSP's successfully gapped in prelim test: 181
Number of HSP's that attempted gapping in prelim test: 2467
Number of HSP's gapped (non-prelim): 2053
length of query: 862
length of database: 9,014,727
effective HSP length: 104
effective length of query: 758
effective length of database: 5,169,223
effective search space: 3918271034
effective search space used: 3918271034
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0476b.3


