

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0437b.7
(824 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
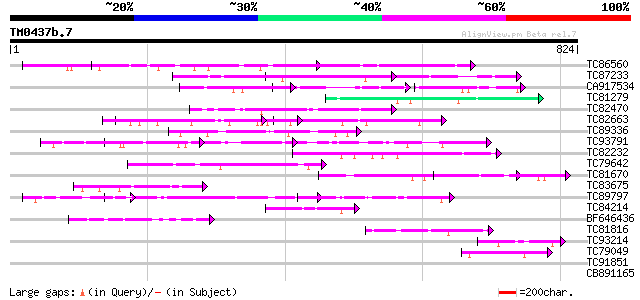
Score E
Sequences producing significant alignments: (bits) Value
TC86560 similar to GP|8843737|dbj|BAA97285.1 myosin heavy chain-... 77 3e-14
TC87233 similar to PIR|T06048|T06048 kinesin-related protein kat... 68 2e-11
CA917534 67 4e-11
TC81279 similar to PIR|T47949|T47949 hypothetical protein F2A19.... 62 9e-10
TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical prot... 58 1e-08
TC82663 similar to PIR|T51505|T51505 hypothetical protein F5E19_... 53 5e-07
TC89336 weakly similar to GP|21554135|gb|AAM63215.1 unknown {Ara... 52 7e-07
TC93791 similar to GP|15810373|gb|AAL07074.1 unknown protein {Ar... 51 2e-06
TC82232 similar to GP|10177316|dbj|BAB10642. kinesin-like protei... 46 5e-05
TC79642 similar to PIR|T07397|T07397 kinesin heavy chain-like pr... 45 9e-05
TC81670 weakly similar to PIR|E86293|E86293 hypothetical protein... 45 9e-05
TC83675 similar to PIR|T14321|T14321 nuclear matrix constituent ... 45 1e-04
TC89797 GP|13775543|gb|AAK39351.1 Hypothetical protein Y73B3A.17... 45 1e-04
TC84214 weakly similar to GP|10176794|dbj|BAB09933. kinesin-like... 45 1e-04
BF646436 weakly similar to GP|9828634|gb F1N21.5 {Arabidopsis th... 45 1e-04
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 45 1e-04
TC93214 similar to GP|10177918|dbj|BAB11329. kinesin-like protei... 42 7e-04
TC79049 weakly similar to PIR|T10798|T10798 pherophorin-S - Volv... 42 7e-04
TC91851 weakly similar to PIR|F96673|F96673 hypothetical protein... 41 0.002
CB891165 homologue to PIR|H82068|H820 RNA polymerase-associated ... 41 0.002
>TC86560 similar to GP|8843737|dbj|BAA97285.1 myosin heavy chain-like
{Arabidopsis thaliana}, partial (28%)
Length = 2450
Score = 77.0 bits (188), Expect = 3e-14
Identities = 121/572 (21%), Positives = 242/572 (42%), Gaps = 14/572 (2%)
Frame = +2
Query: 119 EKVVEGRNALRQAVKILEKRIENLES-ENKKLKKDIQEEQAQRKIE-IEGKLEKSNAFAA 176
E+V+ L A K L K E +++ E K + ++ E+AQR ++ + KL+ +
Sbjct: 473 ERVLAKETQLHVAEKELNKLKEQVKNAETTKAQALVELERAQRTVDDLTQKLKLITE--S 646
Query: 177 LENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEISNA---FAALENEVSALKSENKKL 233
E+ V A ++ + K+ E V K E+ NA +A++ E+ A K + +K
Sbjct: 647 RESAVKATEAAKSQAKQKYGESDG---VNGAWKEELENAVQRYASIMTELDAAKQDLRKT 817
Query: 234 KQDILDEQAQGKFCDRLKKKCEKVVEGRNALRQ---AVKILEKGIENLESENKKLKKDIQ 290
+Q+ D ++ E NA+++ V L K I ++ ++ K
Sbjct: 818 RQEYDSSS------DARVSAVKRTEEAENAMKENTERVSELSKEISAVKESIEQTKLAYV 979
Query: 291 EEHAQRKVEIEGKLEISNAF-AALENEVSALKSENKKLKKDILEEQAQRKVAMEGKL-EI 348
E Q+ + + K + ++ A LE L + K+ +I + ++E +L E
Sbjct: 980 ESQQQQALVLTEKDALRQSYKATLEQSKKKLLALKKEFNPEITK-------SLEAQLAET 1138
Query: 349 SNAFAALENEVSALKSENKKLKQDILEEQAQGKFCDQLKKKCEKVVEGRNALRQAVKILE 408
N AAL E+ +S + + + E D K+ +KVV+ N LR V+ L+
Sbjct: 1139 MNEIAALHTELENKRSSDLDSVKTVTSE------LDGAKESLQKVVDEENTLRSLVETLK 1300
Query: 409 KGIENLESENKKLK-KDIQEEQAQRKIEIEGKLEISNAFAALENEVSALKSESTKLKKDI 467
+EN++ E+ +LK K+ + E + ++ + S A +E S ++ S ++ +
Sbjct: 1301 VELENVKKEHSELKEKESELESTVGNLHVKLRKSKSELEACSADE-SKVRGASEEMILTL 1477
Query: 468 LEEQAQIKVAIEEKLEISNAFAALENEVSALKSEIAALQQKCGAGSREGNGDVEVLKAGI 527
++ + A E ++ N L+ E A K + ++K +E + E KA
Sbjct: 1478 SRLTSETEEARREVEDMKNKTDELKKEAEATKLALEEAEKKL----KEATEEAEAAKAAE 1645
Query: 528 SDTEKEVNTLKKELVEKEKIVADSERKTAVDERKKAAAEARKLLEAAKKIAPEK--AVIP 585
+ +++ T+ E + SE A+ + ++ +E + K+A K A
Sbjct: 1646 ASAIEQI-TVLTERTSARRASTSSESGAAMTISTEEFESLKRKVEESDKLADMKVDAAKA 1822
Query: 586 EPANCCSKCDELKKKCEKVAVGRNALRQA-VKILEKGIENLESENKKLKKENEVSALKSE 644
+ + +E+ KK E + G LEKG + S+ ++V+ ++E
Sbjct: 1823 QVEAVKASENEVLKKLEATSEGD*RYENCNTGGLEKGRDGRSSKEGS*G*ASKVAGARTE 2002
Query: 645 ISALQQKCGAGAREGNGDVEVLKAGISDTKKE 676
S + C + + ++ + + D+K E
Sbjct: 2003 KSG--RSCSSDTSRNTDVIAIISSAL*DSKAE 2092
Score = 73.2 bits (178), Expect = 4e-13
Identities = 103/464 (22%), Positives = 191/464 (40%), Gaps = 29/464 (6%)
Frame = +2
Query: 19 EELKKKCEQVVVGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQRKVAIEGKLEIS 78
+EL K EQV QA+ LE+ ++ +KLK + ++ K K +
Sbjct: 515 KELNKLKEQVKNAETTKAQALVELERAQRTVDDLTQKLKLITESRESAVKATEAAKSQAK 694
Query: 79 NTFA-----------ALENEV---SALISENKKLKQDI--LEEQAQGKICDQLKKCEKVV 122
+ LEN V +++++E KQD+ ++ ++ ++
Sbjct: 695 QKYGESDGVNGAWKEELENAVQRYASIMTELDAAKQDLRKTRQEYDSSSDARVSAVKRTE 874
Query: 123 EGRNALR---QAVKILEKRIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAF-AALE 178
E NA++ + V L K I ++ ++ K E Q Q+ + + K ++ A LE
Sbjct: 875 EAENAMKENTERVSELSKEISAVKESIEQTKLAYVESQQQQALVLTEKDALRQSYKATLE 1054
Query: 179 NEVSALKSENKKLKKDILEEQAQRKVAMEGKL-EISNAFAALENEVSALKSENKKLKQDI 237
L + K+ +I + ++E +L E N AAL E+ ENK+ D+
Sbjct: 1055QSKKKLLALKKEFNPEITK-------SLEAQLAETMNEIAALHTEL-----ENKR-SSDL 1195
Query: 238 LDEQAQGKFCDRLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKK---DIQEEHA 294
+ D K+ +KVV+ N LR V+ L+ +EN++ E+ +LK+ +++
Sbjct: 1196DSVKTVTSELDGAKESLQKVVDEENTLRSLVETLKVELENVKKEHSELKEKESELESTVG 1375
Query: 295 QRKVEIEGKLEISNAFAALENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEISNAFAA 354
V++ A +A E++V E + E + + +E ++ N
Sbjct: 1376NLHVKLRKSKSELEACSADESKVRGASEEMILTLSRLTSETEEARREVE---DMKNKTDE 1546
Query: 355 LENEVSALK---SENKKLKQDILEEQAQGKFCDQLKKKCEKVVEGRNALRQAVKILEKGI 411
L+ E A K E +K ++ EE K + + V+ R + R+A E G
Sbjct: 1547LKKEAEATKLALEEAEKKLKEATEEAEAAKAAEASAIEQITVLTERTSARRASTSSESGA 1726
Query: 412 ENLES--ENKKLKKDIQEEQAQRKIEIEGKLEISNAFAALENEV 453
S E + LK+ ++E ++++ A A ENEV
Sbjct: 1727AMTISTEEFESLKRKVEESDKLADMKVDAAKAQVEAVKASENEV 1858
Score = 28.9 bits (63), Expect = 8.4
Identities = 23/90 (25%), Positives = 39/90 (42%), Gaps = 3/90 (3%)
Frame = +3
Query: 695 ERKTAVDE---RKNAAAEARKLLEAPKKIAAEVEKQIAKVELRQVHLEKQVNERKMKLAF 751
+ KTA E + A A++ +E + E E++ A ++ E Q++ + +
Sbjct: 1896 DMKTATQEALKKAEMAEAAKRAVEGELRRWREREQKKAAEAAARILAETQMSSQSSPQHY 2075
Query: 752 ELSKTKEATKRFEAEKKKLLVEKINAESKI 781
+ K R E KKL EKI+ KI
Sbjct: 2076 RIQKQNVPPPRTTLEVKKLDKEKISVSKKI 2165
>TC87233 similar to PIR|T06048|T06048 kinesin-related protein katB -
Arabidopsis thaliana, partial (18%)
Length = 1488
Score = 67.8 bits (164), Expect = 2e-11
Identities = 64/340 (18%), Positives = 153/340 (44%), Gaps = 14/340 (4%)
Frame = +3
Query: 237 ILDEQAQGKFCDRLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEHAQR 296
+L+E+++ K K++CE +++ L+ ++ + + E +KLK + E Q+
Sbjct: 417 LLNEKSKRKDRFNYKERCENMIDYIKRLKVCIRWFQDLELSYSLEQEKLKSSL-ELSQQK 593
Query: 297 KVEIEGKLEISNAFAALENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEISNAFAALE 356
VEIE L+I L + ++ ++ L++ +++E+ ++ AME ++ A E
Sbjct: 594 CVEIELLLKIKE--EELNSIITEMRRNCTSLQEKLVKEETEKAAAMESLIKEREARLDFE 767
Query: 357 NEVSALKSENKKLKQDILEEQAQGKFCDQLKKKCEKVV----EGRNALRQAVKILEKGIE 412
+ L + + ++++ + + + K+ ++ + + L + +E ++
Sbjct: 768 RSQTTLSEDLGRAQRELETANQKIVSLNDMYKRLQEYITSLQQYNGKLHSELSTVEGDLK 947
Query: 413 NLESENKKLKKDIQEEQAQRKIEIEGKLEISNAFAALENEVSALKSESTKLKKDILEEQA 472
+E E + +++ + Q + + + E + A +EV++L+SE +++ D + +
Sbjct: 948 RVEKEKATVVENLTMLKGQLALSMASQEEATKQKDAFASEVTSLRSELHQVRDDRDRQIS 1127
Query: 473 QIKVAIEEKLEISNAFAALENEVSALKSEIAALQQKCGAGSRE----------GNGDVEV 522
Q++ E ++ ++ +EV+ L + L+ KC +EV
Sbjct: 1128QVQTLSTEIVKFKDSSEKSGSEVNNLTMKTNELETKCTLQDNHIKELQEKLTLAENKLEV 1307
Query: 523 LKAGISDTEKEVNTLKKELVEKEKIVADSERKTAVDERKK 562
+T E KK + E +K +AD+E K E+ K
Sbjct: 1308CDISAIETRTEFEGQKKLVNELQKRLADAEYKLIEGEKLK 1427
Score = 57.8 bits (138), Expect = 2e-08
Identities = 74/379 (19%), Positives = 162/379 (42%), Gaps = 7/379 (1%)
Frame = +3
Query: 373 ILEEQAQGKFCDQLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQR 432
+L E+++ K K++CE +++ L+ ++ + + E +KLK ++ Q Q+
Sbjct: 417 LLNEKSKRKDRFNYKERCENMIDYIKRLKVCIRWFQDLELSYSLEQEKLKSSLELSQ-QK 593
Query: 433 KIEIEGKLEISNAFAALENEVSALKSESTKLKKDILEEQAQIKVAIEEKLEISNAFAALE 492
+EIE L+I L + ++ ++ T L++ +++E+ + A+E ++ A E
Sbjct: 594 CVEIELLLKIKE--EELNSIITEMRRNCTSLQEKLVKEETEKAAAMESLIKEREARLDFE 767
Query: 493 NEVSALKSEIAALQQKCGAGSRE---GNGDVEVLKAGISDTEKEVNTLKKELVEKEKIVA 549
+ L ++ Q++ +++ N + L+ I+ ++ L EL +
Sbjct: 768 RSQTTLSEDLGRAQRELETANQKIVSLNDMYKRLQEYITSLQQYNGKLHSEL---STVEG 938
Query: 550 DSERKTAVDERKKAAAEARKLL--EAAKKIAPEKAVIPEPANCCSKCDELKKKCEKVAVG 607
D +R V++ K E +L + A +A ++ + S+ L+ + +V
Sbjct: 939 DLKR---VEKEKATVVENLTMLKGQLALSMASQEEATKQKDAFASEVTSLRSELHQVRDD 1109
Query: 608 RNALRQAVKILEKGIENLESENKKLKKENEVSALKSEISALQQKCGAGAREGNGDVEVLK 667
R+ RQ ++ E ++ ++ K +EV+ L + + L+ KC
Sbjct: 1110RD--RQISQVQTLSTEIVKFKDSSEKSGSEVNNLTMKTNELETKC--------------- 1238
Query: 668 AGISDTKKEVNRLKKEHVEE--ERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVE 725
L+ H++E E++ + D +A E R E KK+ E++
Sbjct: 1239-----------TLQDNHIKELQEKLTLAENKLEVCD---ISAIETRTEFEGQKKLVNELQ 1376
Query: 726 KQIAKVELRQVHLEKQVNE 744
K++A E + + EK E
Sbjct: 1377KRLADAEYKLIEGEKLKKE 1433
>CA917534
Length = 616
Score = 66.6 bits (161), Expect = 4e-11
Identities = 67/190 (35%), Positives = 90/190 (47%), Gaps = 29/190 (15%)
Frame = +1
Query: 589 NCCSKCDELKKKCEKVAVGRNALRQAVKILEKGIENLESENKKLKKENEVSALKSEISAL 648
N C C++ K K K RNALRQAVK LE I ++S N N+V +K E
Sbjct: 79 NTC--CEQWKNKYLKAQESRNALRQAVKFLELKINEIQSLNNH-NNNNKVCDVKIETGER 249
Query: 649 QQKCGAGA-------------------REGNGDVEV------LKAGISDTKKEVNRLKKE 683
++ AGA R NGD + L+A I++ +E+ RL KE
Sbjct: 250 LEEFNAGAPAVNNGLSSFKSQTVTSAHRGCNGDGDENEKALDLQACIAERDREICRL-KE 426
Query: 684 HVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVEK-QIAKVELRQV---HLE 739
+E E+ ADS+RK AE KLLE K A++ K ++ K E +V LE
Sbjct: 427 LLESEKRRADSKRK--------RVAETWKLLEQEKNKGAQIAKLKVEKAEGYRVQIGQLE 582
Query: 740 KQVNERKMKL 749
KQV+E K KL
Sbjct: 583 KQVSEAKQKL 612
Score = 54.7 bits (130), Expect = 1e-07
Identities = 60/201 (29%), Positives = 86/201 (41%), Gaps = 1/201 (0%)
Frame = +1
Query: 383 CDQLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQRKIEIEGKLEI 442
C+Q K K K E RNALRQAVK LE I ++S N + + KIE +LE
Sbjct: 88 CEQWKNKYLKAQESRNALRQAVKFLELKINEIQSLN---NHNNNNKVCDVKIETGERLEE 258
Query: 443 SNAFA-ALENEVSALKSESTKLKKDILEEQAQIKVAIEEKLEISNAFAALENEVSALKSE 501
NA A A+ N +S+ KS++
Sbjct: 259 FNAGAPAVNNGLSSFKSQT----------------------------------------- 315
Query: 502 IAALQQKCGAGSREGNGDVEVLKAGISDTEKEVNTLKKELVEKEKIVADSERKTAVDERK 561
+ + + C G + N L+A I++ ++E+ L KEL+E EK ADS+RK + K
Sbjct: 316 VTSAHRGCN-GDGDENEKALDLQACIAERDREICRL-KELLESEKRRADSKRKRVAETWK 489
Query: 562 KAAAEARKLLEAAKKIAPEKA 582
E K + A K+ EKA
Sbjct: 490 LLEQEKNKGAQIA-KLKVEKA 549
Score = 48.1 bits (113), Expect = 1e-05
Identities = 54/179 (30%), Positives = 80/179 (44%), Gaps = 10/179 (5%)
Frame = +1
Query: 247 CDRLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEI 306
C++ K K K E RNALRQAVK LE I ++S N + + K+E +LE
Sbjct: 88 CEQWKNKYLKAQESRNALRQAVKFLELKINEIQSLN---NHNNNNKVCDVKIETGERLEE 258
Query: 307 SNAFA-ALENEVSALKSE-----NKKLKKDILEEQAQ---RKVAMEGKLEISNAFAALEN 357
NA A A+ N +S+ KS+ ++ D E + + E EI LE+
Sbjct: 259 FNAGAPAVNNGLSSFKSQTVTSAHRGCNGDGDENEKALDLQACIAERDREICRLKELLES 438
Query: 358 EVSALKSENKKLKQD-ILEEQAQGKFCDQLKKKCEKVVEGRNALRQAVKILEKGIENLE 415
E S+ K++ + L EQ + K K K EK R + Q K + + + LE
Sbjct: 439 EKRRADSKRKRVAETWKLLEQEKNKGAQIAKLKVEKAEGYRVQIGQLEKQVSEAKQKLE 615
Score = 37.7 bits (86), Expect = 0.018
Identities = 46/184 (25%), Positives = 83/184 (45%), Gaps = 9/184 (4%)
Frame = +1
Query: 13 NCCSKCEELKKKCEQVVVGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQRKVAIE 72
N C CE+ K K + RNALRQAVK LE I ++S N + + K+
Sbjct: 79 NTC--CEQWKNKYLKAQESRNALRQAVKFLELKINEIQSLN---NHNNNNKVCDVKIETG 243
Query: 73 GKLEISNTFA-ALENEVSALISEN-KKLKQDILEEQAQGKICDQLKKCEKVVEGRNALRQ 130
+LE N A A+ N +S+ S+ + + + + L+ C + E + +
Sbjct: 244 ERLEEFNAGAPAVNNGLSSFKSQTVTSAHRGCNGDGDENEKALDLQAC--IAERDREICR 417
Query: 131 AVKILEKRIENLESENKKLK---KDIQEEQAQRKIEIEGKLEKSNAF----AALENEVSA 183
++LE +S+ K++ K +++E+ + + K+EK+ + LE +VS
Sbjct: 418 LKELLESEKRRADSKRKRVAETWKLLEQEKNKGAQIAKLKVEKAEGYRVQIGQLEKQVSE 597
Query: 184 LKSE 187
K +
Sbjct: 598 AKQK 609
>TC81279 similar to PIR|T47949|T47949 hypothetical protein F2A19.170 -
Arabidopsis thaliana, partial (31%)
Length = 1205
Score = 62.0 bits (149), Expect = 9e-10
Identities = 84/341 (24%), Positives = 135/341 (38%), Gaps = 24/341 (7%)
Frame = +2
Query: 459 ESTKLKKDILEEQAQIKVAIEEKLEISNAFAALENEVSALKSEIAALQ-QKCGAGSREGN 517
E KL K EE +K+ E+ + + L E++ SEI LQ + S+E +
Sbjct: 68 EEQKLNKSFQEELKLLKL---ERDKTTTEVRQLHKELNEKVSEIKRLQLELTRQRSKEAS 238
Query: 518 GDVEVLKAGISDTEKEVNTLKKELVEKEKIVADSERKTAVDERK-------------KAA 564
++ K I EKE TLK E E E V S D K K +
Sbjct: 239 NAMDSSKRLIETLEKENTTLKMEKSELEAAVKASSASDLSDPSKSFPGKEDMEISLQKMS 418
Query: 565 AEARKLLEAAKKIAPE-----KAVIPEPANCCSKCDELKKKCEKVAVGRNALRQAVKILE 619
+ +K + K E + ++ + K DE K E++ N LR + LE
Sbjct: 419 NDLKKTQQERDKAVQELTRLKQHLLEKENEESEKMDEDTKVIEELRDSNNYLRAQISHLE 598
Query: 620 KGIENLESENKKLKKEN--EVSALKSEISALQQK---CGAGAREGNGDVEVLKAGISDTK 674
+ +E S+ +KLK N E+ + I L +K C + N ++ L+ +
Sbjct: 599 RALEQATSDQEKLKSANNSEILTSREVIDDLNKKLTNCISTIDAKNIELINLQTALGQYY 778
Query: 675 KEVNRLKKEHVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVEKQIAKVELR 734
E+ KEH+EEE A E K+A + L ++I A++ Q KV+
Sbjct: 779 AEIE--AKEHLEEELARAREETANLSQLLKDADSRVDILSGEKEEILAKL-SQSEKVQSE 949
Query: 735 QVHLEKQVNERKMKLAFELSKTKEATKRFEAEKKKLLVEKI 775
++ ER KL L ++ R + L+ +I
Sbjct: 950 WRSRVSKLEERNAKLRRALEQSMTRLNRMSVDSDFLVDRRI 1072
>TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (8%)
Length = 1064
Score = 58.2 bits (139), Expect = 1e-08
Identities = 68/305 (22%), Positives = 135/305 (43%), Gaps = 5/305 (1%)
Frame = +2
Query: 262 NALRQAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEISNAFAALENEVSALK 321
+ L++ + ++EK E++ S K ++E Q K +I LEN+ ++
Sbjct: 113 STLKEKLVLVEKSFEDIRS-----KTQVEERRLQSI-----KRDIEECCEDLENKKKEIR 262
Query: 322 SENKKLKKDILEEQAQRKVAMEGKL-EISNAFAALENEVSALKS----ENKKLKQDILEE 376
+ + +A++K M+GK+ E F A E ++ ++ K+LK LE
Sbjct: 263 DVGRII-------EARKK--MQGKIDECVKDFVAKEGQLGLMEDLIGEHKKELKTKELEL 415
Query: 377 QAQGKFCDQLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQRKIEI 436
+ + K+ +V E N L K E I+ LES+ ++L+ ++E + + K E
Sbjct: 416 RQVMDNISKQKELESQVKELVNDLVSKQKHFESHIKELESKERQLEGRLKEHELEEK-EF 592
Query: 437 EGKLEISNAFAALENEVSALKSESTKLKKDILEEQAQIKVAIEEKLEISNAFAALENEVS 496
EG++ LE++ KSE ++ ++ + QIK ++ +++ LE++ +
Sbjct: 593 EGRMN------ELESKERHFKSEVEEINAKLMPLKGQIKELASKEKQLNGQVKELESKKN 754
Query: 497 ALKSEIAALQQKCGAGSREGNGDVEVLKAGISDTEKEVNTLKKELVEKEKIVADSERKTA 556
++ I L+ K EG K +++ KK+L E + +S+
Sbjct: 755 QFENRIKELESK--EKQHEGRVKEHASKEREFESQVMEQQFKKKLFEIQVKALESKENQL 928
Query: 557 VDERK 561
VD+ K
Sbjct: 929 VDQMK 943
>TC82663 similar to PIR|T51505|T51505 hypothetical protein F5E19_70 -
Arabidopsis thaliana, partial (20%)
Length = 914
Score = 52.8 bits (125), Expect = 5e-07
Identities = 73/301 (24%), Positives = 130/301 (42%), Gaps = 30/301 (9%)
Frame = +3
Query: 155 EEQAQRKIE-IEGKLEKSNAF--------AALENEVSALKSENKKLKKDILEEQAQRKVA 205
E A +E + +LE+SN A+L+ +V L+ + K D+ + + + +A
Sbjct: 24 ERSASESLESVMNQLEESNYLLHDAESVAASLKEKVGLLEMTIVRQKADLEDSERRLLMA 203
Query: 206 MEGKLEISNAFAALENEVSALKSENKKLKQDILDEQAQGKFCDRLKKKCEKVVEGRNALR 265
E LE S ALE+E+ +K E Q + +EQ L ++ K++ + R
Sbjct: 204 KEENLEKSKKVEALESELETVKEEKD---QALNNEQLAASHVQTLLEEKNKLINELDNSR 374
Query: 266 QAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEISNAFAALENEVS----ALK 321
+ + +K +E+L S + E A+ + E L + EN++ LK
Sbjct: 375 EEEEKSKKAMESLASA-------LHEVSAESREAKENFLSTQAERESYENQIEDLKLVLK 533
Query: 322 SENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENE--------VSALKS---ENKKLK 370
N+K + + E Q + V ++ N F + E VS+LK EN +
Sbjct: 534 GTNEKYESMLDEAQHEIDVLIDSIENSKNVFENSKAEWDQRELHLVSSLKKTEEENAAAE 713
Query: 371 QDI-----LEEQAQGKF-CDQLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKD 424
++I L +Q + +F C Q KK + R L++ ++ + SEN KLK++
Sbjct: 714 KEINRLVYLLKQTEEEF*CQQGGKKLS*RKI*KEV*RPRQFHLQEALKEVTSENVKLKEN 893
Query: 425 I 425
I
Sbjct: 894 I 896
Score = 52.0 bits (123), Expect = 9e-07
Identities = 61/265 (23%), Positives = 124/265 (46%), Gaps = 26/265 (9%)
Frame = +3
Query: 135 LEKRIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVSALKSE------N 188
L++++ LE + K D+++ + + + E LEKS ALE+E+ +K E N
Sbjct: 117 LKEKVGLLEMTIVRQKADLEDSERRLLMAKEENLEKSKKVEALESELETVKEEKDQALNN 296
Query: 189 KKLKKDILEEQAQRKVAMEGKLEIS--------NAFAALENEVSALKSENKKLKQDILDE 240
++L ++ + K + +L+ S A +L + + + +E+++ K++ L
Sbjct: 297 EQLAASHVQTLLEEKNKLINELDNSREEEEKSKKAMESLASALHEVSAESREAKENFLST 476
Query: 241 QAQGKFCDRLKKKCEKVVEGRN--------ALRQAVKILEKGIENLES--ENKKLKKDIQ 290
QA+ + + + + V++G N + + +L IEN ++ EN K + D +
Sbjct: 477 QAERESYENQIEDLKLVLKGTNEKYESMLDEAQHEIDVLIDSIENSKNVFENSKAEWDQR 656
Query: 291 EEHAQRKVEIEGKLEISNAFAALENEVSALKSENKKLKKDILEEQAQRKVAMEG--KLEI 348
E H ++ K E N AA E E++ L K+ +++ +Q +K++ K
Sbjct: 657 ELHLVSSLK---KTEEEN--AAAEKEINRLVYLLKQTEEEF*CQQGGKKLS*RKI*KEV* 821
Query: 349 SNAFAALENEVSALKSENKKLKQDI 373
L+ + + SEN KLK++I
Sbjct: 822 RPRQFHLQEALKEVTSENVKLKENI 896
Score = 46.2 bits (108), Expect = 5e-05
Identities = 63/285 (22%), Positives = 116/285 (40%), Gaps = 34/285 (11%)
Frame = +3
Query: 384 DQLKKKCEKVVEGRNALRQAVKI---LEKGIENLESENKKLKKDIQEEQAQRKIEIEGKL 440
+ L+ ++ E L A + L++ + LE + K D+++ + + + E L
Sbjct: 39 ESLESVMNQLEESNYLLHDAESVAASLKEKVGLLEMTIVRQKADLEDSERRLLMAKEENL 218
Query: 441 EISNAFAALENEVSALKSESTKLKKDILEEQAQIKVAIEEK----LEISNAFAALENEVS 496
E S ALE+E+ +K E + + + ++ +EEK E+ N+ E
Sbjct: 219 EKSKKVEALESELETVKEEKDQALNNEQLAASHVQTLLEEKNKLINELDNSREEEEKSKK 398
Query: 497 ALKSEIAALQQKCGAGSREG--------------NGDVEVLKAGISDTEKEVNTLKKELV 542
A++S +AL + A SRE +E LK + T ++ ++ E
Sbjct: 399 AMESLASALHE-VSAESREAKENFLSTQAERESYENQIEDLKLVLKGTNEKYESMLDEAQ 575
Query: 543 EKEKIVADS-ERKTAVDERKKAAAEARK--LLEAAKKIAPEKAVIPEPANCCSK------ 593
+ ++ DS E V E KA + R+ L+ + KK E A + N
Sbjct: 576 HEIDVLIDSIENSKNVFENSKAEWDQRELHLVSSLKKTEEENAAAEKEINRLVYLLKQTE 755
Query: 594 ----CDELKKKCEKVAVGRNALRQAVKILEKGIENLESENKKLKK 634
C + KK + + R L++ ++ + SEN KLK+
Sbjct: 756 EEF*CQQGGKKLS*RKI*KEV*RPRQFHLQEALKEVTSENVKLKE 890
>TC89336 weakly similar to GP|21554135|gb|AAM63215.1 unknown {Arabidopsis
thaliana}, partial (7%)
Length = 1241
Score = 52.4 bits (124), Expect = 7e-07
Identities = 72/306 (23%), Positives = 136/306 (43%), Gaps = 26/306 (8%)
Frame = +1
Query: 232 KLKQDILDEQAQGKFC--------DRLKKKCEKVVEGRNALRQAVKILEKGIENLESENK 283
KLK+D+ D + Q +F D L KK K E +K+ E+ I L+++N+
Sbjct: 394 KLKEDLADTERQSQFVGEGENKSYDELLKKFIKNEEELRVSNLKLKLSEEEIIKLKNQNE 573
Query: 284 KLK---KDIQEEHAQRKVEIEGKLEISNAFAALENEVSALKSENKKLKKDILEEQAQRKV 340
K + +Q+E E+E K + +V L+ + +L+ + Q +V
Sbjct: 574 KSEGQLDSVQKELTLNMDELEHK----------KGQVLELQKQKAELETHVPNLVEQLEV 723
Query: 341 AMEGKLEISN-AFAALENEVSALKSENKKLKQDILEEQAQGKFCDQLKKKCEKVVEGRNA 399
A E L+ISN A L E+ + +E ++L+ + E AQ + + K ++ GR
Sbjct: 724 ANE-HLKISNDEVARLRKELESNLAETRQLQDQL--EVAQ----ENVTKLEWQLDSGRKQ 882
Query: 400 LRQA---VKILEKGIENLESENKKLKKDIQEEQAQRKIEIEGKLEISNAFAALENEVSAL 456
+R+ + + NLE E +KLK ++ + QAQ E + L +++++L
Sbjct: 883 IRELEDRITWFKTNETNLEVEVQKLKDEMHDVQAQFSFEKD----------QLHSDIASL 1032
Query: 457 KSESTKLKKDILEEQA-------QIKVAIEEKLEISNAFA----ALENEVSALKSEIAAL 505
T+L + E ++ +++ E LE +A ++ E S+ K ++
Sbjct: 1033SEIKTQLTSTLEEWESRSHSLANKLRRCEAENLEKEEVYATQKMVMQGENSSFKGRTSSK 1212
Query: 506 QQKCGA 511
+ CG+
Sbjct: 1213KA*CGS 1230
Score = 41.2 bits (95), Expect = 0.002
Identities = 59/270 (21%), Positives = 122/270 (44%), Gaps = 21/270 (7%)
Frame = +1
Query: 55 KLKKDIQEEQAQRKVAIEGKL----EISNTFAALENE--VSAL---ISENKKLKQDILEE 105
KLK+D+ + + Q + EG+ E+ F E E VS L +SE + +K E
Sbjct: 394 KLKEDLADTERQSQFVGEGENKSYDELLKKFIKNEEELRVSNLKLKLSEEEIIKLKNQNE 573
Query: 106 QAQGKICDQLKKCEKVVEGRNALRQAVKILEKRIENLESENKKLKKDIQEEQAQRKIEIE 165
+++G++ K+ ++ + V L+K+ LE+ L + ++ KI
Sbjct: 574 KSEGQLDSVQKELTLNMDELEHKKGQVLELQKQKAELETHVPNLVEQLEVANEHLKIS-- 747
Query: 166 GKLEKSNAFAALENEVSALKSENKKLKKDILEEQAQRKVAMEGKL--------EISNAFA 217
++ A L E+ + +E ++L+ D LE + +E +L E+ +
Sbjct: 748 -----NDEVARLRKELESNLAETRQLQ-DQLEVAQENVTKLEWQLDSGRKQIRELEDRIT 909
Query: 218 ALENEVSALKSENKKLKQDILDEQAQGKF-CDRLKKKCEKVVEGRNALRQAVKILEKGIE 276
+ + L+ E +KLK ++ D QAQ F D+L + E + L ++ E
Sbjct: 910 WFKTNETNLEVEVQKLKDEMHDVQAQFSFEKDQLHSDIASLSEIKTQLTSTLEEWESRSH 1089
Query: 277 NLESENKKLKK---DIQEEHAQRKVEIEGK 303
+L ++ ++ + + +E +A +K+ ++G+
Sbjct: 1090SLANKLRRCEAENLEKEEVYATQKMVMQGE 1179
Score = 40.4 bits (93), Expect = 0.003
Identities = 56/232 (24%), Positives = 104/232 (44%), Gaps = 16/232 (6%)
Frame = +1
Query: 19 EELKKKCEQVVVGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQRKVAIEG---KL 75
EE+ K Q L K L ++ LE + ++ ++Q+++A+ + + +L
Sbjct: 541 EEIIKLKNQNEKSEGQLDSVQKELTLNMDELEHKKGQVL-ELQKQKAELETHVPNLVEQL 717
Query: 76 EISNTFAALENEVSALISENKKLKQDILE-EQAQGKICDQLKKCEKVVEGRNALRQAVKI 134
E++N + N+ A + K+L+ ++ E Q Q ++ + K+ ++ R+ ++
Sbjct: 718 EVANEHLKISNDEVARL--RKELESNLAETRQLQDQLEVAQENVTKLEWQLDSGRKQIRE 891
Query: 135 LEKRIE-------NLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVSALKSE 187
LE RI NLE E +KLK ++ + QAQ E + L +++++L SE
Sbjct: 892 LEDRITWFKTNETNLEVEVQKLKDEMHDVQAQFSFEKD----------QLHSDIASL-SE 1038
Query: 188 NKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSA-----LKSENKKLK 234
K LEE R ++ KL A + EV A ++ EN K
Sbjct: 1039IKTQLTSTLEEWESRSHSLANKLRRCEAENLEKEEVYATQKMVMQGENSSFK 1194
>TC93791 similar to GP|15810373|gb|AAL07074.1 unknown protein {Arabidopsis
thaliana}, partial (40%)
Length = 866
Score = 50.8 bits (120), Expect = 2e-06
Identities = 60/268 (22%), Positives = 129/268 (47%), Gaps = 31/268 (11%)
Frame = +1
Query: 46 IENLESENKKLKKDIQE-----EQAQRKVAIEGKLEISNTFAALENEVSALISENKKLKQ 100
+ LESEN+K+K +++E + + A +LE LE ++ + E ++K
Sbjct: 25 LSELESENRKMKVELEEFRTEATHLENQEATIRRLE--ERSRQLEQQMEEKVKEIAEIKH 198
Query: 101 DILEEQAQGKICDQLKKCEKVVEGR-NALRQAVKILEKRIENLESENKKLKKDIQEEQAQ 159
L E+ Q K + LK+ E++++ + + +++V ++K E +++ +L+ +EE+A
Sbjct: 199 RNLAEENQ-KTLEILKEREQLLQDQLQSAKESVSNMKKLHELAQNQLFELRAQSEEERAA 375
Query: 160 RKIE---IEGKLEKSNA-FAALENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEISNA 215
++ E + ++E++ LE E L+S+ + +D + ++ ++ + N+
Sbjct: 376 KQAEANLLMDEVERAQTMLLGLEREKGVLRSQLQTANED---SETKKSDNLDSNNALENS 546
Query: 216 FAALENEVSALKSENKKLKQDILDEQAQ-----GKFCDRLKKK---------------CE 255
A E +S L E ++ + +E+ + KF L +K E
Sbjct: 547 LIAKEKLISELNMELHNIETTLSNEREEHINDVKKFTAMLNEKEASIVAMKKELQTRPTE 726
Query: 256 KVVEGRNALRQAVKILEK-GIENLESEN 282
K+V+ LR+ VKIL+ G ++E+E+
Sbjct: 727 KIVDD---LRKKVKILQAVGYNSIEAED 801
Score = 48.5 bits (114), Expect = 1e-05
Identities = 65/292 (22%), Positives = 130/292 (44%), Gaps = 11/292 (3%)
Frame = +1
Query: 138 RIENLESENKKLKKDIQEEQAQ------RKIEIEGKLEKSNAFAALENEVSALKSENKKL 191
++ LESEN+K+K +++E + + ++ I E+S LE ++ E ++
Sbjct: 22 KLSELESENRKMKVELEEFRTEATHLENQEATIRRLEERSR---QLEQQMEEKVKEIAEI 192
Query: 192 KKDILEEQAQR--KVAMEGKLEISNAFAALENEVSALKSENKKLKQDILDEQAQGKFCDR 249
K L E+ Q+ ++ E + + + + + VS +K ++ + + + +AQ + +R
Sbjct: 193 KHRNLAEENQKTLEILKEREQLLQDQLQSAKESVSNMKKLHELAQNQLFELRAQSEE-ER 369
Query: 250 LKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEISNA 309
K+ E N L V+ + + LE E L+ +Q + + + L+ +N
Sbjct: 370 AAKQAE-----ANLLMDEVERAQTMLLGLEREKGVLRSQLQTANEDSETKKSDNLDSNN- 531
Query: 310 FAALENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKL 369
ALEN + A+ K+ E +E+ N L NE ++ KK
Sbjct: 532 --ALENSLI-----------------AKEKLISELNMELHNIETTLSNEREEHINDVKKF 654
Query: 370 KQDILEEQAQ--GKFCDQLKKKCEKVVEGRNALRQAVKILEK-GIENLESEN 418
+ E++A + + EK+V+ LR+ VKIL+ G ++E+E+
Sbjct: 655 TAMLNEKEASIVAMKKELQTRPTEKIVDD---LRKKVKILQAVGYNSIEAED 801
Score = 44.7 bits (104), Expect = 1e-04
Identities = 68/305 (22%), Positives = 124/305 (40%), Gaps = 15/305 (4%)
Frame = +1
Query: 411 IENLESENKKLKKDIQEEQAQRKIEIEGKLEISNAFAALENEVSALKSESTKLKKDILEE 470
+ LESEN+K+K +++E + + LEN+ + ++ + ++ LE+
Sbjct: 25 LSELESENRKMKVELEEFRTEA--------------THLENQEATIRRLEERSRQ--LEQ 156
Query: 471 QAQIKVAIEEKLEISNAFAALENE--VSALKSEIAALQQKCGAGSREGNGDVEVLKAGIS 528
Q + KV +E EI + A EN+ + LK LQ + ++ K +S
Sbjct: 157 QMEEKV--KEIAEIKHRNLAEENQKTLEILKEREQLLQDQ-----------LQSAKESVS 297
Query: 529 DTEKEVNTLKKELVEKEKIVADSERKTAVDERKKAAAEARKLLEAAKK-------IAPEK 581
+ +K + +L E + A SE +ER AEA L++ ++ + EK
Sbjct: 298 NMKKLHELAQNQLFE---LRAQSE-----EERAAKQAEANLLMDEVERAQTMLLGLEREK 453
Query: 582 AVIPEPANCCSKCDELKKKCEKVAVGRNALRQAVKILEKGIENLES----ENKKLKKENE 637
V+ ++ E KK +NL+S EN + KE
Sbjct: 454 GVLRSQLQTANEDSETKKS----------------------DNLDSNNALENSLIAKEKL 567
Query: 638 VSALKSEISALQQKCGAGAREGNGDVEVLKAGISDTKKEVNRLKKEHVEE--ERIVADSE 695
+S L E+ ++ E DV+ A +++ + + +KKE E+IV D
Sbjct: 568 ISELNMELHNIETTLSNEREEHINDVKKFTAMLNEKEASIVAMKKELQTRPTEKIVDDLR 747
Query: 696 RKTAV 700
+K +
Sbjct: 748 KKVKI 762
>TC82232 similar to GP|10177316|dbj|BAB10642. kinesin-like protein
{Arabidopsis thaliana}, partial (21%)
Length = 1181
Score = 46.2 bits (108), Expect = 5e-05
Identities = 77/350 (22%), Positives = 141/350 (40%), Gaps = 46/350 (13%)
Frame = +3
Query: 411 IENLESENKKLKKDIQEEQAQRKI-EIEGKLEISNAFAALENEVSALKSESTKLKKDILE 469
+E +E K+L+ +++ R++ E++ KLE A L N S LK K ++
Sbjct: 150 VEPVEIHEKELEHSSAQQKLDRELKELDKKLEQKEAEMKLVNNASVLKQHYEKKLNELEH 329
Query: 470 EQAQIKVAIEE------------KLEISNAFAALENEVSALK------SEIAALQQKCGA 511
E+ ++ IEE K E ALE++VS LK + + +QK
Sbjct: 330 EKKFLQREIEELKSTSGDSTHKLKEEYLQKLNALESQVSELKKKQDAQAHLLRQKQKGDE 509
Query: 512 GSREGNGDVEVLKA------------------GISDTEKEVNTLKKE----LVEKEKIVA 549
++ +++ +K+ + EKEV LKKE E+ K++A
Sbjct: 510 AAKRLQDEIQRIKSQKVQLQHKIKQESEQFRLWKASREKEVLQLKKEGRKNEYERHKLLA 689
Query: 550 DSERKTAVDERKK-----AAAEARKLLEAAKKIAPEKAVIPEPANCCSKCDELKKKCEKV 604
++R V +RK A ++LLE+ K + E + + ++ + E V
Sbjct: 690 LNQRTKMVLQRKTEEASLATKRLKELLESRKASSRETGISGNGPGIQALMQTIEHELE-V 866
Query: 605 AVGRNALRQAVKILEKGIENLESENKKLKKENEVSALKSEISALQQKCGAGAREGNGDVE 664
V + +R + + + E+ +LK+E E+ L + +R +E
Sbjct: 867 TVRVHEVRSEYQRQMEVRAEMAKESARLKEEAEMMKLNNTSDVSMSPAARSSR--IFALE 1040
Query: 665 VLKAGISDTKKEVNRLKKEHVEEERIVADSERKTAVDERKNAAAEARKLL 714
+ A S T + E E ER+ + R V + A+A+ L+
Sbjct: 1041NMLATSSTTLVSMASQLSEAEERERVFSGKGRWNQV----RSLADAKNLM 1178
>TC79642 similar to PIR|T07397|T07397 kinesin heavy chain-like protein
(clone PKCBP) - potato, partial (42%)
Length = 2321
Score = 45.4 bits (106), Expect = 9e-05
Identities = 63/304 (20%), Positives = 127/304 (41%), Gaps = 15/304 (4%)
Frame = +2
Query: 172 NAFAALENEVSALKSENKKLKK-DILEEQAQRKVAMEGK--LEISNAFAALENEVSALKS 228
N A++ ++ L+S K+++ L E++QR K E ++ ++ LK
Sbjct: 164 NGDASINSKPPNLESYEKRIQDLSKLVEESQRSADQLHKDLHEKQEKEVKMQEQLEGLKE 343
Query: 229 ENKKLKQDILDEQAQGKFCDRLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKD 288
K KQ++ QA C+RL+ C K + A+ K ++L N ++ +
Sbjct: 344 SLKANKQNL---QAVTSDCERLRSLCNKKDQALQAIESTSK------KDLVETNNQVLQK 496
Query: 289 IQEEHAQRKVEIEGKLE----ISNAFAALENEVSALKSENKKLKKDILEEQAQRKVAMEG 344
++ E K E++ E + + A LE ++S L+ N + +L + Q + A++
Sbjct: 497 LKYELKYCKGELDSAEETIKTLRSEKAILEQKLSVLEKRNSEESSSLLRKLEQERKAVKS 676
Query: 345 KL-EISNAFAALENEVSALKSENKKLKQDILEEQAQGKFCDQLKKKCEKVVEGRNALRQA 403
++ ++ E+ A KS ++ Q K ++L++ E +
Sbjct: 677 EVYDLERKIEGYRQELMAAKSIISVKDSELSALQNNFKELEELREMKEDIDRKNEQTASI 856
Query: 404 VKILEKGIENLESENKKLKKDIQEEQAQRK------IEIEGKLEISNAFAAL-ENEVSAL 456
+K+ + +E K EEQ RK +++GK+ + + E EVS
Sbjct: 857 LKMQRAQLAEMEGLYK-------EEQVLRKRYFNVIEDMKGKIRVYCRLRPISEKEVSEK 1015
Query: 457 KSES 460
+ E+
Sbjct: 1016EREA 1027
>TC81670 weakly similar to PIR|E86293|E86293 hypothetical protein AAF18488.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 1797
Score = 45.4 bits (106), Expect = 9e-05
Identities = 65/306 (21%), Positives = 129/306 (41%), Gaps = 11/306 (3%)
Frame = +2
Query: 449 LENEVSALKSESTKLKKDILEEQAQIKVAIEEKLEISNAFAALENEVSALKSEIAALQQK 508
L NE+ K++ E+ + +EEK + NAF E ++Q+K
Sbjct: 830 LANEIDITNENLNKMQYKYNEKTMSLSRMLEEKDRLHNAFV----------EESRSMQRK 979
Query: 509 CGAGSREGNGDVEVLKAGISDTEKEVNTLKKELVEKEKIVADSERKTAVDERKKAAAEAR 568
R + E L+ + + ++++T ++L K +++ D ER+ +++KK +
Sbjct: 980 AREEVRRILEEQEKLRNELDEKMRKLDTWSRDL-NKREVLTDQERQKLEEDKKKKDSRNE 1156
Query: 569 KLLEAAK--KIAPEKAV------IPEPANCCSKCDELKKKC---EKVAVGRNALRQAVKI 617
L+ A+K KIA E E +K +L+K+ +K+ + LR +++
Sbjct: 1157 SLMLASKEQKIADENVFRLVEEQKREKEEALNKILQLEKQLDAKQKLEMEIEELRGKLQV 1336
Query: 618 LEKGIENLESENKKLKKENEVSALKSEISALQQKCGAGAREGNGDVEVLKAGISDTKKEV 677
+ K + + + K K E S L+ +I +L+ E ++K + E+
Sbjct: 1337 M-KHLGDQDDTAIKKKMEEMNSELEDKIESLED------MESMNSTLIVKE--RQSNDEL 1489
Query: 678 NRLKKEHVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVEKQIAKVELRQVH 737
+KE +E + + KT + ++ + + + A KK + E I VEL +
Sbjct: 1490 QEARKELIEGLNEML-TGAKTNIGTKRMGDLDQKVFVNACKKRFSSDEAGIKAVELCSLW 1666
Query: 738 LEKQVN 743
E N
Sbjct: 1667 QENVKN 1684
Score = 43.1 bits (100), Expect = 4e-04
Identities = 44/208 (21%), Positives = 97/208 (46%), Gaps = 9/208 (4%)
Frame = +2
Query: 616 KILEKGIENLESENKKLKKENEV-SALKSEISALQQKCGAGAREGNGDVEVLKAGISDTK 674
K+ K E S ++ L++++ + +A E ++Q+K R + E L+ + +
Sbjct: 869 KMQYKYNEKTMSLSRMLEEKDRLHNAFVEESRSMQRKAREEVRRILEEQEKLRNELDEKM 1048
Query: 675 KEVNRLKKEHVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVEKQIAKVEL- 733
++++ ++ + + ++ D ER+ +++K + L+ A K E++IA +
Sbjct: 1049 RKLDTWSRD-LNKREVLTDQERQKLEEDKKKKDSRNESLMLASK------EQKIADENVF 1207
Query: 734 RQVHLEKQVNERKMKLAFELSKTKEATKRFEAE----KKKLLVEKI---NAESKIKKAQE 786
R V +K+ E + +L K +A ++ E E + KL V K ++ IKK E
Sbjct: 1208 RLVEEQKREKEEALNKILQLEKQLDAKQKLEMEIEELRGKLQVMKHLGDQDDTAIKKKME 1387
Query: 787 RSESELDKKTADMEKQQAEEQKKLAEDK 814
SEL+ K +E ++ + +++
Sbjct: 1388 EMNSELEDKIESLEDMESMNSTLIVKER 1471
>TC83675 similar to PIR|T14321|T14321 nuclear matrix constituent protein 1 -
carrot, partial (17%)
Length = 905
Score = 45.1 bits (105), Expect = 1e-04
Identities = 57/215 (26%), Positives = 98/215 (45%), Gaps = 20/215 (9%)
Frame = +3
Query: 93 SENKKLKQDILE-----EQAQGKICDQLKKCEKVVEG-RNAL---RQAVKILEKRIENLE 143
S +L QDI+E EQ + L + EK E R AL ++ V LEK + +
Sbjct: 6 SNYNELSQDIVEVKDALEQEKAAHLFALSEAEKREENLRKALGVEKECVLDLEKALREMR 185
Query: 144 SENKKLKKDIQEEQA----------QRKIEIEGKLEKSNAFAALENEVSALKSENKKLKK 193
SE+ K+K + A ++ +E+E KL ++A A E+S SE +
Sbjct: 186 SEHAKIKFAADSKLAEANALIASVEEKSLEVEAKLRSADAKLA---EISRKSSEIDRKSH 356
Query: 194 DI-LEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQDILDEQAQGKFCDRLKK 252
D+ +E A R+ + E + + L + L+ KKL QD + A+G+ L +
Sbjct: 357 DLESQESALRRERLSFIAEQESHESTLSKQREDLREWEKKL-QDGEERLAKGQRI--LNE 527
Query: 253 KCEKVVEGRNALRQAVKILEKGIENLESENKKLKK 287
+ ++ + RQ K LE+ +N+++ N +K
Sbjct: 528 REQRANDIDKICRQKEKDLEEAQKNIDAANVTFEK 632
Score = 36.2 bits (82), Expect = 0.053
Identities = 38/116 (32%), Positives = 60/116 (50%), Gaps = 6/116 (5%)
Frame = +3
Query: 695 ERKTAVDERKNA----AAEARKLLEAPKKIAAEVEKQIAKVELRQVHLEKQVNERKMKLA 750
E K A+++ K A +EA K E +K A VEK+ ++L + E + K+K A
Sbjct: 39 EVKDALEQEKAAHLFALSEAEKREENLRK-ALGVEKECV-LDLEKALREMRSEHAKIKFA 212
Query: 751 FELSKTKEATKRFEA-EKKKLLVEKINAESKIKKAQ-ERSESELDKKTADMEKQQA 804
+ SK EA + E+K L VE + K A+ R SE+D+K+ D+E Q++
Sbjct: 213 AD-SKLAEANALIASVEEKSLEVEAKLRSADAKLAEISRKSSEIDRKSHDLESQES 377
>TC89797 GP|13775543|gb|AAK39351.1 Hypothetical protein Y73B3A.17
{Caenorhabditis elegans}, partial (1%)
Length = 958
Score = 45.1 bits (105), Expect = 1e-04
Identities = 52/241 (21%), Positives = 115/241 (47%), Gaps = 13/241 (5%)
Frame = -2
Query: 419 KKLKKDIQEEQAQRKIEIEGKLEISNAFAALENEVSALKSESTKLKKDILEEQAQIKVAI 478
+K+ ++ E + + ++ E ++ + E V ++ ++ KL + E ++++
Sbjct: 675 EKIVEEKNEIEYEVSVQREKMKDLGLCLESEERNVEKMRLDARKLFDEKAERESKV---- 508
Query: 479 EEKLEISNAFAALENEVSALKSEIAALQQKCGAGSREGNGDVEVLKAGISDTEK-EVNTL 537
E+LE + A++ + ++K I L++K +E N D E G++ T+ +++ L
Sbjct: 507 -EELE-KDRDLAVKKSIESVKV-IDELKEKIDLVVKEKN-DAE----GVNSTQDVKISNL 352
Query: 538 KKELVEKEKIVADSERKTAVDERKKAAAEARKLLEAAKKIAPEKAVIPEPANCCSKCDEL 597
EL + +++ S + AV K E + +E A + E+ ++ E + + E+
Sbjct: 351 GLELQQLNEVLKSSRNEEAVMRGK--IREMEETIEVA--LEKEREMMVENSKLVGEKKEM 184
Query: 598 KKKCEKVAVGRNALRQAVKILEKGIENL------------ESENKKLKKENEVSALKSEI 645
+K E + GR+ + + + ++++ +EN E E K+ ENEV L+ EI
Sbjct: 183 EKSIENLTEGRDTVYRTLDMVKRELENRQREVDEARRARDEVEKVKVGYENEVVKLQGEI 4
Query: 646 S 646
S
Sbjct: 3 S 1
Score = 44.7 bits (104), Expect = 1e-04
Identities = 61/318 (19%), Positives = 139/318 (43%), Gaps = 1/318 (0%)
Frame = -2
Query: 138 RIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVSALKSENKKLKKDILE 197
+I+NL++ N L K E + I+ L+ ++A +E++ AL +N +
Sbjct: 870 QIQNLKNLNNVLLK----ETTNHRNRIQLLLKANHAAMDVEDKNLALDLQNDVFFVFVKT 703
Query: 198 EQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQDILDEQAQGKFCDRLKKKCEKV 257
+ + E +E N +E EVS + +K+K L +++ + ++++ K+
Sbjct: 702 QITELGFLFEKIVEEKNE---IEYEVSV---QREKMKDLGLCLESEERNVEKMRLDARKL 541
Query: 258 VEGRNALRQAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEISNAFAALENEV 317
+ + V+ LEK + ++ + K I E + + ++ K + + + ++
Sbjct: 540 FDEKAERESKVEELEKDRDLAVKKSIESVKVIDELKEKIDLVVKEKNDAEGVNSTQDVKI 361
Query: 318 SALKSENKKLKKDILEEQAQRKVAMEGKL-EISNAFAALENEVSALKSENKKLKQDILEE 376
S L E ++L ++L+ + M GK+ E+ + + EN KL + E
Sbjct: 360 SNLGLELQQLN-EVLKSSRNEEAVMRGKIREMEETIEVALEKEREMMVENSKLVGEKKE- 187
Query: 377 QAQGKFCDQLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQRKIEI 436
++K E + EGR+ + + + ++++ +EN + +++ E + R
Sbjct: 186 ---------MEKSIENLTEGRDTVYRTLDMVKRELENRQ-------REVDEARRARDEVE 55
Query: 437 EGKLEISNAFAALENEVS 454
+ K+ N L+ E+S
Sbjct: 54 KVKVGYENEVVKLQGEIS 1
Score = 42.7 bits (99), Expect = 6e-04
Identities = 48/171 (28%), Positives = 83/171 (48%), Gaps = 7/171 (4%)
Frame = -2
Query: 19 EELKKKCEQVVVGRNALR-----QAVKILEKGIENLESENKKLKKDIQEEQAQRKVAIEG 73
+ELK+K + VV +N Q VKI G+E L+ N+ LK EE R G
Sbjct: 444 DELKEKIDLVVKEKNDAEGVNSTQDVKISNLGLE-LQQLNEVLKSSRNEEAVMR-----G 283
Query: 74 KL-EISNTF-AALENEVSALISENKKLKQDILEEQAQGKICDQLKKCEKVVEGRNALRQA 131
K+ E+ T ALE E ++ EN KL + E + K E + EGR+ + +
Sbjct: 282 KIREMEETIEVALEKEREMMV-ENSKLVGEKKEME---------KSIENLTEGRDTVYRT 133
Query: 132 VKILEKRIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVS 182
+ ++++ +EN + E + ++ ++E + K+ E ++ K L+ E+S
Sbjct: 132 LDMVKRELENRQREVDEARR-ARDEVEKVKVGYENEVVK------LQGEIS 1
>TC84214 weakly similar to GP|10176794|dbj|BAB09933. kinesin-like protein
{Arabidopsis thaliana}, partial (19%)
Length = 650
Score = 45.1 bits (105), Expect = 1e-04
Identities = 34/143 (23%), Positives = 69/143 (47%), Gaps = 7/143 (4%)
Frame = +2
Query: 373 ILEEQAQGKFCDQLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQR 432
+L E+A+ K K++CE +V+ L+ ++ + + + +KLK + E Q+
Sbjct: 212 LLNEKAKKKERFNYKERCENMVDYIKRLKVCIRWFQDLEMSYSIDQEKLKNSL-EMTKQK 388
Query: 433 KIEIEGKLEISNAFAALENEVSALKSESTKLKKDILEEQAQIKVAIEE-------KLEIS 485
IEIE L+I L + ++ T L++ +++E+A+ A+E +L+I
Sbjct: 389 SIEIEMLLKIKE--EELNLIIIEMRKNCTSLQEKLIKEEAEKSAAVESLSKEREARLDIE 562
Query: 486 NAFAALENEVSALKSEIAALQQK 508
+ L ++ + EI + QK
Sbjct: 563 RSHTTLSEDLGKAEREIQSANQK 631
Score = 40.8 bits (94), Expect = 0.002
Identities = 33/141 (23%), Positives = 72/141 (50%), Gaps = 4/141 (2%)
Frame = +2
Query: 237 ILDEQAQGKFCDRLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEHAQR 296
+L+E+A+ K K++CE +V+ L+ ++ + + + +KLK + E Q+
Sbjct: 212 LLNEKAKKKERFNYKERCENMVDYIKRLKVCIRWFQDLEMSYSIDQEKLKNSL-EMTKQK 388
Query: 297 KVEIEGKLEISNAFAALENEVSALKSENKK----LKKDILEEQAQRKVAMEGKLEISNAF 352
+EIE L+I E E++ + E +K L++ +++E+A++ A+E + A
Sbjct: 389 SIEIEMLLKIK------EEELNLIIIEMRKNCTSLQEKLIKEEAEKSAAVESLSKEREAR 550
Query: 353 AALENEVSALKSENKKLKQDI 373
+E + L + K +++I
Sbjct: 551 LDIERSHTTLSEDLGKAEREI 613
>BF646436 weakly similar to GP|9828634|gb F1N21.5 {Arabidopsis thaliana},
partial (11%)
Length = 659
Score = 44.7 bits (104), Expect = 1e-04
Identities = 50/214 (23%), Positives = 104/214 (48%), Gaps = 2/214 (0%)
Frame = +1
Query: 86 NEVSALISENKKLKQDILEEQAQGKICDQLKKCEKVVEGRNALRQAVKILEKRIENLESE 145
N S L+ E L+Q + + D LK+ + E +N L+Q +IL+ + ++E
Sbjct: 70 NRKSXLMKEADDLRQQKETFEREWDELD-LKRADVEKELKNVLQQKEEILKLQ----QNE 234
Query: 146 NKKLKKDIQ--EEQAQRKIEIEGKLEKSNAFAALENEVSALKSENKKLKKDILEEQAQRK 203
++LKK+ Q E+ QR++E +L K + A +E E S+L + + K +L + R+
Sbjct: 235 EERLKKEKQATEDYLQRELETL-QLAKESFAAEMELEKSSLAEKAQNEKNQLLLDFEMRR 411
Query: 204 VAMEGKLEISNAFAALENEVSALKSENKKLKQDILDEQAQGKFCDRLKKKCEKVVEGRNA 263
+E A ++N++ ++ ++D+ + + ++K E + N
Sbjct: 412 KELE---------ADMQNQL-------EQKEKDLFESRR------LFEEKRESELNNINF 525
Query: 264 LRQAVKILEKGIENLESENKKLKKDIQEEHAQRK 297
LR+ + +G+E ++ + KL+++ Q+ RK
Sbjct: 526 LRE---VANRGMEEMKHQRSKLEREKQDADENRK 618
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 44.7 bits (104), Expect = 1e-04
Identities = 53/189 (28%), Positives = 86/189 (45%), Gaps = 4/189 (2%)
Frame = +3
Query: 518 GDVEVLKAGISDTEKEVNTLKKELVEKEKIVADSERKTAVDERKKAAAEARKLLEAAKKI 577
GDV+ K I + + E+ +++K+ +KEK + + KT VDE K +K E KK
Sbjct: 111 GDVKEDKVEI-EKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKD-----KKDKEKKKKE 272
Query: 578 APEKAVIPEPANCCSKCDELKKKCEKVAVGRNALRQAVKILEKGIENLESENKKLKKEN- 636
E+ V E + K D+ KKK EK G+ K E+ + E KK K E+
Sbjct: 273 KKEENVKGEEEDGDEKKDKEKKKKEKKEKGK---EDKDKDGEEKKSKKDKEKKKDKNEDD 443
Query: 637 ---EVSALKSEISALQQKCGAGAREGNGDVEVLKAGISDTKKEVNRLKKEHVEEERIVAD 693
E + K + ++K + G V V I +T KE KK+ ++E +
Sbjct: 444 DEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRDIDIEETAKEGKEKKKKKEDKE----E 611
Query: 694 SERKTAVDE 702
++K+ D+
Sbjct: 612 KKKKSGKDK 638
Score = 41.2 bits (95), Expect = 0.002
Identities = 52/227 (22%), Positives = 101/227 (43%), Gaps = 7/227 (3%)
Frame = +3
Query: 69 VAIEGKLEISNTFAALENEVSALISENKKLKQDILEEQAQGKICDQLKKCEK-------V 121
+ + K+E SN ++++ + + E+K + LE ++ K ++ +K +K V
Sbjct: 48 ITVSSKMEESNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDV 227
Query: 122 VEGRNALRQAVKILEKRIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEV 181
EG++ + K EK+ EN++ E + + +E+ +++ + +GK +K
Sbjct: 228 DEGKDKKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDK--------- 380
Query: 182 SALKSENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQDILDEQ 241
E KK KKD +++ + + EG E+ K+++KK K+ DE+
Sbjct: 381 ---DGEEKKSKKDKEKKKDKNEDDDEG-----------EDGSKKKKNKDKKEKKKEEDEK 518
Query: 242 AQGKFCDRLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKD 288
+GK R E EG+ +K E+ E + KK KD
Sbjct: 519 EEGKVSVRDIDIEETAKEGKEK--------KKKKEDKEEKKKKSGKD 635
>TC93214 similar to GP|10177918|dbj|BAB11329. kinesin-like protein
{Arabidopsis thaliana}, partial (22%)
Length = 715
Score = 42.4 bits (98), Expect = 7e-04
Identities = 38/139 (27%), Positives = 64/139 (45%), Gaps = 11/139 (7%)
Frame = +3
Query: 681 KKEHVEEERIVADSERKTAVDERKNAAA----EARKLLEAPKKIAAEVEKQIAKVELRQV 736
K +E+E+ ER + E +N AA + +KL + + +E QI ++ +Q
Sbjct: 36 KITELEDEKRTVQQERDCLLAEVENLAANSDGQTQKLEDTHSQKLKALEAQILDLKKKQ- 212
Query: 737 HLEKQVNERKMKLAFELSKTKEATKRFEAEKKKLLVEKINAESKIKKAQER-------SE 789
E QV K K K+ EATKR + E + + +K+ + KIK+ E+ E
Sbjct: 213 --ESQVQLMKQK-----QKSDEATKRLQDEIQSIKAQKVQLQQKIKQEAEQFRQWKASRE 371
Query: 790 SELDKKTADMEKQQAEEQK 808
EL + + K + E+ K
Sbjct: 372 KELLQLRKEGRKSEYEKHK 428
Score = 33.5 bits (75), Expect = 0.34
Identities = 42/197 (21%), Positives = 85/197 (42%), Gaps = 15/197 (7%)
Frame = +3
Query: 252 KKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEISNAFA 311
+K ++ + + ++Q L +ENL + + + +++ H+Q+
Sbjct: 33 RKITELEDEKRTVQQERDCLLAEVENLAANSDGQTQKLEDTHSQK-------------LK 173
Query: 312 ALENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQ 371
ALE ++ LK + E Q Q M+ K + A L++E+ ++K++ +L+Q
Sbjct: 174 ALEAQILDLKKKQ--------ESQVQ---LMKQKQKSDEATKRLQDEIQSIKAQKVQLQQ 320
Query: 372 DILEEQAQGKFCDQLKKKCEKVV-----EGR---------NALRQAVK-ILEKGIENLES 416
I +E Q + Q K EK + EGR AL Q + +L++ E
Sbjct: 321 KIKQEAEQFR---QWKASREKELLQLRKEGRKSEYEKHKLQALNQRQRMVLQRKTEEAAM 491
Query: 417 ENKKLKKDIQEEQAQRK 433
K+LK+ ++ + +
Sbjct: 492 ATKRLKELLEARKTSSR 542
>TC79049 weakly similar to PIR|T10798|T10798 pherophorin-S - Volvox carteri,
partial (6%)
Length = 957
Score = 42.4 bits (98), Expect = 7e-04
Identities = 42/147 (28%), Positives = 67/147 (45%), Gaps = 15/147 (10%)
Frame = -3
Query: 657 REGNGDVEVLK---AGISDTKKEVNRLKKEHVEEERIVADSERKTAVDERKNAAAEARKL 713
+E G EVLK + + KEV LKK E E V D E++ V E K +++
Sbjct: 919 KEAEGLKEVLKDKEGRVEELVKEVEGLKKVKAESEARVKDLEKRIGVLEMKEIEERNKRI 740
Query: 714 L--EAPKKIAAEVEKQIAKVELRQVHLEKQVNERK--------MKLAFE--LSKTKEATK 761
E + E +++I + LEK E+K KL++E L +++E K
Sbjct: 739 RVEEELRDTIGEKDREIDGFRNKVEELEKVGAEKKDEAGDWLNEKLSYEKALRESEEKAK 560
Query: 762 RFEAEKKKLLVEKINAESKIKKAQERS 788
FE++ +LL E + IK E++
Sbjct: 559 GFESQIVQLLEEVGESGKMIKSLNEKA 479
Score = 33.9 bits (76), Expect = 0.26
Identities = 38/148 (25%), Positives = 61/148 (40%), Gaps = 4/148 (2%)
Frame = -3
Query: 514 REGNGDVEVLK---AGISDTEKEVNTLKKELVEKEKIVADSERKTAVDERKKAAAEARKL 570
+E G EVLK + + KEV LKK E E V D E++ V E +++
Sbjct: 919 KEAEGLKEVLKDKEGRVEELVKEVEGLKKVKAESEARVKDLEKRIGV-------LEMKEI 761
Query: 571 LEAAKKIAPEKAVIPEPANCCSKCDELKKKCEKV-AVGRNALRQAVKILEKGIENLESEN 629
E K+I E+ + + D + K E++ VG +A L E L E
Sbjct: 760 EERNKRIRVEEELRDTIGEKDREIDGFRNKVEELEKVGAEKKDEAGDWLN---EKLSYEK 590
Query: 630 KKLKKENEVSALKSEISALQQKCGAGAR 657
+ E + +S+I L ++ G +
Sbjct: 589 ALRESEEKAKGFESQIVQLLEEVGESGK 506
Score = 32.3 bits (72), Expect = 0.76
Identities = 31/160 (19%), Positives = 71/160 (44%), Gaps = 20/160 (12%)
Frame = -3
Query: 119 EKVVEGRNALRQAVKILEKRIENLESENKKLK----------KDIQE----------EQA 158
E++ + L++ +K E R+E L E + LK KD+++ E+
Sbjct: 931 EEMKKEAEGLKEVLKDKEGRVEELVKEVEGLKKVKAESEARVKDLEKRIGVLEMKEIEER 752
Query: 159 QRKIEIEGKLEKSNAFAALENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEISNAFAA 218
++I +E +L + + E+ +++ ++L+K E++ + + KL A
Sbjct: 751 NKRIRVEEELR--DTIGEKDREIDGFRNKVEELEKVGAEKKDEAGDWLNEKLSYEKALRE 578
Query: 219 LENEVSALKSENKKLKQDILDEQAQGKFCDRLKKKCEKVV 258
E + +S+ +L +++ + GK L +K ++V
Sbjct: 577 SEEKAKGFESQIVQLLEEVGE---SGKMIKSLNEKAAEIV 467
>TC91851 weakly similar to PIR|F96673|F96673 hypothetical protein F13O11.30
[imported] - Arabidopsis thaliana, partial (12%)
Length = 981
Score = 41.2 bits (95), Expect = 0.002
Identities = 47/200 (23%), Positives = 86/200 (42%), Gaps = 3/200 (1%)
Frame = +3
Query: 449 LENEVSALKSESTKLKKDILEEQAQIKVAIEEKLEISNAFAALENEVSALKSEIAALQQK 508
L+N+++ + + K K+ IL+ + + AI+E + A E L+ + A +Q
Sbjct: 366 LQNQLNVAQEDLKKAKEQILQAEKEKVKAIDE---LKEAQRVAEEANEKLQEALVAQKQA 536
Query: 509 CGAGSREGNGDVEVLKAGISDTEKEVNTLKKEL--VEKEKIVADSERKTAVDERKKAAAE 566
E VE+ +AGI K+ +KEL V + + + + +E ++ E
Sbjct: 537 KEESEIEKFRAVELEQAGIETVNKKEEEWQKELESVRNQHALDVASLASTTEELERVKQE 716
Query: 567 ARKLLEAAKKIAPEKAVIPEPANCCSKCDELKKKCEKVAVGRNALRQAVKILEKGIENLE 626
+ +A + A+ +K E+ EKV + L Q +L+ E
Sbjct: 717 LTMMCDAKNQAL-------NHADDAAKVAEV--HAEKVEIYAAELTQLKALLDSTQETKA 869
Query: 627 SENKK-LKKENEVSALKSEI 645
S+N LK + E+ ALK E+
Sbjct: 870 SDNNLILKLKAEIEALKKEL 929
Score = 39.3 bits (90), Expect = 0.006
Identities = 47/194 (24%), Positives = 82/194 (42%), Gaps = 22/194 (11%)
Frame = +3
Query: 19 EELKKKCEQVVVGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQRKVAIEGKLEIS 78
E+LKK EQ++ +A+ L++ E N+KL++ + AQ++ E ++E
Sbjct: 393 EDLKKAKEQILQAEKEKVKAIDELKEAQRVAEEANEKLQEAL---VAQKQAKEESEIE-- 557
Query: 79 NTFAALENEVSALISENKK-----------LKQDILEEQAQGKICDQLKK--------CE 119
F A+E E + + + NKK Q L+ + ++L++ C+
Sbjct: 558 -KFRAVELEQAGIETVNKKEEEWQKELESVRNQHALDVASLASTTEELERVKQELTMMCD 734
Query: 120 KVVEGRNALRQAVKILEKRIENLE---SENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAA 176
+ N A K+ E E +E +E +LK + Q E K +N
Sbjct: 735 AKNQALNHADDAAKVAEVHAEKVEIYAAELTQLKALLDSTQ-------ETKASDNNLILK 893
Query: 177 LENEVSALKSENKK 190
L+ E+ ALK E K
Sbjct: 894 LKAEIEALKKELDK 935
>CB891165 homologue to PIR|H82068|H820 RNA polymerase-associated protein HepA
VC2506 [imported] - Vibrio cholerae, partial (1%)
Length = 692
Score = 40.8 bits (94), Expect = 0.002
Identities = 36/134 (26%), Positives = 67/134 (49%), Gaps = 3/134 (2%)
Frame = -2
Query: 678 NRLKKEHVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVEKQIAKVELRQVH 737
N + ++V E+ I A +R+ V ++ + + +++ + E ++ + + E +
Sbjct: 529 NFVMTKNVVEDHINALRKREKLVQLMRDGKKQKKSMMKEKSQFEDEAKRLVEEGEKVEEK 350
Query: 738 LEKQVNERKMKLAFELSKTKEATKRFEAEKKKLLVEKINAESKIKKAQERSESELDKKT- 796
+ V E+K + E K KE+ +R E EKKKL E A++KI +++E S KT
Sbjct: 349 IRILV-EQKNSIELEKIKLKESMERHEDEKKKLEDE---AKNKITESKELMSSIKKSKTS 182
Query: 797 --ADMEKQQAEEQK 808
A + KQQ + K
Sbjct: 181 YDAALSKQQKMKDK 140
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.305 0.124 0.307
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,028,457
Number of Sequences: 36976
Number of extensions: 140372
Number of successful extensions: 1132
Number of sequences better than 10.0: 144
Number of HSP's better than 10.0 without gapping: 671
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1093
length of query: 824
length of database: 9,014,727
effective HSP length: 104
effective length of query: 720
effective length of database: 5,169,223
effective search space: 3721840560
effective search space used: 3721840560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0437b.7


