

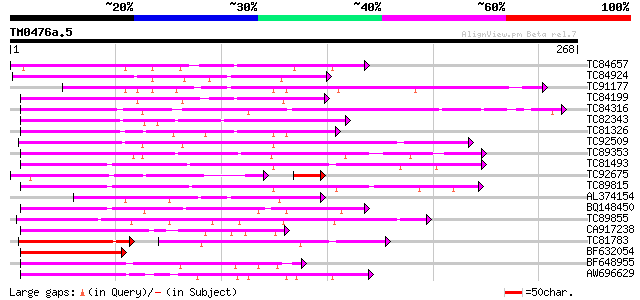
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476a.5
(268 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84657 weakly similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.... 129 9e-31
TC84924 weakly similar to PIR|F96545|F96545 hypothetical protein... 120 7e-28
TC91177 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 119 2e-27
TC84199 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 117 5e-27
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 115 2e-26
TC82343 similar to GP|10177628|dbj|BAB10775. gb|AAF30317.1~gene_... 112 1e-25
TC81326 weakly similar to PIR|T45651|T45651 hypothetical protein... 106 8e-24
TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein... 98 3e-21
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 97 7e-21
TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [i... 92 3e-19
TC92675 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 80 1e-18
TC89815 86 2e-17
AL374154 79 1e-15
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 76 2e-14
TC89855 71 5e-13
CA917238 weakly similar to PIR|A84538|A845 hypothetical protein ... 70 9e-13
TC81783 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 59 5e-12
BF632054 similar to GP|10177628|dbj gb|AAF30317.1~gene_id:K6M13.... 67 6e-12
BF648955 similar to GP|18043667|gb| Unknown (protein for IMAGE:3... 66 1e-11
AW696629 65 4e-11
>TC84657 weakly similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 884
Score = 129 bits (325), Expect = 9e-31
Identities = 85/197 (43%), Positives = 114/197 (57%), Gaps = 27/197 (13%)
Frame = +1
Query: 1 MEKLT---LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAP-THR 56
MEK T LPHELI +I+LRLPV+SL+RFKCVCKS LISD FAKSHF+L+ A T+R
Sbjct: 304 MEKKTGLYLPHELIIQIMLRLPVKSLIRFKCVCKSLLALISDHNFAKSHFELSTATHTNR 483
Query: 57 LLLRCLFDLD--------SLGDNSAVVTLNL---PPPCMSCIQNSLYFLGSCRGFMLLAN 105
++ L+ SL D+SA +LNL PP S SL SCRGF++L
Sbjct: 484 IVFMSTLALETRSIDFEASLNDDSASTSLNLNFMPPESYS----SLEIKSSCRGFIVLTC 651
Query: 106 EYQRVIVWNPSTGFHKQILTSCDFIIGS----LYGFGYDNSTDDYFLVLI--------VL 153
+ +WNPSTG HKQI + LYGFGYD+ DDY +V + V
Sbjct: 652 S-SNIYLWNPSTGHHKQIPFPASNLDAKYSCCLYGFGYDHLRDDYLVVSVSYNTSIDPVD 828
Query: 154 ASMNAKIHAYSVKTNSW 170
++++ + +S++ N+W
Sbjct: 829 DNISSHLKFFSLRANTW 879
>TC84924 weakly similar to PIR|F96545|F96545 hypothetical protein F8A12.9
[imported] - Arabidopsis thaliana, partial (9%)
Length = 575
Score = 120 bits (300), Expect = 7e-28
Identities = 74/160 (46%), Positives = 91/160 (56%), Gaps = 9/160 (5%)
Frame = +1
Query: 2 EKLTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRC 61
+ + LP ELI + LLRLPV+SLL FKC+CK WF +ISDP FA SHF LN A R L C
Sbjct: 49 QSMDLPQELIIQFLLRLPVKSLLVFKCICKLWFSIISDPHFANSHFQLNHAKHTRRFL-C 225
Query: 62 LFDLD-SLGDNSAVVTLNLPP--PCMSCIQNSLYF----LGSCRGFMLLANEYQRVIVWN 114
+ L + LN P P +C YF GSCRGF+ + + + +WN
Sbjct: 226 ISALSPEIRSIDFDAFLNDAPASPNFNCSLPDSYFPFEIKGSCRGFIFM-YRHPNIYIWN 402
Query: 115 PSTGFHKQILTSC--DFIIGSLYGFGYDNSTDDYFLVLIV 152
PSTG +QIL S +LYGFGYD S DDY +V IV
Sbjct: 403 PSTGSKRQILMSAFNTKAYINLYGFGYDQSRDDYVVVFIV 522
>TC91177 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 1298
Score = 119 bits (297), Expect = 2e-27
Identities = 95/268 (35%), Positives = 128/268 (47%), Gaps = 39/268 (14%)
Frame = +2
Query: 26 FKCVCKSWFYLISDPQFAKSHFDLNAAP-THRLLL------RCLFDLD-SLGDNSAVVTL 77
FKCVCK W LIS P FA SHF L A T+R++L DL+ SL D+SAV T
Sbjct: 74 FKCVCKLWLSLISQPHFANSHFQLTTATHTNRIMLITPYLQSLSIDLELSLNDDSAVYTT 253
Query: 78 --------------------NLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPST 117
+L PP I L F GSCRGF+LL N Y + +WNPST
Sbjct: 254 DISFLIDDEDYYSSSSSDMDDLSPPKSFFI---LDFKGSCRGFILL-NCYSSLCIWNPST 421
Query: 118 GFHKQI-LTSCDF--IIGSLYGFGYDNSTDDYFLVLIVLA------SMNAKIHAYSVKTN 168
GFHK+I T+ D YGFGYD STDDY ++ + M + + +S++ N
Sbjct: 422 GFHKRIPFTTIDSNPDANYFYGFGYDESTDDYLVISMSYEPSPSSDGMLSHLGIFSLRAN 601
Query: 169 SWDYNHVHVLYMHLGCDYRHGV--FLNNSLHWLVISKVTSLQVVIAYDLSETSLSEIPLL 226
W L ++ + V N ++HWL S+ V++A+ L E L E+ L
Sbjct: 602 VWTRVEGGNLLLYSQNSLLNLVESLSNGAIHWLAFRNDISMPVIVAFHLMERKLLELRLP 781
Query: 227 PELAEPVLKAECMPTFYQVRVLGGCLSL 254
E+ +A Y + V GCL+L
Sbjct: 782 NEIINGPSRA------YDLWVYRGCLAL 847
Score = 35.0 bits (79), Expect = 0.030
Identities = 23/56 (41%), Positives = 30/56 (53%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRC 61
LP ELI +ILLRLPV+SL+R S+ YL+ + H N P H + C
Sbjct: 15 LPFELIIQILLRLPVKSLIRLNAFV-SYGYLLYLNLTLQIHI-FNLPPLHTRIELC 176
>TC84199 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 673
Score = 117 bits (293), Expect = 5e-27
Identities = 74/161 (45%), Positives = 92/161 (56%), Gaps = 15/161 (9%)
Frame = +1
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLL------ 59
LP ELI +ILL LPV+SLLRFKCVCKSWF LISD FA SHF + A + R+L
Sbjct: 109 LPLELIIQILLWLPVKSLLRFKCVCKSWFSLISDTHFANSHFQITAKHSRRVLFMLNHVP 288
Query: 60 -RCLFDLDSLGDNSAVVTLNLP------PPCMSCIQNSLYFLGSCRGFMLLANEYQRVIV 112
D ++L ++AV + P PPC S NS SCRGF+ L N+ + +
Sbjct: 289 TTLSLDFEALHCDNAVSEIPNPIPNFVEPPCDSLDTNS----SSCRGFIFLHND-PDLFI 453
Query: 113 WNPSTGFHKQILTSCD--FIIGSLYGFGYDNSTDDYFLVLI 151
WNPST +KQI S + LYGFGYD DDY +V +
Sbjct: 454 WNPSTRVYKQIPLSPNDSNSFHCLYGFGYDQLRDDYLVVSV 576
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 115 bits (287), Expect = 2e-26
Identities = 86/273 (31%), Positives = 130/273 (47%), Gaps = 15/273 (5%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRC---- 61
LP ELI ILLRLPVRSLLRFKCVCKSW L SD FA +HF ++ + L+ C
Sbjct: 42 LPEELIVIILLRLPVRSLLRFKCVCKSWKTLFSDTHFANNHFLIST--VYPQLVACESVS 215
Query: 62 --------LFDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVI-V 112
+ ++SL +NS+ + P + LGSC GF+ L + YQR + +
Sbjct: 216 AYRTWEIKTYPIESLLENSSTTVI----PVSNTGHQRYTILGSCNGFLCLYDNYQRCVRL 383
Query: 113 WNPSTGFHKQILTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASMNAKIHAYSVKTNSWDY 172
WNPS + + D I YGFGYD Y L+ + S + Y+ NS
Sbjct: 384 WNPSINLKSKSSPTIDRFI--YYGFGYDQVNHKYKLLAVKAFSRITETMIYTFGENSCKN 557
Query: 173 NHVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLSETSLSEIPLLPELAEP 232
V + G F++ +L+W+V + ++++D+ + + ++ LLP+
Sbjct: 558 VEVKDFPRYPPNRKHLGKFVSGTLNWIVDER-DGRATILSFDIEKETYRQV-LLPQHGYA 731
Query: 233 VLKAECMPTFYQVRVLGGCLSLC--YTGSRWDM 263
V P Y VL C+ +C + +RW +
Sbjct: 732 VYS----PGLY---VLSNCICVCTSFLDTRWQL 809
>TC82343 similar to GP|10177628|dbj|BAB10775.
gb|AAF30317.1~gene_id:K6M13.17~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 112 bits (280), Expect = 1e-25
Identities = 75/166 (45%), Positives = 92/166 (55%), Gaps = 10/166 (6%)
Frame = +2
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCL--- 62
LP ELI +IL+RLPV+SL+RFK VCKSWF LISD FA SHF + AA TH L + L
Sbjct: 65 LPDELITKILVRLPVKSLIRFKSVCKSWFSLISDNHFANSHFQVTAA-THTLRILFLTAT 241
Query: 63 -----FDLDSL--GDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNP 115
+DSL D + V LN P + + L SCRGF + + +WNP
Sbjct: 242 PEFRYIAVDSLFTDDYNEPVPLN-PNFPLPEFEFDLEIKASCRGF-IYVHTCSEAYIWNP 415
Query: 116 STGFHKQILTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASMNAKIH 161
STGF KQI + YGFGYD STDDY +V + L + H
Sbjct: 416 STGFLKQIPFPPNVSNLIFYGFGYDESTDDYLVVSVYLVIIMNLFH 553
>TC81326 weakly similar to PIR|T45651|T45651 hypothetical protein F13I12.200
- Arabidopsis thaliana, partial (12%)
Length = 838
Score = 106 bits (265), Expect = 8e-24
Identities = 76/164 (46%), Positives = 90/164 (54%), Gaps = 13/164 (7%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LPHELIF+ILLRLPV+SL RFK V KSWF LIS P FA SHF L +A R +F +
Sbjct: 168 LPHELIFQILLRLPVKSLTRFKSVRKSWFSLISAPHFANSHFQLTSA--KHAASRIMF-I 338
Query: 66 DSLGDNSAVVT----LNLPPPCMSCIQNSL-------YFLGSCRGFMLLANEYQRVIVWN 114
+L + + LN P I SL GSCRGF+LL + +WN
Sbjct: 339 STLSHETRSIDFKAFLNDDDPASLNITFSLTRSHFPVEIRGSCRGFILLYRP-PDIYIWN 515
Query: 115 PSTGFHKQI-LTSCDF-IIGSLYGFGYDNSTDDYFLVLIVLASM 156
PSTGF K I L+ D + GFGYD S DDY IVL S+
Sbjct: 516 PSTGFKKHIHLSPVDSKSVAQCQGFGYDQSRDDYLGGFIVL*SI 647
Score = 35.4 bits (80), Expect = 0.023
Identities = 21/80 (26%), Positives = 35/80 (43%), Gaps = 5/80 (6%)
Frame = +1
Query: 143 TDDYFLVLIVLASMNAKIHA-----YSVKTNSWDYNHVHVLYMHLGCDYRHGVFLNNSLH 197
T + L+V S N + +SV+ N+W + + R G+ N +H
Sbjct: 598 TSQEMITLVVSLSYNPSAFSTHLKFFSVRDNTWKEIEGNYFPYGVLSSCREGLLFNGVIH 777
Query: 198 WLVISKVTSLQVVIAYDLSE 217
WL + + L V+ A+DL E
Sbjct: 778 WLALRRDLGLTVIGAFDLME 837
>TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein F26G5.80 -
Arabidopsis thaliana, partial (8%)
Length = 858
Score = 98.2 bits (243), Expect = 3e-21
Identities = 70/232 (30%), Positives = 106/232 (45%), Gaps = 17/232 (7%)
Frame = +3
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRC--- 61
TLP EL+ EIL RLPV+ L + +C+CKS+ LISDP+FAK H + P H L+LR
Sbjct: 171 TLPFELVAEILCRLPVKLLXQLRCLCKSFNSLISDPKFAKKHLHSSTTP-HHLILRSNNG 347
Query: 62 --LFDLDSLGDNSAVVTLNLP--------PPCMSCIQNSLYFLGSCRGFMLLANEYQRVI 111
F L S + T +P P C++ S Y SC G + L +Y +
Sbjct: 348 SGRFALIVSPIQSVLSTSTVPVPQTQLTYPTCLTEEFASPYEWCSCDGIICLTTDYSSAV 527
Query: 112 VWNPSTGFHKQI----LTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASMNAKIHAYSVKT 167
+WNP K + S L+ FGYD D+Y + I + +++ T
Sbjct: 528 LWNPFINKFKTLPPLKYISLKRSPSCLFTFGYDPFADNYKVFAITFCVKRTTVEVHTMGT 707
Query: 168 NSWDYNHVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLSETS 219
+SW + + G+F+ +HWL S + +++ DL + S
Sbjct: 708 SSWRRIEDFPSWSFIP---DSGIFVAGYVHWLTYDGPGSQREIVSLDLEDES 854
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 97.1 bits (240), Expect = 7e-21
Identities = 76/245 (31%), Positives = 112/245 (45%), Gaps = 25/245 (10%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRL-----LLR 60
+P E+I EILLRLPVRSLL+F+CVCK W LISDPQFAK H ++ A + + +
Sbjct: 105 MPEEIIVEILLRLPVRSLLQFRCVCKLWKTLISDPQFAKKHVSISTAYPQLVSVFVSIAK 284
Query: 61 C---LFDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPST 117
C + L L DN + + P S+ +GSC G + L++ YQ +WNPS
Sbjct: 285 CNLVSYPLKPLLDNPSAHRVE-PADFEMIHTTSMTIIGSCNGLLCLSDFYQ-FTLWNPSI 458
Query: 118 GFHKQ------ILTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASMN---AKIHAYSVKTN 168
+ S D GFGYD D Y ++ +V N K Y+
Sbjct: 459 KLKSKPSPTIIAFDSFDSKRFLYRGFGYDQVNDRYKVLAVVQNCYNLDETKTLIYTFGGK 638
Query: 169 SWDYNHVHVLYMHLGCDYRH--------GVFLNNSLHWLVISKVTSLQVVIAYDLSETSL 220
W CD G F++ +L+W+V S +V++ +D+ + +
Sbjct: 639 DW------TTIQKFPCDPSRCDLGRLGVGKFVSGNLNWIV-----SKKVIVFFDIEKETY 785
Query: 221 SEIPL 225
E+ L
Sbjct: 786 GEMSL 800
>TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 728
Score = 91.7 bits (226), Expect = 3e-19
Identities = 75/242 (30%), Positives = 108/242 (43%), Gaps = 22/242 (9%)
Frame = +1
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LP E+ EIL R+P + LLR + CK W LI F H L+ + ++LR L
Sbjct: 16 LPTEVTTEILSRVPAKPLLRLRSTCKWWRNLIDSTDFIFLH--LSKSRDSVIILRQHSRL 189
Query: 66 DSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQI-- 123
L NS L P M C N + LGSC G + + N + WNP+ H+ I
Sbjct: 190 YELDLNSMDRVKELDHPLM-CYSNRIKVLGSCNGLLCICNIADDIAFWNPTIRKHRIIPS 366
Query: 124 -----------LTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASMNAKIHAYSVKTNSWDY 172
T + +YGFGYD++TDDY LV I S +H S ++ Y
Sbjct: 367 EPLIRKETNENNTITTLLAAHVYGFGYDSATDDYKLVSI---SNFVDLHNRSYDSHVTIY 537
Query: 173 NHVHVLYMHLG-------CDYRHGVFLNNSLHWLV--ISKVTSLQVVIAYDLSETSLSEI 223
++M L C GVF++ +LHW+V + S +++A+DL E+
Sbjct: 538 TMGSDVWMPLPGVPYALCCARPMGVFVSGALHWVVPRALEPDSRDLIVAFDLRFEVFREV 717
Query: 224 PL 225
L
Sbjct: 718 AL 723
>TC92675 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (17%)
Length = 630
Score = 79.7 bits (195), Expect(2) = 1e-18
Identities = 52/125 (41%), Positives = 66/125 (52%), Gaps = 3/125 (2%)
Frame = +3
Query: 1 MEKLTLPH---ELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRL 57
ME TLPH EL+ +ILLRLPV+SL+RFK VCKSWF L+SD FA SHF N P H L
Sbjct: 48 METTTLPHMPNELMIQILLRLPVKSLIRFKSVCKSWFSLVSDTHFAYSHF--NFPPQHTL 221
Query: 58 LLRCLFDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPST 117
+ R LF + + S + L+L + +SLY +WNP
Sbjct: 222 VGRVLF-ISTSARKSRSIDLDLEASPLDDDDSSLY-------------------LWNPFI 341
Query: 118 GFHKQ 122
HK+
Sbjct: 342 RVHKE 356
Score = 30.0 bits (66), Expect(2) = 1e-18
Identities = 11/15 (73%), Positives = 13/15 (86%)
Frame = +1
Query: 135 YGFGYDNSTDDYFLV 149
YGFGYD ST+DY +V
Sbjct: 403 YGFGYDQSTNDYLVV 447
>TC89815
Length = 807
Score = 85.5 bits (210), Expect = 2e-17
Identities = 68/243 (27%), Positives = 110/243 (44%), Gaps = 24/243 (9%)
Frame = +1
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
+P E+ +IL LP LLRF+ KS +I F H L + L++R +L
Sbjct: 46 IPPEIFIDILSLLPPHPLLRFRSTSKSLKSIIDSHTFTNLH--LKNSNNFYLIIRHNANL 219
Query: 66 DSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQI-- 123
L + + L P MS N + GSC G + ++N + WNP+ H+ I
Sbjct: 220 YQLDFPNLTPPIPLNHPLMS-YSNRITLFGSCNGLICISNIADDIAFWNPNIRKHRIIPY 396
Query: 124 -------LTSCDFIIGSLYGFGYDNSTDDYFLVLIVL------ASMNAKIHAYSVKTNSW 170
+ ++GFGYD+ DY LV I S ++++ +S+K NSW
Sbjct: 397 LPTTPRSESDTTLFAARVHGFGYDSFAGDYKLVRISYFVDLQNRSFDSQVRVFSLKMNSW 576
Query: 171 DYNHVHVLYMHLGCDYRHGVFL-------NNSLHWLVISKVTSLQ--VVIAYDLSETSLS 221
+ + L C GVF+ +NSLHW+V K+ Q +++A++L+ +
Sbjct: 577 --KELPSMNYALCCARTMGVFVEDSNNLNSNSLHWVVTRKLEPFQPDLIVAFNLTLEIFN 750
Query: 222 EIP 224
E+P
Sbjct: 751 EVP 759
>AL374154
Length = 389
Score = 79.3 bits (194), Expect = 1e-15
Identities = 57/129 (44%), Positives = 64/129 (49%), Gaps = 10/129 (7%)
Frame = +2
Query: 31 KSWFYLISDPQFAKSHFDLNAAP------THRLLLRCLFDLDSLGDNSAV-VTLNLPPPC 83
KSWF LIS FA SHF + P T R + SL D +V +LNL
Sbjct: 5 KSWFSLISXTNFANSHFQITHTPRVLFISTSTCETRSIDFESSLNDXXSVSASLNLDFLL 184
Query: 84 MSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQILTS---CDFIIGSLYGFGYD 140
N L GSCRGF+ L + Y +WNPSTGFHKQI S YGFGYD
Sbjct: 185 PESYLN-LEIKGSCRGFIFL-HCYPNKYLWNPSTGFHKQIPFSPFGSSLYAEYFYGFGYD 358
Query: 141 NSTDDYFLV 149
STDDY LV
Sbjct: 359 PSTDDYLLV 385
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar to
unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 75.9 bits (185), Expect = 2e-14
Identities = 59/189 (31%), Positives = 85/189 (44%), Gaps = 24/189 (12%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCL--- 62
LP E+ EIL RLPV+ L++F+CVCK W IS P F K H L + T L L
Sbjct: 3 LPFEIQVEILSRLPVKYLMQFQCVCKLWKSQISKPDFVKKH--LRVSNTRHLFLLTFSKL 176
Query: 63 --------FDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWN 114
+ L S+ L P ++ S +GSC G + + ++WN
Sbjct: 177 SPELVIKSYPLSSVFTEMTPTFTQLEYP-LNNRDESDSMVGSCHGILCIQCNLSFPVLWN 353
Query: 115 PS-TGFHKQILTSCDF----IIGSLYGFGYDNSTDDYFLVLIVLAS--------MNAKIH 161
PS F K L S +F I Y FGYD+S+D Y +V + S + ++
Sbjct: 354 PSIRKFTK--LPSFEFPQNKFINPTYAFGYDHSSDTYKVVAVFCTSNIDNGVYQLKTLVN 527
Query: 162 AYSVKTNSW 170
+++ TN W
Sbjct: 528 VHTMGTNCW 554
>TC89855
Length = 766
Score = 70.9 bits (172), Expect = 5e-13
Identities = 59/229 (25%), Positives = 105/229 (45%), Gaps = 33/229 (14%)
Frame = +3
Query: 4 LTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHR------- 56
L P +++F IL +LP++ L RF CV KSW +L +P F + F N +
Sbjct: 57 LIYPDDIVFSILSKLPIKHLKRFACVRKSWSHLFENPIF-MNMFRNNLVSKSQTGYDDDD 233
Query: 57 LLLRCLFDLDSLGDNSAV--------VTLNLPPPCMSCIQNSLYFL----GSCRGFMLLA 104
L C + LD + S + + L+LPP + L ++ + G + +
Sbjct: 234 ACLICHWVLDPVKKLSFLTGEKFEKEIKLDLPPQVQIQQNDFLDYISILCSAINGILCIY 413
Query: 105 NEY--QRVIVWNPSTGFHKQILTSCD------FIIGSLYGFGYDNSTDDYFLVLIV---- 152
N + ++++WNP+T + ++ F+ LYGFGYD+ +DDY ++ +V
Sbjct: 414 NWFDPSQIVLWNPTTNEVHVVPSNLPESLPNVFVDQFLYGFGYDHDSDDYKVIRVVRFRE 593
Query: 153 --LASMNAKIHAYSVKTNSWDYNHVHVLYMHLGCDYRHGVFLNNSLHWL 199
+ + YS++++SW V + + G V+L+ HWL
Sbjct: 594 DMFKTHDPFYEIYSLRSHSWRKLDVDIPIVFYG-PLSSEVYLDGVCHWL 737
>CA917238 weakly similar to PIR|A84538|A845 hypothetical protein At2g16220
[imported] - Arabidopsis thaliana, partial (6%)
Length = 603
Score = 70.1 bits (170), Expect = 9e-13
Identities = 52/152 (34%), Positives = 71/152 (46%), Gaps = 25/152 (16%)
Frame = +2
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LP ELI E+L R V+SL+R +CV + W +ISDP+F K H +A H L C +
Sbjct: 146 LPDELITEVLSRGDVKSLMRMRCVSEYWNSMISDPRFVKLHMKRSARNAHLTLSLCKSGI 325
Query: 66 DSLGDNSAVVTLNLPPPCMSCIQNSL-----------------YFLGSCRGFMLL----- 103
D GDN+ V P P I+N L Y +GSC G++ L
Sbjct: 326 D--GDNNVV-----PYPVRGLIENGLITLPSDPYYRLKDKECQYVIGSCNGWLCLLGFSS 484
Query: 104 ANEYQRV--IVWNPSTGFHKQIL-TSCDFIIG 132
Y+ + WNP+ G Q L CD ++G
Sbjct: 485 IGAYRHIWFRFWNPAMGKMTQKLGYICDNVLG 580
>TC81783 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (6%)
Length = 1225
Score = 59.3 bits (142), Expect(2) = 5e-12
Identities = 29/55 (52%), Positives = 40/55 (72%)
Frame = +2
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLL 59
TLP +L+ EIL RLPV+ L++ +C+CK + LISDP FAK HF +A HRL++
Sbjct: 131 TLPFDLLPEILCRLPVKLLIQLRCLCKFFNSLISDPNFAKKHFQF-SAKHHRLMV 292
Score = 28.1 bits (61), Expect(2) = 5e-12
Identities = 28/116 (24%), Positives = 47/116 (40%), Gaps = 6/116 (5%)
Frame = +3
Query: 71 NSAVVTLNLPPPCMSCIQN----SLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQILTS 126
+++V L PP +N + + SC G VWNPS K +
Sbjct: 360 STSVTQTQLDPPFTLARRNYVNATAIAMCSCDGIFCGELNLGYCFVWNPSIRKFKLLPPY 539
Query: 127 CDFIIGSLYG--FGYDNSTDDYFLVLIVLASMNAKIHAYSVKTNSWDYNHVHVLYM 180
+ + G + FGYD+ D+Y +V I S ++ ++ T+ W H L +
Sbjct: 540 KNPLEGDPFSISFGYDHFIDNYKVVAI---SSKYEVFVNALGTDYWRRIREHPLLL 698
>BF632054 similar to GP|10177628|dbj gb|AAF30317.1~gene_id:K6M13.17~similar
to unknown protein {Arabidopsis thaliana}, partial (8%)
Length = 462
Score = 67.4 bits (163), Expect = 6e-12
Identities = 28/50 (56%), Positives = 37/50 (74%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTH 55
LP ELI EIL++LP++SLLRF+CVCKSW ++IS+P F K + TH
Sbjct: 144 LPEELILEILIKLPIKSLLRFRCVCKSWLHIISNPYFIKKQLHFSTQNTH 293
>BF648955 similar to GP|18043667|gb| Unknown (protein for IMAGE:3661715) {Mus
musculus}, partial (3%)
Length = 514
Score = 66.2 bits (160), Expect = 1e-11
Identities = 50/154 (32%), Positives = 76/154 (48%), Gaps = 19/154 (12%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LP ELI EI+ LPV+ L++F+CV K + LISDP F + H + +A H L ++
Sbjct: 57 LPSELIVEIISWLPVKYLMQFRCVSKFYKTLISDPYFVQMHLEKSARNPH---LALMWQD 227
Query: 66 DSLGDNSAVVTLNL---------PPPCMSCIQNSLYFLGSCRGFMLLAN-------EYQR 109
D L ++ +++ L++ PP S +GSC G + L + Y
Sbjct: 228 DLLREDGSIIFLSVSRLLGNKYTTPPFQSGTFYHSCIIGSCNGLLCLVDFHCPDYYYYYY 407
Query: 110 VIVWNPS--TGFHKQILT-SCDFIIGSLYGFGYD 140
+ WNP+ T F ++T S DF + FGYD
Sbjct: 408 LYFWNPATRTKFXNILITLSXDF----KFSFGYD 497
>AW696629
Length = 663
Score = 64.7 bits (156), Expect = 4e-11
Identities = 62/196 (31%), Positives = 85/196 (42%), Gaps = 29/196 (14%)
Frame = +2
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LP +LI +IL LPV+ L+RF V K W LI DP FAK H + TH ++L L D
Sbjct: 65 LPSDLIMQILSWLPVKLLIRFTSVSKHWKSLILDPNFAKLHLQKSPKNTH-MILTALDD- 238
Query: 66 DSLGDNSAVVTLNLPPPCMSCI---------------QNSLYFLGSCRGFMLLANE---- 106
D++ VVT P P S + +S + +GS G + LA E
Sbjct: 239 ---EDDTWVVT---PYPVRSLLLEQSSFSDEECCCFDYHSYFIVGSTNGLVCLAVEKSLE 400
Query: 107 ---YQRVI-VWNPSTGFHKQILTSCDF-IIGSL-YGFGYDNSTDDYFLVLI----VLASM 156
Y+ I WNPS + S + + G+ GFGYD+ D Y V + M
Sbjct: 401 NRKYELFIKFWNPSLRLRSKKAPSLNIGLYGTARLGFGYDDLNDTYKAVAVFWDHTTHKM 580
Query: 157 NAKIHAYSVKTNSWDY 172
++H DY
Sbjct: 581 EGRVHCMGDSCWKKDY 628
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.142 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,168,869
Number of Sequences: 36976
Number of extensions: 197464
Number of successful extensions: 1538
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 1452
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1479
length of query: 268
length of database: 9,014,727
effective HSP length: 94
effective length of query: 174
effective length of database: 5,538,983
effective search space: 963783042
effective search space used: 963783042
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0476a.5


