

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
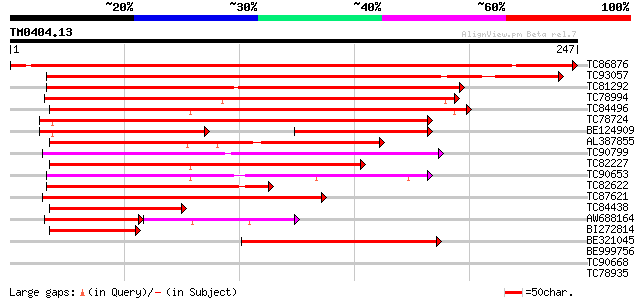
Query= TM0404.13
(247 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box pro... 297 3e-81
TC93057 homologue to GP|23194453|gb|AAN15183.1 MADS box protein ... 239 5e-64
TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalf... 159 9e-40
TC78994 similar to SP|O64645|SOC1_ARATH SUPPRESSOR OF CONSTANS O... 151 3e-37
TC84496 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 149 1e-36
TC78724 similar to GP|4322475|gb|AAD16052.1| putative MADS box t... 140 5e-34
BE124909 similar to GP|4322475|gb| putative MADS box transcripti... 106 9e-31
AL387855 similar to GP|1483230|emb MADS4 protein {Betula pendula... 120 4e-28
TC90799 similar to GP|7672991|gb|AAF66690.1| MADS-box transcript... 119 1e-27
TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 119 1e-27
TC90653 similar to GP|10835358|gb|AAC13695.2 PTD protein {Populu... 110 7e-25
TC82622 similar to GP|21739238|emb|CAD40993. OSJNBa0072F16.17 {O... 106 9e-24
TC87621 similar to SP|Q9FVC1|SVP_ARATH SHORT VEGETATIVE PHASE pr... 99 2e-21
TC84438 homologue to GP|1483232|emb|CAA67969.1 MADS5 protein {Be... 94 6e-20
AW688164 similar to GP|3114588|gb|A MADS box protein {Eucalyptus... 69 2e-19
BI272814 PIR|S71757|S71 MADS box protein DEFH200 - garden snapdr... 74 5e-14
BE321045 similar to PIR|T09700|T097 MADS-box protein - alfalfa (... 63 1e-10
BE999756 weakly similar to GP|9758603|dbj| gene_id:MDF20.13~unkn... 36 0.012
TC90668 similar to PIR|H86295|H86295 hypothetical protein AAF185... 31 0.39
TC78935 similar to GP|6760451|gb|AAF28357.1| S-ribonuclease bind... 31 0.50
>TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box protein {Malus
x domestica}, partial (85%)
Length = 2050
Score = 297 bits (760), Expect = 3e-81
Identities = 154/247 (62%), Positives = 189/247 (76%)
Frame = +2
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
M PN+ S Q+KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV
Sbjct: 44 MELPNEG--GEGSSQKKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 217
Query: 61 ALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISN 120
AL+VFS RGRLYEYANNSV+A+IERYKKAC+ S+ S S AN QFYQQE++KLR QI +
Sbjct: 218 ALVVFSTRGRLYEYANNSVRATIERYKKACAASTNAESVSEANTQFYQQESSKLRRQIRD 397
Query: 121 LQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNS 180
+QN NR +LGEAL +++ ++LKNLE +LEKG+SR+RS+K+E LFA++E+MQKREI+L N
Sbjct: 398 IQNLNRHILGEALGSLSLKELKNLEGRLEKGLSRVRSRKHETLFADVEFMQKREIELQNH 577
Query: 181 NQLLRAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQD 240
N LRAKIAE + + N++P T + S Q +D R FF V L + Q +RQD
Sbjct: 578 NNYLRAKIAEHERAQQQQHNLMPDQTMCDQSLPSSQAYD-RNFFPVNLLGSDQQQYSRQD 754
Query: 241 QISLQFV 247
Q +LQ V
Sbjct: 755 QTALQLV 775
>TC93057 homologue to GP|23194453|gb|AAN15183.1 MADS box protein GHMADS-2
{Gossypium hirsutum}, partial (95%)
Length = 858
Score = 239 bits (611), Expect = 5e-64
Identities = 127/225 (56%), Positives = 161/225 (71%)
Frame = +3
Query: 17 KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN 76
+ G ++ K IENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+RGRLYEY+N
Sbjct: 27 RWGGERLR*KGIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYSN 206
Query: 77 NSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNM 136
N+++++I+RYKKACSD S +T+ NAQ+YQQE+AKLR QI LQN NR ++G+ALS +
Sbjct: 207 NNIRSTIDRYKKACSDHSSTTTTTEINAQYYQQESAKLRQQIQMLQNSNRHLMGDALSTL 386
Query: 137 NARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKN 196
++LK LE +LE+GI+RIRSKK+EML AEIEY QKREI+L N N LR KI +D +
Sbjct: 387 TVKELKQLENRLERGITRIRSKKHEMLLAEIEYFQKREIELENENLCLRTKI--NDVERL 560
Query: 197 HNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQ 241
NM+ G + Q SR FF ++ + D+
Sbjct: 561 PQVNMVSGQE-----LNAIQALASRNFFNPNMMEDGETSYHQSDK 680
>TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalfa
(fragment), partial (91%)
Length = 800
Score = 159 bits (402), Expect = 9e-40
Identities = 79/182 (43%), Positives = 127/182 (69%)
Frame = +2
Query: 17 KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN 76
+MGRGKI+I+RI+N+T+RQVTF KRRNGLLKKA EL++LCDAEV +++FS+ +LY++A+
Sbjct: 68 EMGRGKIQIRRIDNSTSRQVTFSKRRNGLLKKAKELAILCDAEVGVMIFSSTAKLYDFAS 247
Query: 77 NSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNM 136
S+++ I+RY K + + GS S + +F+Q+EAA LR Q+ NLQ +RQ++GE LS +
Sbjct: 248 TSMRSVIDRYNKTKEEHNQLGS-STSEIKFWQREAAMLRQQLHNLQESHRQIMGEELSGL 424
Query: 137 NARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKN 196
++L+ LE +LE + +R KK ++ EI+ + ++ +H N L K+ + ++
Sbjct: 425 TVKELQGLENQLEISLRGVRMKKEQLFMDEIQELNRKGDIIHQENVELYRKVYGTKDKNG 604
Query: 197 HN 198
N
Sbjct: 605 TN 610
>TC78994 similar to SP|O64645|SOC1_ARATH SUPPRESSOR OF CONSTANS
OVEREXPRESSION 1 protein (Agamous-like MADS box protein
AGL20)., partial (83%)
Length = 946
Score = 151 bits (381), Expect = 3e-37
Identities = 86/189 (45%), Positives = 126/189 (66%), Gaps = 8/189 (4%)
Frame = +3
Query: 16 RKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYA 75
+KM RGK ++KRIEN T+RQVTF KRRNGLLKKA+ELSVLCDAEVALIVFS RGRLYE+A
Sbjct: 48 KKMVRGKTQMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSPRGRLYEFA 227
Query: 76 NNSVKASIERYKKACS-DSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALS 134
++S+ +IERY+ +++ S S N Q ++EA + +I L+ R++LGE L
Sbjct: 228 SSSILETIERYRSHTRINNTPTTSESVENTQQLKEEAENMMKKIDLLETSKRKLLGEGLG 407
Query: 135 NMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKI------ 188
+ + +L+ +E +LEK I++IR KK ++ +I+ ++++E L N L K
Sbjct: 408 SCSIDELQKIEQQLEKSINKIRVKKTKVFREQIDQLKEKEKALVAENVRLSEKYGNYSTQ 587
Query: 189 -AESDERKN 196
+ D+R+N
Sbjct: 588 ESTKDQREN 614
>TC84496 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (74%)
Length = 681
Score = 149 bits (376), Expect = 1e-36
Identities = 75/191 (39%), Positives = 128/191 (66%), Gaps = 7/191 (3%)
Frame = +2
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANN 77
MGRG++++K+IEN NRQVTF KRR+GLLKKA+E+SVLCDAEVALIVFSN+G+LYEY+++
Sbjct: 26 MGRGRVQLKKIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALIVFSNKGKLYEYSSD 205
Query: 78 -SVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNM 136
++ +ERY++ + + + E A+L+ ++ +Q + R +GE L +
Sbjct: 206 PCMEKILERYERYSYAERQHVANDQPQNENWIIEHARLKTRLEVIQKNQRNFMGEELDGL 385
Query: 137 NARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESD---- 192
+ ++L++LE +L+ + +IRS+KN++++ I + K++ L N+LL K+ E +
Sbjct: 386 SMKELQHLEHQLDSALKQIRSRKNQLMYESISELSKKDKALQEKNKLLTTKMKEKEKALA 565
Query: 193 --ERKNHNFNM 201
E++N + N+
Sbjct: 566 QLEQQNEDMNL 598
>TC78724 similar to GP|4322475|gb|AAD16052.1| putative MADS box
transcription factor ETL {Eucalyptus globulus subsp.
globulus}, partial (71%)
Length = 1089
Score = 140 bits (353), Expect = 5e-34
Identities = 76/173 (43%), Positives = 116/173 (66%), Gaps = 2/173 (1%)
Frame = +1
Query: 14 PQRK--MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRL 71
PQR+ M RGK ++KRIEN +NRQVTF KRRNGLLKKA+ELSVLCDAEVALIVFS G+L
Sbjct: 208 PQREFIMVRGKTQMKRIENESNRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSTTGKL 387
Query: 72 YEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGE 131
YE++++S+ ++ERY+ + N Q ++ ++ +L+ R++LGE
Sbjct: 388 YEFSSSSISKTVERYQGKVKELVLSTKGIQENTQHLKECDIDTTKKLEHLELSKRKLLGE 567
Query: 132 ALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLL 184
L + +L+ +E +LE+ +S+IR++KN++L +IE ++ +E L N+ L
Sbjct: 568 ELGSCAFDELQQIENQLERSLSKIRARKNQLLKEQIEKLKDKERLLLEENKRL 726
>BE124909 similar to GP|4322475|gb| putative MADS box transcription factor
ETL {Eucalyptus globulus subsp. globulus}, partial (60%)
Length = 608
Score = 106 bits (264), Expect(2) = 9e-31
Identities = 52/76 (68%), Positives = 66/76 (86%), Gaps = 2/76 (2%)
Frame = +1
Query: 14 PQRK--MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRL 71
PQR+ M RGK ++KRIEN +NRQVTF KRRNGLLKKA+ELSVLCDAEVALIVFS G+L
Sbjct: 130 PQREFIMVRGKTQMKRIENESNRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSTTGKL 309
Query: 72 YEYANNSVKASIERYK 87
YE++++S+ ++ERY+
Sbjct: 310 YEFSSSSISKTVERYQ 357
Score = 44.3 bits (103), Expect(2) = 9e-31
Identities = 21/60 (35%), Positives = 41/60 (68%)
Frame = +2
Query: 125 NRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLL 184
+R++LGE L + +L+ +E +LE+ +S+IR++KN++L +IE ++ +E L N+ L
Sbjct: 407 HRKLLGEELGSCAFDELQQIENQLERSLSKIRARKNQLLKEQIEKLKDKERLLLEENKRL 586
>AL387855 similar to GP|1483230|emb MADS4 protein {Betula pendula}, partial
(51%)
Length = 493
Score = 120 bits (302), Expect = 4e-28
Identities = 63/148 (42%), Positives = 106/148 (71%), Gaps = 2/148 (1%)
Frame = +2
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN- 76
MGRG++++KRIEN T++QVTF KRR GLLKKA E+SVLCDA+VALI+FS +G+L+EY++
Sbjct: 53 MGRGRVQLKRIENKTSQQVTFFKRRTGLLKKANEISVLCDAQVALIMFSTKGKLFEYSSA 232
Query: 77 NSVKASIERYKKA-CSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSN 135
S++ +ERY++ ++ +G + + N F E KL ++ L+ + R +G L
Sbjct: 233 PSMEDILERYERQNHTELTGATNETQGNWSF---EYMKLTAKVQVLERNLRNFVGNDLDP 403
Query: 136 MNARDLKNLETKLEKGISRIRSKKNEML 163
++ ++L++LE +L+ + RIR++KN+++
Sbjct: 404 LSVKELQSLEQQLDTSLKRIRTRKNQVM 487
>TC90799 similar to GP|7672991|gb|AAF66690.1| MADS-box transcription factor
{Canavalia lineata}, partial (76%)
Length = 653
Score = 119 bits (298), Expect = 1e-27
Identities = 67/175 (38%), Positives = 101/175 (57%)
Frame = +2
Query: 15 QRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEY 74
+R M R KI+IK+I+N T RQVTF KRR G+ KKA ELS+LCDAEV L++FS G+LYEY
Sbjct: 62 ERLMARQKIKIKKIDNATARQVTFSKRRRGIFKKAEELSILCDAEVGLVIFSTTGKLYEY 241
Query: 75 ANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALS 134
A++++K I RY + + Q + A+L ++++ R M E
Sbjct: 242 ASSNMKDIITRYGQQSHHIT--KLDKPLQVQVEKNMPAELNKEVADRTQQLRGMKSEDFE 415
Query: 135 NMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIA 189
+N L+ LE LE + R+ K + + EI+ ++ +EI L N+ L+ K+A
Sbjct: 416 GLNLEGLQQLEKSLESXLKRVIEMKEKKILNEIKALRMKEIMLEXENKHLKQKMA 580
>TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (68%)
Length = 681
Score = 119 bits (297), Expect = 1e-27
Identities = 60/139 (43%), Positives = 97/139 (69%), Gaps = 1/139 (0%)
Frame = +1
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANN 77
MGRG++++KRIEN NRQVTF KRR+GLLKKA E+SVLCDAEVALI+FS +G+L+EY+++
Sbjct: 130 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAQEISVLCDAEVALIIFSTKGKLFEYSSD 309
Query: 78 -SVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNM 136
++ +ERY++ ++ + + + E AKL+ ++ LQ + R +GE L +
Sbjct: 310 PCMEKILERYERCSYMERQLVTSEQSPNENWVLEHAKLKARMEVLQRNQRNFMGEDLDGL 489
Query: 137 NARDLKNLETKLEKGISRI 155
++L++LE +L+ + +I
Sbjct: 490 GLKELQSLEQQLDSALKQI 546
>TC90653 similar to GP|10835358|gb|AAC13695.2 PTD protein {Populus
balsamifera subsp. trichocarpa}, partial (59%)
Length = 845
Score = 110 bits (274), Expect = 7e-25
Identities = 69/175 (39%), Positives = 103/175 (58%), Gaps = 7/175 (4%)
Frame = +2
Query: 17 KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN 76
KMGRGKIEIK IEN TNRQVT+ KRRNG+ KKA+ELSVLCDA+V+LI+FS +++EY
Sbjct: 41 KMGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYIT 220
Query: 77 N--SVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEA-- 132
S K I++Y+K D S + + + KL+ L+ R +GE
Sbjct: 221 PGLSTKKIIDQYQKTLGDIDLWRS----HYEKMLENLKKLKDINHKLRRQIRHRIGEGGM 388
Query: 133 -LSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQK--REIDLHNSNQLL 184
L +++ + L++LE + I++IR +K ++ + +K R ++ N N LL
Sbjct: 389 ELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLL 553
>TC82622 similar to GP|21739238|emb|CAD40993. OSJNBa0072F16.17 {Oryza
sativa}, partial (78%)
Length = 847
Score = 106 bits (264), Expect = 9e-24
Identities = 53/99 (53%), Positives = 71/99 (71%)
Frame = +3
Query: 17 KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN 76
+MGRGKI I+RI+N T+RQVTF KRR GL+KKA EL++LCDA+V L++FS+ G+LYEYAN
Sbjct: 135 RMGRGKIVIRRIDNCTSRQVTFSKRRKGLIKKAKELAILCDAQVGLVIFSSTGKLYEYAN 314
Query: 77 NSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLR 115
S+K+ IERY D S +F+Q+EA +
Sbjct: 315 TSMKSVIERYNICKEDQQVTNPES--EVKFWQREAVHFK 425
>TC87621 similar to SP|Q9FVC1|SVP_ARATH SHORT VEGETATIVE PHASE protein.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (65%)
Length = 1322
Score = 99.0 bits (245), Expect = 2e-21
Identities = 54/124 (43%), Positives = 78/124 (62%)
Frame = +2
Query: 15 QRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEY 74
+R M R KI+IK+IEN+T RQVTF KRR GL+KKA ELSVLCDA+VALI+FS+ G+L+EY
Sbjct: 428 RRDMAREKIQIKKIENSTARQVTFSKRRRGLIKKAEELSVLCDADVALIIFSSTGKLFEY 607
Query: 75 ANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALS 134
+N S++ +ER+ + + S ++L +++ + RQM GE L
Sbjct: 608 SNLSMREILERHHLHSKNLAKLEEPSLELQLVENSNCSRLSKEVAQKSHQLRQMRGEDLQ 787
Query: 135 NMNA 138
+
Sbjct: 788 GFES 799
>TC84438 homologue to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (24%)
Length = 702
Score = 93.6 bits (231), Expect = 6e-20
Identities = 42/60 (70%), Positives = 55/60 (91%)
Frame = +1
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANN 77
MGRG++++KRIEN NRQVTF KRR+GLLKKA E+SVLCDAEVALI+FS +G+L+EY+++
Sbjct: 223 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAQEISVLCDAEVALIIFSTKGKLFEYSSD 402
>AW688164 similar to GP|3114588|gb|A MADS box protein {Eucalyptus grandis},
partial (31%)
Length = 549
Score = 69.3 bits (168), Expect(2) = 2e-19
Identities = 32/43 (74%), Positives = 38/43 (87%)
Frame = +1
Query: 16 RKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDA 58
RKMGRG++ ++RIEN NRQVTF KRR+GLLKKA+EL VLCDA
Sbjct: 1 RKMGRGRVVLERIENKINRQVTFSKRRSGLLKKAFELCVLCDA 129
Score = 43.1 bits (100), Expect(2) = 2e-19
Identities = 24/70 (34%), Positives = 42/70 (59%), Gaps = 2/70 (2%)
Frame = +2
Query: 59 EVALIVFSNRGRLYEYANNS-VKASIERYKKACSDSSGGGSTSGAN-AQFYQQEAAKLRV 116
EVALI+FS+RG+L++Y++ + + IERY++ G++ G N +Q Q+ KL+
Sbjct: 131 EVALIIFSSRGKLFQYSSTTDINKIIERYRQCRYSKPQAGNSLGHNESQNLYQDYLKLKA 310
Query: 117 QISNLQNHNR 126
+ +L R
Sbjct: 311 KYESLDRKQR 340
>BI272814 PIR|S71757|S71 MADS box protein DEFH200 - garden snapdragon,
partial (16%)
Length = 320
Score = 73.9 bits (180), Expect = 5e-14
Identities = 34/40 (85%), Positives = 37/40 (92%)
Frame = +1
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCD 57
MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCD
Sbjct: 199 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCD 318
>BE321045 similar to PIR|T09700|T097 MADS-box protein - alfalfa (fragment),
partial (38%)
Length = 423
Score = 62.8 bits (151), Expect = 1e-10
Identities = 30/87 (34%), Positives = 57/87 (65%)
Frame = +1
Query: 102 ANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNE 161
+ +F+Q+EA LR Q+ +LQ ++RQ++GE L ++ R+L++LE++LE + +R KK +
Sbjct: 43 SEVKFWQREADILRQQLQSLQENHRQLMGEQLYGLSIRNLQDLESQLELSLQGVRMKKEK 222
Query: 162 MLFAEIEYMQKREIDLHNSNQLLRAKI 188
+L EI+ + ++ +H N L K+
Sbjct: 223 ILTDEIQELNRKGSIIHQENVELYKKV 303
>BE999756 weakly similar to GP|9758603|dbj| gene_id:MDF20.13~unknown
protein {Arabidopsis thaliana}, partial (15%)
Length = 534
Score = 36.2 bits (82), Expect = 0.012
Identities = 18/48 (37%), Positives = 27/48 (55%)
Frame = +1
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVF 65
M K+++ I N T R+ T+ KR L+KK E++ LC + IVF
Sbjct: 13 MAGNKVKLAFITNHTARRATYKKRVQSLMKKLNEITTLCGVKACGIVF 156
>TC90668 similar to PIR|H86295|H86295 hypothetical protein AAF18507.1
[imported] - Arabidopsis thaliana, partial (27%)
Length = 1243
Score = 31.2 bits (69), Expect = 0.39
Identities = 37/157 (23%), Positives = 72/157 (45%), Gaps = 2/157 (1%)
Frame = +2
Query: 46 LKKAYELSVLCDAEVALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQ 105
+++A ELS L A ++I+F++ L + K + +R+ CS++ G +S
Sbjct: 38 MREANELSGLQSANSSVILFASTPILVPLRRKNCKENSDRFYCKCSEALHGDESSKG--- 208
Query: 106 FYQQEAAKLRVQISNLQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIR-SKKNEMLF 164
Y E+ + +V + L + +G L L L KL + I + R S E LF
Sbjct: 209 IYCDESPRRKVP-AKLVAASGMGIGTVL-----LCLFLLSCKLYQHIKKRRASTHKEKLF 370
Query: 165 AEI-EYMQKREIDLHNSNQLLRAKIAESDERKNHNFN 200
+ Y+ + ++ + + ++ + AE +R N+N
Sbjct: 371 RQNGGYLLQEKLSSYGNGEMAKLFTAEELQRATDNYN 481
>TC78935 similar to GP|6760451|gb|AAF28357.1| S-ribonuclease binding protein
SBP1 {Petunia x hybrida}, partial (93%)
Length = 1304
Score = 30.8 bits (68), Expect = 0.50
Identities = 23/101 (22%), Positives = 45/101 (43%)
Frame = +1
Query: 121 LQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNS 180
L+ Q+ L + A L+++ +K + ++R K+ E +E + KR ++L +
Sbjct: 541 LKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETE-----VENINKRNMELEDQ 705
Query: 181 NQLLRAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSR 221
+ L + +R +N NM+ F Q Q DS+
Sbjct: 706 MEQLSVEAGAWQQRARYNENMI-AALKFNLQQAYLQGRDSK 825
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.128 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,958,074
Number of Sequences: 36976
Number of extensions: 65448
Number of successful extensions: 300
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 294
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 294
length of query: 247
length of database: 9,014,727
effective HSP length: 94
effective length of query: 153
effective length of database: 5,538,983
effective search space: 847464399
effective search space used: 847464399
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0404.13


