

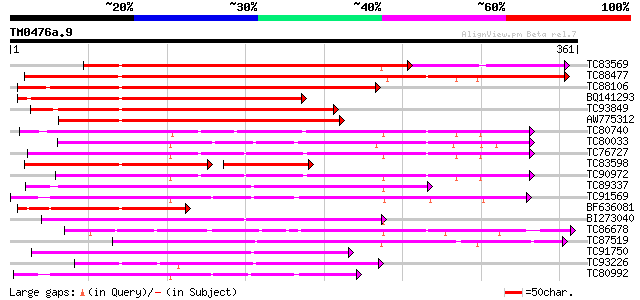
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476a.9
(361 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83569 weakly similar to GP|6733842|emb|CAB69346.1 unnamed prot... 268 1e-94
TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase p... 313 7e-86
TC88106 weakly similar to GP|6733842|emb|CAB69346.1 unnamed prot... 283 7e-77
BQ141293 weakly similar to GP|20466732|gb| unknown protein {Arab... 243 1e-64
TC93849 similar to PIR|A86420|A86420 probable lipase/hydrolase ... 198 2e-51
AW775312 similar to GP|20466732|gb unknown protein {Arabidopsis ... 187 5e-48
TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif lipase/h... 178 3e-45
TC80033 weakly similar to GP|21593518|gb|AAM65485.1 putative GDS... 172 1e-43
TC76727 similar to GP|10638955|emb|CAB81548. putative proline-ri... 165 3e-41
TC83598 similar to PIR|H86419|H86419 probable lipase/hydrolase ... 134 4e-41
TC90972 similar to GP|10638955|emb|CAB81548. putative proline-ri... 163 8e-41
TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif... 160 7e-40
TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protei... 159 1e-39
BF636081 weakly similar to GP|6733842|emb| unnamed protein produ... 153 1e-37
BI273040 similar to GP|6721157|gb| putative GDSL-motif lipase/ac... 146 1e-35
TC86678 similar to PIR|E96579|E96579 hypothetical protein T18A20... 140 1e-33
TC87519 weakly similar to GP|21592967|gb|AAM64916.1 putative GDS... 139 1e-33
TC91750 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif... 132 2e-31
TC93226 weakly similar to PIR|B96737|B96737 hypothetical protein... 127 7e-30
TC80992 weakly similar to GP|21593518|gb|AAM65485.1 putative GDS... 123 1e-28
>TC83569 weakly similar to GP|6733842|emb|CAB69346.1 unnamed protein product
{unidentified}, partial (39%)
Length = 1185
Score = 268 bits (686), Expect(2) = 1e-94
Identities = 134/213 (62%), Positives = 166/213 (77%), Gaps = 4/213 (1%)
Frame = +1
Query: 48 NNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDI 107
NNNNLPTSAK+N+ PYGIDFP PTGR++NGR ID I Q+LGF++FIPPFAN+NGSDI
Sbjct: 1 NNNNLPTSAKSNYKPYGIDFPMG-PTGRFTNGRTSIDIITQLLGFEKFIPPFANINGSDI 177
Query: 108 LKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLY 167
LKGVNYASG+AGIR E+ G ++LGLQL +H+ IVSRIA RL G+DKA +YL++CLY
Sbjct: 178 LKGVNYASGAAGIRIETSITTGFVISLGLQLENHKVIVSRIASRLEGIDKAQEYLSKCLY 357
Query: 168 YVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDR 227
YVNIG+ND+ NYF P + TS++Y+P+QYA+ALIQ+LS L TLH GARK VLVGL
Sbjct: 358 YVNIGSNDYINNYFRPQFYPTSQIYSPEQYAEALIQELSLNLLTLHDIGARKYVLVGLGL 537
Query: 228 LGCIPKLL----VNGSCVEEKNVATFLFNDQLK 256
LGC P + NGSCV+E+N +FN +LK
Sbjct: 538 LGCTPSAIFTHGTNGSCVDEENAPALIFNFKLK 636
Score = 96.3 bits (238), Expect(2) = 1e-94
Identities = 50/103 (48%), Positives = 62/103 (59%), Gaps = 1/103 (0%)
Frame = +2
Query: 255 LKSLVDRLNKKILMDSKFIFINSTAIIH-DKSVGFTVTHHGCCPTNEKGKCIRDGIPCQN 313
L LVD N K DSKFIF+N+T S GF V++ CCP+ CI D PC N
Sbjct: 632 LNFLVDHFNNKFSADSKFIFVNTTLESDAQNSDGFLVSNVPCCPSG----CIPDERPCYN 799
Query: 314 RHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLIQ 356
R EY FWD +H TEA+N + AI SYNS N YP +IK+L++
Sbjct: 800 RSEYAFWDEVHPTEASNQLYAIRSYNSHNSGFTYPMDIKNLVE 928
>TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase putative
{Arabidopsis thaliana}, partial (92%)
Length = 1415
Score = 313 bits (802), Expect = 7e-86
Identities = 161/360 (44%), Positives = 236/360 (64%), Gaps = 13/360 (3%)
Frame = +2
Query: 10 VLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPA 69
V+++ ++V + S V + QVP F+FGDSL D+GNNN L + A+A++LPYGIDF
Sbjct: 107 VINMFALLVVVLGLWSGVGADPQVPWYFIFGDSLVDNGNNNGLQSLARADYLPYGIDFGG 286
Query: 70 TFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLG 129
PTGR+SNG+ +D IA++LGF ++IPP+A+ + ILKGVNYAS +AGIR+E+G QLG
Sbjct: 287 --PTGRFSNGKTTVDAIAELLGFDDYIPPYASASDDAILKGVNYASAAAGIREETGRQLG 460
Query: 130 HNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTS 189
++ Q+ ++++ VS++ + LG D+A +L++C+Y + +G+ND+ NYF+P +NT
Sbjct: 461 ARLSFSAQVQNYQSTVSQVVNILGTEDQAASHLSKCIYSIGLGSNDYLNNYFMPQFYNTH 640
Query: 190 RMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNGS-----CVEEK 244
YTP +YA LIQ + L+TL++ ARK VL G+ ++GC P L S CVEE
Sbjct: 641 DQYTPDEYADDLIQSYTEQLRTLYNN*ARKMVLFGIGQIGCSPNELATRSADGVTCVEEI 820
Query: 245 NVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCC--- 296
N A +FN++LK LVD+ N + L DSK I++NS I D + GF+VT+ GCC
Sbjct: 821 NSANQIFNNKLKGLVDQFNNQ-LPDSKVIYVNSYGIFQDIISNPSAYGFSVTNAGCCGVG 997
Query: 297 PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLIQ 356
N + C+ PC+NR EY+FWD H TEA N+V A +Y++ +P AYP +I HL Q
Sbjct: 998 RNNGQFTCLPLQTPCENRREYLFWDAFHPTEAGNVVVAQRAYSAQSPDDAYPIDISHLAQ 1177
>TC88106 weakly similar to GP|6733842|emb|CAB69346.1 unnamed protein product
{unidentified}, partial (28%)
Length = 788
Score = 283 bits (724), Expect = 7e-77
Identities = 140/232 (60%), Positives = 181/232 (77%), Gaps = 1/232 (0%)
Frame = +2
Query: 6 KTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPT-SAKANFLPYG 64
K WLV +L++ + Q +V G+SQVPC+F+FGDSLSD GNNNNLP S ++N+ PYG
Sbjct: 53 KPWLVFPILLLSANYLQQ--SVNGKSQVPCLFIFGDSLSDGGNNNNLPANSPRSNYNPYG 226
Query: 65 IDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKES 124
IDFP PTGR++NGR ID I Q+LGF++FIPPFAN+NGSDILKGVNYASG AGIR E+
Sbjct: 227 IDFPMG-PTGRFTNGRTTIDIITQLLGFEKFIPPFANINGSDILKGVNYASGGAGIRMET 403
Query: 125 GSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPN 184
S G+ ++LGLQL +HR IVS+IA +LGG+DKA +YLN+CLYYV+IG+ND+ NYFLP
Sbjct: 404 YSAKGYAISLGLQLRNHRVIVSQIASQLGGIDKAQEYLNKCLYYVHIGSNDYINNYFLPQ 583
Query: 185 VFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV 236
++ +S +Y+P+QYA+ LI++LS LQ LH GARK VL GL LGC P ++
Sbjct: 584 LYLSSNVYSPEQYAENLIEELSLNLQALHEIGARKYVLPGLGLLGCTPSAIL 739
>BQ141293 weakly similar to GP|20466732|gb| unknown protein {Arabidopsis
thaliana}, partial (24%)
Length = 583
Score = 243 bits (619), Expect = 1e-64
Identities = 120/184 (65%), Positives = 147/184 (79%)
Frame = +2
Query: 6 KTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGI 65
KTWLV+ L+V +++ V G+SQVPC+F+FGDSLSDSGNNNNLPTSAK+N+ PYGI
Sbjct: 41 KTWLVV--LIVFLSANYFKQCVNGKSQVPCVFIFGDSLSDSGNNNNLPTSAKSNYKPYGI 214
Query: 66 DFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESG 125
DFP PTGR++NGR ID I Q+LGF+ FIPPFAN++GSDILKGVNYASG AGIR E+
Sbjct: 215 DFPMG-PTGRFTNGRTAIDIITQLLGFENFIPPFANISGSDILKGVNYASGGAGIRMETY 391
Query: 126 SQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNV 185
S G+ ++LGLQL +HR IVS+IA RLGG+DKA QYLN+CLYYVNIG+N + N F P
Sbjct: 392 SAKGYAISLGLQLRNHRVIVSQIASRLGGIDKAQQYLNKCLYYVNIGSNXYINNXFXPXF 571
Query: 186 FNTS 189
+ TS
Sbjct: 572 YPTS 583
>TC93849 similar to PIR|A86420|A86420 probable lipase/hydrolase
118270-120144 [imported] - Arabidopsis thaliana, partial
(48%)
Length = 673
Score = 198 bits (504), Expect = 2e-51
Identities = 94/196 (47%), Positives = 137/196 (68%)
Frame = +1
Query: 14 LVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPT 73
+VV+V S+ V QVPC F+FGDSL D+GNNN L + AKAN+LPYGIDFP PT
Sbjct: 91 IVVLVLSLWSGIGVA--QQVPCYFIFGDSLVDNGNNNQLTSIAKANYLPYGIDFPGG-PT 261
Query: 74 GRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVN 133
GR+SNG+ +D IA+ LGF +IP +A+ G IL+GVNYAS +AGIR+E+G QLG ++
Sbjct: 262 GRFSNGKTTVDVIAEQLGFNGYIPSYASARGRQILRGVNYASAAAGIREETGQQLGQRIS 441
Query: 134 LGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYT 193
Q+ +++ VS++ + G + A YL++C+Y + +G+ND+ NYF+P +++TSR +T
Sbjct: 442 FRGQVQNYQRTVSQLVNYFGDENTAANYLSKCIYTIGLGSNDYLNNYFMPTIYSTSRQFT 621
Query: 194 PKQYAKALIQQLSHYL 209
P+QYA L+Q + L
Sbjct: 622 PQQYANVLLQAYAQQL 669
>AW775312 similar to GP|20466732|gb unknown protein {Arabidopsis thaliana},
partial (46%)
Length = 655
Score = 187 bits (475), Expect = 5e-48
Identities = 89/183 (48%), Positives = 126/183 (68%), Gaps = 1/183 (0%)
Frame = +3
Query: 32 QVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQMLG 91
QVPC F+FGDSL D+GNNN + T A+AN+ PYGIDFP PTGR++NGR +D +AQ+LG
Sbjct: 105 QVPCFFIFGDSLVDNGNNNGILTLARANYRPYGIDFPQG-PTGRFTNGRTFVDALAQLLG 281
Query: 92 FQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRIAHR 151
F+ +IPP + G D+L+GVNYASG+AGIR+E+GS LG + ++ Q+ + V +
Sbjct: 282 FRAYIPPNSRARGLDVLRGVNYASGAAGIREETGSNLGAHTSMTEQVTNFGNTVQEMRRL 461
Query: 152 L-GGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQ 210
G D YL++C+YY +G+ND+ NYF+ + ++TS YTPK +A AL+Q + L
Sbjct: 462 FRGDNDALNSYLSKCIYYSGLGSNDYLNNYFMTDFYSTSTQYTPKAFASALLQDYARQLS 641
Query: 211 TLH 213
LH
Sbjct: 642 QLH 650
>TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif
lipase/hydrolase-like protein {Arabidopsis thaliana},
partial (94%)
Length = 1167
Score = 178 bits (451), Expect = 3e-45
Identities = 117/344 (34%), Positives = 179/344 (52%), Gaps = 16/344 (4%)
Frame = +3
Query: 7 TWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGID 66
T+ L+ L++ V + V G+ VP +F+FGDS+ D+ NNNNL T K+NF PYG D
Sbjct: 117 TYNFLTFLLLFVV----FNVVKGQPLVPALFIFGDSVVDARNNNNLYTIVKSNFPPYGRD 284
Query: 67 FPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANL--NGSDILKGVNYASGSAGIRKES 124
F PTGR+ NG+ D A+ LGF + P + NL ++L G N+ASG++G +
Sbjct: 285 FNNQMPTGRFCNGKLAADFTAENLGFTTYPPAYLNLQEKRKNLLNGANFASGASGY-FDP 461
Query: 125 GSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPN 184
++L H ++L QL H++ I + G A+ ++ +Y V G++DF QNY++
Sbjct: 462 TAKLYHAISLEQQLEHYKE-CQNILVGVAGKSNASSIISGAIYLVRAGSSDFVQNYYINP 638
Query: 185 VFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV-----NGS 239
+ +++T Q++ L+Q + ++Q L+ GARK + L LGC+P + +
Sbjct: 639 LL--YKVFTADQFSDILMQHYTIFIQNLYALGARKIGVTTLPPLGCLPAAITLFGSHSNE 812
Query: 240 CVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHG 294
CV+ N FN +L + L K+ L + ++ +HD GF
Sbjct: 813 CVDRLNNDALNFNTKLNTTSQNLQKE-LSNLTLAVLDIYQPLHDLVTKPTENGFYEARKA 989
Query: 295 CCPT---NEKGKCIRDGI-PCQNRHEYVFWDGIHTTEAANLVTA 334
CC T C +D I C N EYVFWDG HT+EAAN V A
Sbjct: 990 CCGTGLIETSILCNKDSIGTCANATEYVFWDGFHTSEAANNVLA 1121
>TC80033 weakly similar to GP|21593518|gb|AAM65485.1 putative GDSL-motif
lipase/hydrolase {Arabidopsis thaliana}, partial (89%)
Length = 1164
Score = 172 bits (437), Expect = 1e-43
Identities = 111/320 (34%), Positives = 167/320 (51%), Gaps = 16/320 (5%)
Frame = +1
Query: 31 SQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQML 90
++VP I VFGDS D+GNNN +PT A++NF PYG DF TGR+SNGR P D IA+
Sbjct: 121 AKVPAIIVFGDSSVDAGNNNFIPTVARSNFQPYGRDFQGGKATGRFSNGRIPTDFIAESF 300
Query: 91 GFQEFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRI 148
G +E +P + + N SD GV++AS + G + L + L QL +++ +
Sbjct: 301 GIKESVPAYLDPKYNISDFATGVSFASAATGYDNATSDVLS-VIPLWKQLEYYKDYQKNL 477
Query: 149 AHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHY 208
+ LG KA + +++ ++ +++GTNDF +NY+ + + YTP+QY L ++
Sbjct: 478 SSYLGEA-KAKETISESVHLMSMGTNDFLENYY--TMPGRASQYTPQQYQTFLAGIAENF 648
Query: 209 LQTLHHFGARKSVLVGLDRLGCIP-----KLLVNGSCVEEKNVATFLFNDQLKSLVDRLN 263
++ L+ GARK L GL +GC+P + CV N FND+LK++ +LN
Sbjct: 649 IRNLYALGARKISLGGLPPMGCLPLERTTNFMGQNGCVANFNNIALEFNDKLKNITTKLN 828
Query: 264 KKILMDSKFIFINSTAII-----HDKSVGFTVTHHGCCPTN--EKGKCIRDG--IPCQNR 314
++ L D K +F N I+ GF CC T E G G C +
Sbjct: 829 QE-LPDMKLVFSNPYYIMLHIIKKPDLYGFESASVACCATGMFEMGYACSRGSMFSCTDA 1005
Query: 315 HEYVFWDGIHTTEAANLVTA 334
++VFWD H TE N + A
Sbjct: 1006SKFVFWDSFHPTEKTNNIVA 1065
>TC76727 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (92%)
Length = 1919
Score = 165 bits (417), Expect = 3e-41
Identities = 108/340 (31%), Positives = 169/340 (48%), Gaps = 17/340 (5%)
Frame = -2
Query: 12 SLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATF 71
+L++++V+ + + ++ VP I FGDS D GNN+ LPT KAN+ PYG DF
Sbjct: 1156 TLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQ 977
Query: 72 PTGRYSNGRNPIDKIAQMLGFQEFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQLG 129
PTGR+ NG+ D A+ LGF F P + + +G ++L G N+AS ++G E + L
Sbjct: 976 PTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGY-DEKAATLN 800
Query: 130 HNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTS 189
H + L QL + + ++A ++ G KA + LY ++ G++DF QNY+ N
Sbjct: 799 HAIPLSQQLEYFKEYQGKLA-QVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWIN-- 629
Query: 190 RMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV-----NGSCVEEK 244
+ T QY+ L+ +++++ ++ GARK + L LGC+P CV
Sbjct: 628 QAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARI 449
Query: 245 NVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCCPT- 298
N FN ++ S L K+ L K + + ++D + GF GCC T
Sbjct: 448 NTDAQGFNKKVSSAASNLQKQ-LPGLKIVIFDIYKPLYDLVQNPSNFGFAEAGKGCCGTG 272
Query: 299 ---NEKGKCIRDGI-PCQNRHEYVFWDGIHTTEAANLVTA 334
C + C N +YVFWD +H +EAAN V A
Sbjct: 271 LVETTSLLCNPKSLGTCSNATQYVFWDSVHPSEAANQVLA 152
>TC83598 similar to PIR|H86419|H86419 probable lipase/hydrolase
114382-116051 [imported] - Arabidopsis thaliana, partial
(42%)
Length = 686
Score = 134 bits (338), Expect(2) = 4e-41
Identities = 66/121 (54%), Positives = 90/121 (73%), Gaps = 1/121 (0%)
Frame = +3
Query: 10 VLSLLVVMVASI-MQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFP 68
+++L V+MV + + S V + QVPC F+FGDSL D GNNNNL + AKAN+LPYGIDF
Sbjct: 102 IMNLCVMMVVVLGLWSSKVEADPQVPCYFIFGDSLVDDGNNNNLNSLAKANYLPYGIDFN 281
Query: 69 ATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQL 128
PTGR+SNG+ +D IA++LGF+ +I P++ +IL+GVNYAS +AGIR+E+G QL
Sbjct: 282 GG-PTGRFSNGKTTVDVIAELLGFEGYISPYSTARDQEILQGVNYASAAAGIREETGQQL 458
Query: 129 G 129
G
Sbjct: 459 G 461
Score = 51.2 bits (121), Expect(2) = 4e-41
Identities = 21/57 (36%), Positives = 38/57 (65%)
Frame = +1
Query: 137 QLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYT 193
Q+ +++ VS++ + LG D A+ YLN+C+Y + +G+ND+ NYF+P + +YT
Sbjct: 484 QVQNYQKTVSQVVNLLGDEDTASNYLNKCIYSIGLGSNDYLNNYFMPCLSQW*TVYT 654
>TC90972 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (88%)
Length = 1258
Score = 163 bits (413), Expect = 8e-41
Identities = 106/322 (32%), Positives = 164/322 (50%), Gaps = 17/322 (5%)
Frame = +3
Query: 30 ESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQM 89
++ VP I FGDS D GNN+ L T KAN+ PYG DF PTGR+ NG+ D A+
Sbjct: 93 DTVVPAIVTFGDSAVDVGNNDYLFTLFKANYPPYGRDFVXHKPTGRFCNGKLATDITAET 272
Query: 90 LGFQEFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSR 147
LGF+ + P + + G ++L G N+AS ++G E + L H + L QL +++ S+
Sbjct: 273 LGFKSYAPAYLSPQATGKNLLIGANFASAASGY-DEKAAILNHAIPLSQQLKYYKEYQSK 449
Query: 148 IAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSH 207
++ ++ G KA + LY ++ G++DF QNY++ + N ++ TP QY+ L+ S
Sbjct: 450 LS-KIAGSKKAASIIKGALYLLSGGSSDFIQNYYVNPLIN--KVVTPDQYSAYLVDTYSS 620
Query: 208 YLQTLHHFGARKSVLVGLDRLGCIPKLLV-----NGSCVEEKNVATFLFNDQLKSLVDRL 262
+++ L+ GARK + L LGC+P CV N FN ++ S +L
Sbjct: 621 FVKDLYKLGARKIGVTSLPPLGCLPATRTLFGFHEKGCVTRINNDAQGFNKKINSATVKL 800
Query: 263 NKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCCPT----NEKGKCIRDGI-PCQ 312
K+ L K + N +++ GF GCC T C + + C
Sbjct: 801 QKQ-LPGLKIVVFNIYKPLYELVQSPSKFGFAEARKGCCGTGIVETTSLLCNQKSLGTCS 977
Query: 313 NRHEYVFWDGIHTTEAANLVTA 334
N +YVFWD +H +EAAN + A
Sbjct: 978 NATQYVFWDSVHPSEAANQILA 1043
>TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(65%)
Length = 881
Score = 160 bits (405), Expect = 7e-40
Identities = 100/266 (37%), Positives = 149/266 (55%), Gaps = 7/266 (2%)
Frame = +1
Query: 11 LSLLVVMVASIMQHSTVLGESQVPCIF-VFGDSLSDSGNNNNLPTSAKANFLPYGIDF-P 68
++L++++V I H +P F VFGDSL D+GNNN L T+A+A+ PYGID+ P
Sbjct: 55 VALVILVVGGIFVHEI----EAIPRTFLVFGDSLVDNGNNNYLATTARADAPPYGIDYQP 222
Query: 69 ATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFA-NLNGSDILKGVNYASGSAGIRKESGSQ 127
+ PTGR+SNG N D I+Q LG + +P + L G +L G N+AS GI ++G Q
Sbjct: 223 SHRPTGRFSNGYNIPDIISQKLGAEPTLPYLSPELRGEKLLVGANFASAGIGILNDTGIQ 402
Query: 128 LGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFN 187
+ + + Q + + SR++ +G +A +NQ L + +G NDF NY+L
Sbjct: 403 FINIIRMYRQYEYFQEYQSRLSALIGA-SQAKSRVNQALVLITVGGNDFVNNYYLVPYSA 579
Query: 188 TSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV----NGSCVEE 243
SR Y +Y K LI + LQ L+ GAR+ ++ G +GC+P + NG C E
Sbjct: 580 RSRQYPLPEYVKYLISEYQKLLQKLYDLGARRVLVTGTGPMGCVPSEIAQRGRNGQCSTE 759
Query: 244 KNVATFLFNDQLKSLVDRLNKKILMD 269
A+ LFN QL++++ LNKKI D
Sbjct: 760 LQRASSLFNPQLENMLLGLNKKIGRD 837
>TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protein
{Arabidopsis thaliana}, partial (60%)
Length = 1398
Score = 159 bits (403), Expect = 1e-39
Identities = 107/349 (30%), Positives = 174/349 (49%), Gaps = 17/349 (4%)
Frame = +1
Query: 1 MGCESKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANF 60
MG WL+L ++MV T E+ VP + VFGDS DSGNNN + T K+NF
Sbjct: 52 MGNICIVWLILITQMIMV-------TCNNENYVPAVIVFGDSSVDSGNNNMISTFLKSNF 210
Query: 61 LPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFAN--LNGSDILKGVNYASGSA 118
PYG D PTGR+SNGR P D I++ G + IP + + D + GV +AS
Sbjct: 211 RPYGRDIDGGRPTGRFSNGRIPPDFISEAFGIKSLIPAYLDPAYTIDDFVTGVCFASAGT 390
Query: 119 GIRKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQ 178
G + + L + + L ++ ++ ++ +G +K+ + +++ LY +++GTNDF
Sbjct: 391 GYDNATSAIL-NVIPLWKEVEFYKEYQDKLKAHIGE-EKSIEIISEALYIISLGTNDFLG 564
Query: 179 NYFLPNVFNTSRM-YTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIP-KLLV 236
NY+ F T R YT QY LI ++++ L+ GARK + GL +GC+P + +
Sbjct: 565 NYY---GFTTLRFRYTISQYQDYLIGIAENFIRQLYSLGARKLAITGLIPMGCLPLERAI 735
Query: 237 N-----GSCVEEKNVATFLFNDQLKSLVDRLNKKI----LMDSKFIFINSTAIIHDKSVG 287
N C E+ N+ FN +L++++ +LNK++ + + + + I G
Sbjct: 736 NIFGGFHRCYEKYNIVALEFNVKLENMISKLNKELPQLKALSANVYDLFNDIITRPSFYG 915
Query: 288 FTVTHHGCCPTNE---KGKCIRDGI-PCQNRHEYVFWDGIHTTEAANLV 332
CC T C + + C++ +Y+FWD H TE N +
Sbjct: 916 IEEVEKACCSTGTIEMSYLCNKMNLMTCKDASKYMFWDAFHPTEKTNRI 1062
>BF636081 weakly similar to GP|6733842|emb| unnamed protein product
{unidentified}, partial (22%)
Length = 348
Score = 153 bits (386), Expect = 1e-37
Identities = 77/110 (70%), Positives = 91/110 (82%)
Frame = +1
Query: 6 KTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGI 65
K WLV+ L+V A+ +Q V G+SQVPC+F+FGDSLSDSGNNNNLPTSAK+N+ PYGI
Sbjct: 28 KKWLVV-LIVFFSANYLQQC-VNGKSQVPCLFIFGDSLSDSGNNNNLPTSAKSNYNPYGI 201
Query: 66 DFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYAS 115
DFP PTGR++NGR ID I Q+LGF+ FIPPFAN+NGSDILKGVNYAS
Sbjct: 202 DFPMG-PTGRFTNGRTSIDIITQLLGFENFIPPFANINGSDILKGVNYAS 348
>BI273040 similar to GP|6721157|gb| putative GDSL-motif lipase/acylhydrolase
{Arabidopsis thaliana}, partial (60%)
Length = 692
Score = 146 bits (369), Expect = 1e-35
Identities = 83/225 (36%), Positives = 124/225 (54%), Gaps = 5/225 (2%)
Frame = +2
Query: 21 IMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTGRYSNGR 80
++ +TV+ + + FVFGDSL D+GNNN L T+A+A+ PYGID+P TGR+SNG
Sbjct: 5 LVNFNTVVPQVEARAFFVFGDSLVDNGNNNYLATTARADSYPYGIDYPTHRATGRFSNGL 184
Query: 81 NPIDKIAQMLGFQEFIPPFA-NLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLL 139
N D I++ +G Q +P + LNG +L G N+AS GI ++G Q + + + QL
Sbjct: 185 NMPDLISERIGSQPTLPYLSPELNGEALLVGANFASAGIGILNDTGIQFFNIIRITRQLQ 364
Query: 140 HHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAK 199
+ R++ L G ++ + +N+ LY + +G NDF NYFL SR + Y
Sbjct: 365 YFEQYQQRVS-ALIGEEETVRLVNEALYLMTLGGNDFVNNYFLVPFSARSRQFXLPDYVV 541
Query: 200 ALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV----NGSC 240
LI + L L+ GAR+ ++ G LGC+P L NG C
Sbjct: 542 YLISEYRKILARLYELGARRVLVTGTXPLGCVPXXLXXHXKNGEC 676
>TC86678 similar to PIR|E96579|E96579 hypothetical protein T18A20.15
[imported] - Arabidopsis thaliana, partial (43%)
Length = 1341
Score = 140 bits (352), Expect = 1e-33
Identities = 110/345 (31%), Positives = 165/345 (46%), Gaps = 20/345 (5%)
Frame = +2
Query: 36 IFVFGDSLSDSGNNN--NLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQ 93
+F+FGDS D+GNNN N T +ANFLPYG + FPTGR+S+GR D IA+ +
Sbjct: 185 LFIFGDSFLDAGNNNYINTTTFDQANFLPYGETY-FNFPTGRFSDGRLISDFIAEYVNI- 358
Query: 94 EFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLG 153
+PPF + + GVN+ASG AG E+ G + Q ++ + + + + H+LG
Sbjct: 359 PLVPPFLQPDNNKYYNGVNFASGGAGALVETFQ--GSVIPFKTQAINFKKVTTWLRHKLG 532
Query: 154 GLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLH 213
D T L+ +Y +IG+ND+ + FL N + + Y+ +Y +I + ++ +H
Sbjct: 533 SSDSKT-LLSNAVYMFSIGSNDY-LSPFLTN-SDVLKHYSHTEYVAMVIGNFTSTIKEIH 703
Query: 214 HFGARKSVLVGLDRLGCIPKLLV-----NGSCVEEKNVATFLFNDQLKSLVDRLNKKILM 268
GA+K V++ L LGC+P + GSC+EE + + N L ++ L K+ L
Sbjct: 704 KRGAKKFVILNLPPLGCLPGTRIIQSQGKGSCLEELSSLASIHNQALYEVLLELQKQ-LR 880
Query: 269 DSKFIFIN-----STAIIHDKSVGFTVTHHGCCPTNE-KGKCIRDGIP-------CQNRH 315
KF + S I H GF CC + +G+ G C +
Sbjct: 881 GFKFSLYDFNSDLSHMINHPLKYGFKEGKSACCGSGPFRGEYSCGGKRGEKHFELCDKPN 1060
Query: 316 EYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLIQSNTI 360
E VFWD H TE+A Y + PT H I S TI
Sbjct: 1061ESVFWDSYHLTESA--------YKQLAAQMWSPTGNSHTIGSYTI 1171
>TC87519 weakly similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(76%)
Length = 1084
Score = 139 bits (351), Expect = 1e-33
Identities = 91/299 (30%), Positives = 146/299 (48%), Gaps = 9/299 (3%)
Frame = +2
Query: 66 DFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFA-NLNGSDILKGVNYASGSAGIRKES 124
DFP PTGR+SNG N D I++ LG +P + L G +L G N+AS GI ++
Sbjct: 2 DFPTRQPTGRFSNGLNVPDLISKELGSSPPLPYLSPKLRGHRMLNGANFASAGIGILNDT 181
Query: 125 GSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPN 184
G Q + + QL R++ +G + A + +N L + G NDF NY+L
Sbjct: 182 GFQFIEVIRMYKQLDFFEEYQKRVSDLIGKKE-AKKLINGALILITCGGNDFVNNYYLVP 358
Query: 185 VFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLL----VNGSC 240
SR Y +Y L+ + L+ L+H GAR+ ++ G +GC P L +G C
Sbjct: 359 NSLRSRQYALPEYVTYLLSEYKKILRRLYHLGARRVLVSGTGPMGCAPAALAIGGTDGEC 538
Query: 241 VEEKNVATFLFNDQLKSLVDRLNKKILMD-SKFIFINSTAIIHDKSVGFTVTHHGCC--- 296
E +A L+N +L L+ LN++I D + I++ ++ ++ F + CC
Sbjct: 539 APELQLAASLYNPKLVQLITELNQQIGSDVFSVLNIDALSLFGNE---FKTSKVACCGQG 709
Query: 297 PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLI 355
P N G C CQNR +++FWD H +E AN + + + + + YP N+ ++
Sbjct: 710 PYNGIGLCTLASSICQNRDDHLFWDAFHPSERANKM-IVKQIMTGSTDVIYPMNLSTIL 883
>TC91750 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(50%)
Length = 686
Score = 132 bits (333), Expect = 2e-31
Identities = 76/206 (36%), Positives = 113/206 (53%), Gaps = 1/206 (0%)
Frame = +3
Query: 15 VVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTG 74
VV++ + S G FVFGDSL D+GNNN L T+A+A+ PYGID+P PTG
Sbjct: 54 VVIILMVALTSCFKGTVAQRAFFVFGDSLVDNGNNNYLATTARADAPPYGIDYPTRRPTG 233
Query: 75 RYSNGRNPIDKIAQMLGFQEFIPPFA-NLNGSDILKGVNYASGSAGIRKESGSQLGHNVN 133
R+SNG N D I+Q LG + +P + LNG +L G N+AS GI ++G Q + +
Sbjct: 234 RFSNGYNIPDFISQALGAEPTLPYLSPELNGEALLVGANFASAGIGILNDTGIQFINIIR 413
Query: 134 LGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYT 193
+ QL + + R++ +G ++ +N L + +G NDF NY+L SR Y
Sbjct: 414 IFRQLEYFQQYQQRVSGLIGP-EQTQSLVNGALVLITLGGNDFVNNYYLVPFSARSRQYN 590
Query: 194 PKQYAKALIQQLSHYLQTLHHFGARK 219
Y + +I + L+ L+H GAR+
Sbjct: 591 LPDYVRYIISEYKKILRRLYHLGARR 668
>TC93226 weakly similar to PIR|B96737|B96737 hypothetical protein F3I17.10
[imported] - Arabidopsis thaliana, partial (60%)
Length = 847
Score = 127 bits (319), Expect = 7e-30
Identities = 76/199 (38%), Positives = 107/199 (53%), Gaps = 2/199 (1%)
Frame = +2
Query: 42 SLSDSGNNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFAN 101
SL + GNNN L T AK+NF PYGID+ PTGR+SNG++ ID I MLG PPF +
Sbjct: 2 SLVEVGNNNFLSTFAKSNFYPYGIDYNGR-PTGRFSNGKSLIDFIGDMLGVPS-PPPFLD 175
Query: 102 LNGSD--ILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKAT 159
++ +L GVNYASGS GI +SG G ++ QL + +++ + +
Sbjct: 176 PTSTENKLLNGVNYASGSGGILDDSGRHYGDRHSMSRQLQNFERTLNQYKKMMNET-ALS 352
Query: 160 QYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARK 219
Q+L + + V G+ND+ NY P + TSR Y+ Q+ L+ + L+ RK
Sbjct: 353 QFLAKSIVIVVTGSNDYINNYLRPEYYGTSRNYSVPQFGNLLLNTFGRQILALYSLRLRK 532
Query: 220 SVLVGLDRLGCIPKLLVNG 238
L G+ LGCIP NG
Sbjct: 533 FFLAGVGPLGCIPNQRANG 589
>TC80992 weakly similar to GP|21593518|gb|AAM65485.1 putative GDSL-motif
lipase/hydrolase {Arabidopsis thaliana}, partial (53%)
Length = 812
Score = 123 bits (309), Expect = 1e-28
Identities = 76/225 (33%), Positives = 118/225 (51%), Gaps = 3/225 (1%)
Frame = +1
Query: 3 CESKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLP 62
C WL+L ++MV T ++ VP + VFGDS DSGNNN + T K+NF P
Sbjct: 64 CICIAWLILITQIIMV-------TCKTKNHVPAVIVFGDSSVDSGNNNRIATLLKSNFKP 222
Query: 63 YGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFAN--LNGSDILKGVNYASGSAGI 120
YG DF PTGR+ NGR P D IA+ G + IP + + D + GV +AS G
Sbjct: 223 YGRDFEGGRPTGRFCNGRTPPDFIAEAFGVKRNIPAYLDPAYTIDDFVTGVCFASAGTGY 402
Query: 121 RKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNY 180
+ L + + L ++ + ++ + G KA + +++ LY +++GTNDF +NY
Sbjct: 403 DNATSDVL-NVIPLWKEIEFFKEYQEKLRVHV-GKKKANEIISEALYLISLGTNDFLENY 576
Query: 181 FLPNVFNTSRM-YTPKQYAKALIQQLSHYLQTLHHFGARKSVLVG 224
+ +F T ++ +T QY L+ +++ LH GARK + G
Sbjct: 577 Y---IFPTRQLHFTVSQYQDFLVDIAEDFVRKLHSLGARKLSITG 702
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,226,334
Number of Sequences: 36976
Number of extensions: 178490
Number of successful extensions: 1086
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 985
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 994
length of query: 361
length of database: 9,014,727
effective HSP length: 97
effective length of query: 264
effective length of database: 5,428,055
effective search space: 1433006520
effective search space used: 1433006520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0476a.9


