

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0404.11
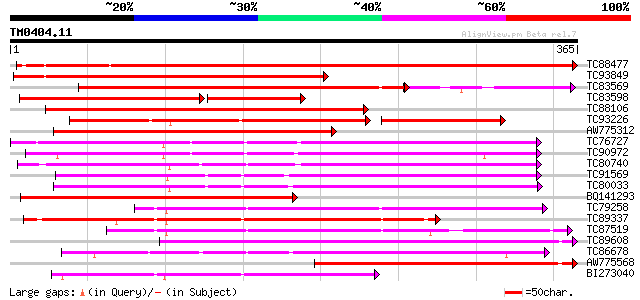
(365 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase p... 603 e-173
TC93849 similar to PIR|A86420|A86420 probable lipase/hydrolase ... 312 1e-85
TC83569 weakly similar to GP|6733842|emb|CAB69346.1 unnamed prot... 225 5e-75
TC83598 similar to PIR|H86419|H86419 probable lipase/hydrolase ... 181 1e-69
TC88106 weakly similar to GP|6733842|emb|CAB69346.1 unnamed prot... 233 1e-61
TC93226 weakly similar to PIR|B96737|B96737 hypothetical protein... 169 7e-61
AW775312 similar to GP|20466732|gb unknown protein {Arabidopsis ... 228 3e-60
TC76727 similar to GP|10638955|emb|CAB81548. putative proline-ri... 208 2e-54
TC90972 similar to GP|10638955|emb|CAB81548. putative proline-ri... 206 1e-53
TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif lipase/h... 200 8e-52
TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protei... 199 2e-51
TC80033 weakly similar to GP|21593518|gb|AAM65485.1 putative GDS... 199 2e-51
BQ141293 weakly similar to GP|20466732|gb| unknown protein {Arab... 195 3e-50
TC79258 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif... 179 2e-45
TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif... 178 3e-45
TC87519 weakly similar to GP|21592967|gb|AAM64916.1 putative GDS... 160 6e-40
TC89608 similar to PIR|T01143|T01143 probable GDSL-motif lipase/... 159 1e-39
TC86678 similar to PIR|E96579|E96579 hypothetical protein T18A20... 159 1e-39
AW775568 similar to GP|20466732|gb unknown protein {Arabidopsis ... 159 2e-39
BI273040 similar to GP|6721157|gb| putative GDSL-motif lipase/ac... 156 1e-38
>TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase putative
{Arabidopsis thaliana}, partial (92%)
Length = 1415
Score = 603 bits (1556), Expect = e-173
Identities = 295/361 (81%), Positives = 325/361 (89%)
Frame = +2
Query: 5 VNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDF 64
+NM A+LV+VLGLW G VGA PQVP YFIFGDSLVDNGNNN L+SLARADY+PYGIDF
Sbjct: 110 INMF--ALLVVVLGLWSG-VGADPQVPWYFIFGDSLVDNGNNNGLQSLARADYLPYGIDF 280
Query: 65 PGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQL 124
GGP+GRFSNGKTTVD IAELLGFDD+IPPY S S D IL+GVN+ASAAAGIREETG+QL
Sbjct: 281 -GGPTGRFSNGKTTVDAIAELLGFDDYIPPYASASDDAILKGVNYASAAAGIREETGRQL 457
Query: 125 GGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYST 184
G R+SFS QVQNYQSTVSQVVNILG EDQAA++LSKCIYSIGLGSNDYLNNYFMPQFY+T
Sbjct: 458 GARLSFSAQVQNYQSTVSQVVNILGTEDQAASHLSKCIYSIGLGSNDYLNNYFMPQFYNT 637
Query: 185 SRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSD 244
QYT D+YAD LIQ+YTEQL+TLYN ARKMVLFGIGQIGCSPNELA RS DG TCV +
Sbjct: 638 HDQYTPDEYADDLIQSYTEQLRTLYNN*ARKMVLFGIGQIGCSPNELATRSADGVTCVEE 817
Query: 245 INDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVG 304
IN ANQIFNNKLK +VDQFNNQLPD++VIY+N+YGIFQDII++P+ YGFS TN GCCGVG
Sbjct: 818 INSANQIFNNKLKGLVDQFNNQLPDSKVIYVNSYGIFQDIISNPSAYGFSVTNAGCCGVG 997
Query: 305 RNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQ 364
RNNGQ TCLP+QTPCENRREYLFWDAFHP+EAGNVV+AQRAYSAQSP DAYPIDI LAQ
Sbjct: 998 RNNGQFTCLPLQTPCENRREYLFWDAFHPTEAGNVVVAQRAYSAQSPDDAYPIDISHLAQ 1177
Query: 365 I 365
+
Sbjct: 1178L 1180
>TC93849 similar to PIR|A86420|A86420 probable lipase/hydrolase
118270-120144 [imported] - Arabidopsis thaliana, partial
(48%)
Length = 673
Score = 312 bits (799), Expect = 1e-85
Identities = 151/203 (74%), Positives = 175/203 (85%)
Frame = +1
Query: 3 LNVNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGI 62
L+++MM + ++V+VL LW G +G A QVPCYFIFGDSLVDNGNNN L S+A+A+Y+PYGI
Sbjct: 64 LDLSMMNIGIVVLVLSLWSG-IGVAQQVPCYFIFGDSLVDNGNNNQLTSIAKANYLPYGI 240
Query: 63 DFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQ 122
DFPGGP+GRFSNGKTTVDVIAE LGF+ +IP Y S G ILRGVN+ASAAAGIREETGQ
Sbjct: 241 DFPGGPTGRFSNGKTTVDVIAEQLGFNGYIPSYASARGRQILRGVNYASAAAGIREETGQ 420
Query: 123 QLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFY 182
QLG RISF GQVQNYQ TVSQ+VN G+E+ AANYLSKCIY+IGLGSNDYLNNYFMP Y
Sbjct: 421 QLGQRISFRGQVQNYQRTVSQLVNYFGDENTAANYLSKCIYTIGLGSNDYLNNYFMPTIY 600
Query: 183 STSRQYTTDQYADVLIQAYTEQL 205
STSRQ+T QYA+VL+QAY +QL
Sbjct: 601 STSRQFTPQQYANVLLQAYAQQL 669
>TC83569 weakly similar to GP|6733842|emb|CAB69346.1 unnamed protein product
{unidentified}, partial (39%)
Length = 1185
Score = 225 bits (573), Expect(2) = 5e-75
Identities = 109/213 (51%), Positives = 153/213 (71%)
Frame = +1
Query: 45 NNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDIL 104
NNN L + A+++Y PYGIDFP GP+GRF+NG+T++D+I +LLGF+ FIPP+ + +G DIL
Sbjct: 1 NNNNLPTSAKSNYKPYGIDFPMGPTGRFTNGRTSIDIITQLLGFEKFIPPFANINGSDIL 180
Query: 105 RGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYS 164
+GVN+AS AAGIR ET G IS Q++N++ VS++ + L D+A YLSKC+Y
Sbjct: 181 KGVNYASGAAGIRIETSITTGFVISLGLQLENHKVIVSRIASRLEGIDKAQEYLSKCLYY 360
Query: 165 IGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQI 224
+ +GSNDY+NNYF PQFY TS+ Y+ +QYA+ LIQ + L TL++ GARK VL G+G +
Sbjct: 361 VNIGSNDYINNYFRPQFYPTSQIYSPEQYAEALIQELSLNLLTLHDIGARKYVLVGLGLL 540
Query: 225 GCSPNELAQRSQDGRTCVSDINDANQIFNNKLK 257
GC+P+ + +G +CV + N IFN KLK
Sbjct: 541 GCTPSAIFTHGTNG-SCVDEENAPALIFNFKLK 636
Score = 74.3 bits (181), Expect(2) = 5e-75
Identities = 42/114 (36%), Positives = 62/114 (53%), Gaps = 3/114 (2%)
Frame = +2
Query: 254 NKLKSVVDQFNNQLP-DARVIYINAYGIFQDIIASPA--TYGFSNTNEGCCGVGRNNGQI 310
+ L +VD FNN+ D++ I++N + S A + GF +N CC G
Sbjct: 626 SSLNFLVDHFNNKFSADSKFIFVNT------TLESDAQNSDGFLVSNVPCCPSG------ 769
Query: 311 TCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQ 364
C+P + PC NR EY FWD HP+EA N + A R+Y++ + YP+DIK L +
Sbjct: 770 -CIPDERPCYNRSEYAFWDEVHPTEASNQLYAIRSYNSHNSGFTYPMDIKNLVE 928
>TC83598 similar to PIR|H86419|H86419 probable lipase/hydrolase
114382-116051 [imported] - Arabidopsis thaliana, partial
(42%)
Length = 686
Score = 181 bits (458), Expect(2) = 1e-69
Identities = 87/119 (73%), Positives = 101/119 (84%)
Frame = +3
Query: 7 MMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPG 66
M + M+V+VLGLW V A PQVPCYFIFGDSLVD+GNNN L SLA+A+Y+PYGIDF G
Sbjct: 105 MNLCVMMVVVLGLWSSKVEADPQVPCYFIFGDSLVDDGNNNNLNSLAKANYLPYGIDFNG 284
Query: 67 GPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLG 125
GP+GRFSNGKTTVDVIAELLGF+ +I PY + +IL+GVN+ASAAAGIREETGQQLG
Sbjct: 285 GPTGRFSNGKTTVDVIAELLGFEGYISPYSTARDQEILQGVNYASAAAGIREETGQQLG 461
Score = 100 bits (250), Expect(2) = 1e-69
Identities = 49/63 (77%), Positives = 53/63 (83%)
Frame = +1
Query: 128 ISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQ 187
ISFSGQVQNYQ TVSQVVN+LG+ED A+NYL+KCIYSIGLGSNDYLNNYFMP
Sbjct: 469 ISFSGQVQNYQKTVSQVVNLLGDEDTASNYLNKCIYSIGLGSNDYLNNYFMPCLSQW*TV 648
Query: 188 YTT 190
YTT
Sbjct: 649 YTT 657
>TC88106 weakly similar to GP|6733842|emb|CAB69346.1 unnamed protein product
{unidentified}, partial (28%)
Length = 788
Score = 233 bits (593), Expect = 1e-61
Identities = 111/209 (53%), Positives = 152/209 (72%), Gaps = 1/209 (0%)
Frame = +2
Query: 24 VGAAPQVPCYFIFGDSLVDNGNNNALRSLA-RADYMPYGIDFPGGPSGRFSNGKTTVDVI 82
V QVPC FIFGDSL D GNNN L + + R++Y PYGIDFP GP+GRF+NG+TT+D+I
Sbjct: 110 VNGKSQVPCLFIFGDSLSDGGNNNNLPANSPRSNYNPYGIDFPMGPTGRFTNGRTTIDII 289
Query: 83 AELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVS 142
+LLGF+ FIPP+ + +G DIL+GVN+AS AGIR ET G IS Q++N++ VS
Sbjct: 290 TQLLGFEKFIPPFANINGSDILKGVNYASGGAGIRMETYSAKGYAISLGLQLRNHRVIVS 469
Query: 143 QVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYT 202
Q+ + LG D+A YL+KC+Y + +GSNDY+NNYF+PQ Y +S Y+ +QYA+ LI+ +
Sbjct: 470 QIASQLGGIDKAQEYLNKCLYYVHIGSNDYINNYFLPQLYLSSNVYSPEQYAENLIEELS 649
Query: 203 EQLQTLYNFGARKMVLFGIGQIGCSPNEL 231
LQ L+ GARK VL G+G +GC+P+ +
Sbjct: 650 LNLQALHEIGARKYVLPGLGLLGCTPSAI 736
>TC93226 weakly similar to PIR|B96737|B96737 hypothetical protein F3I17.10
[imported] - Arabidopsis thaliana, partial (60%)
Length = 847
Score = 169 bits (428), Expect(2) = 7e-61
Identities = 81/196 (41%), Positives = 129/196 (65%), Gaps = 2/196 (1%)
Frame = +2
Query: 39 SLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVST 98
SLV+ GNNN L + A++++ PYGID+ G P+GRFSNGK+ +D I ++LG PP++
Sbjct: 2 SLVEVGNNNFLSTFAKSNFYPYGIDYNGRPTGRFSNGKSLIDFIGDMLGVPS-PPPFLDP 178
Query: 99 SGDD--ILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAAN 156
+ + +L GVN+AS + GI +++G+ G R S S Q+QN++ T++Q ++ NE +
Sbjct: 179 TSTENKLLNGVNYASGSGGILDDSGRHYGDRHSMSRQLQNFERTLNQYKKMM-NETALSQ 355
Query: 157 YLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKM 216
+L+K I + GSNDY+NNY P++Y TSR Y+ Q+ ++L+ + Q+ LY+ RK
Sbjct: 356 FLAKSIVIVVTGSNDYINNYLRPEYYGTSRNYSVPQFGNLLLNTFGRQILALYSLRLRKF 535
Query: 217 VLFGIGQIGCSPNELA 232
L G+G +GC PN+ A
Sbjct: 536 FLAGVGPLGCIPNQRA 583
Score = 82.8 bits (203), Expect(2) = 7e-61
Identities = 37/80 (46%), Positives = 50/80 (62%)
Frame = +3
Query: 240 TCVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEG 299
TCV +N +N L+S+V+QFN DA+ +Y N YG+F DI+ +PA Y FS +
Sbjct: 606 TCVDSVNQMVGTYNGGLRSMVEQFNRDHSDAKFVYGNTYGVFGDILNNPAAYAFSVIDRA 785
Query: 300 CCGVGRNNGQITCLPMQTPC 319
C +GR QI+CLPMQ PC
Sbjct: 786 CLCLGRK*SQISCLPMQFPC 845
>AW775312 similar to GP|20466732|gb unknown protein {Arabidopsis thaliana},
partial (46%)
Length = 655
Score = 228 bits (581), Expect = 3e-60
Identities = 111/183 (60%), Positives = 135/183 (73%), Gaps = 1/183 (0%)
Frame = +3
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGF 88
QVPC+FIFGDSLVDNGNNN + +LARA+Y PYGIDFP GP+GRF+NG+T VD +A+LLGF
Sbjct: 105 QVPCFFIFGDSLVDNGNNNGILTLARANYRPYGIDFPQGPTGRFTNGRTFVDALAQLLGF 284
Query: 89 DDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNIL 148
+IPP G D+LRGVN+AS AAGIREETG LG S + QV N+ +TV ++ +
Sbjct: 285 RAYIPPNSRARGLDVLRGVNYASGAAGIREETGSNLGAHTSMTEQVTNFGNTVQEMRRLF 464
Query: 149 -GNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQT 207
G+ D +YLSKCIY GLGSNDYLNNYFM FYSTS QYT +A L+Q Y QL
Sbjct: 465 RGDNDALNSYLSKCIYYSGLGSNDYLNNYFMTDFYSTSTQYTPKAFASALLQDYARQLSQ 644
Query: 208 LYN 210
L++
Sbjct: 645 LHS 653
>TC76727 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (92%)
Length = 1919
Score = 208 bits (530), Expect = 2e-54
Identities = 120/347 (34%), Positives = 185/347 (52%), Gaps = 5/347 (1%)
Frame = -2
Query: 1 MDLNVNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPY 60
M++N +V ++V L G VP FGDS VD GNN+ L +L +A+Y PY
Sbjct: 1177 MNMNSKETLVLLIVSCF-LTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPY 1001
Query: 61 GIDFPGG-PSGRFSNGKTTVDVIAELLGFDDFIPPYVS--TSGDDILRGVNFASAAAGIR 117
G DF P+GRF NGK D AE LGF F P Y+S SG ++L G NFASAA+G
Sbjct: 1000 GRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGY- 824
Query: 118 EETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYF 177
+E L I S Q++ ++ ++ + G++ +AA+ + +Y + GS+D++ NY+
Sbjct: 823 DEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSK-KAASIIKDSLYVLSAGSSDFVQNYY 647
Query: 178 MPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQD 237
+ + + T DQY+ L+ ++T ++ +Y GARK+ + + +GC P
Sbjct: 646 TNPWINQA--ITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYH 473
Query: 238 GRTCVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTN 297
CV+ IN Q FN K+ S QLP +++ + Y D++ +P+ +GF+
Sbjct: 472 ENGCVARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNFGFAEAG 293
Query: 298 EGCCGVGR-NNGQITCLPMQT-PCENRREYLFWDAFHPSEAGNVVIA 342
+GCCG G + C P C N +Y+FWD+ HPSEA N V+A
Sbjct: 292 KGCCGTGLVETTSLLCNPKSLGTCSNATQYVFWDSVHPSEAANQVLA 152
>TC90972 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (88%)
Length = 1258
Score = 206 bits (523), Expect = 1e-53
Identities = 119/339 (35%), Positives = 184/339 (54%), Gaps = 7/339 (2%)
Frame = +3
Query: 11 AMLVMVLGLWGGWVGAAPQ--VPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG- 67
A V+ ++ GW A VP FGDS VD GNN+ L +L +A+Y PYG DF
Sbjct: 39 AFFVVFAFVFLGWGNAQDDTVVPAIVTFGDSAVDVGNNDYLFTLFKANYPPYGRDFVXHK 218
Query: 68 PSGRFSNGKTTVDVIAELLGFDDFIPPYVS--TSGDDILRGVNFASAAAGIREETGQQLG 125
P+GRF NGK D+ AE LGF + P Y+S +G ++L G NFASAA+G +E L
Sbjct: 219 PTGRFCNGKLATDITAETLGFKSYAPAYLSPQATGKNLLIGANFASAASGY-DEKAAILN 395
Query: 126 GRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTS 185
I S Q++ Y+ S++ I G++ +AA+ + +Y + GS+D++ NY++ +
Sbjct: 396 HAIPLSQQLKYYKEYQSKLSKIAGSK-KAASIIKGALYLLSGGSSDFIQNYYVNPLIN-- 566
Query: 186 RQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDI 245
+ T DQY+ L+ Y+ ++ LY GARK+ + + +GC P + CV+ I
Sbjct: 567 KVVTPDQYSAYLVDTYSSFVKDLYKLGARKIGVTSLPPLGCLPATRTLFGFHEKGCVTRI 746
Query: 246 NDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVG- 304
N+ Q FN K+ S + QLP +++ N Y +++ SP+ +GF+ +GCCG G
Sbjct: 747 NNDAQGFNKKINSATVKLQKQLPGLKIVVFNIYKPLYELVQSPSKFGFAEARKGCCGTGI 926
Query: 305 -RNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIA 342
+ C N +Y+FWD+ HPSEA N ++A
Sbjct: 927 VETTSLLCNQKSLGTCSNATQYVFWDSVHPSEAANQILA 1043
>TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif
lipase/hydrolase-like protein {Arabidopsis thaliana},
partial (94%)
Length = 1167
Score = 200 bits (508), Expect = 8e-52
Identities = 118/342 (34%), Positives = 179/342 (51%), Gaps = 5/342 (1%)
Frame = +3
Query: 6 NMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFP 65
N + +L +V + V P VP FIFGDS+VD NNN L ++ ++++ PYG DF
Sbjct: 123 NFLTFLLLFVVFNV----VKGQPLVPALFIFGDSVVDARNNNNLYTIVKSNFPPYGRDFN 290
Query: 66 GG-PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGD--DILRGVNFASAAAGIREETGQ 122
P+GRF NGK D AE LGF + P Y++ ++L G NFAS A+G + T +
Sbjct: 291 NQMPTGRFCNGKLAADFTAENLGFTTYPPAYLNLQEKRKNLLNGANFASGASGYFDPTAK 470
Query: 123 QLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFM-PQF 181
L IS Q+++Y+ + +V + G + A++ +S IY + GS+D++ NY++ P
Sbjct: 471 -LYHAISLEQQLEHYKECQNILVGVAGKSN-ASSIISGAIYLVRAGSSDFVQNYYINPLL 644
Query: 182 YSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTC 241
Y +T DQ++D+L+Q YT +Q LY GARK+ + + +GC P + C
Sbjct: 645 YKV---FTADQFSDILMQHYTIFIQNLYALGARKIGVTTLPPLGCLPAAITLFGSHSNEC 815
Query: 242 VSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCC 301
V +N+ FN KL + +L + + ++ Y D++ P GF + CC
Sbjct: 816 VDRLNNDALNFNTKLNTTSQNLQKELSNLTLAVLDIYQPLHDLVTKPTENGFYEARKACC 995
Query: 302 GVGRNNGQITCLPMQT-PCENRREYLFWDAFHPSEAGNVVIA 342
G G I C C N EY+FWD FH SEA N V+A
Sbjct: 996 GTGLIETSILCNKDSIGTCANATEYVFWDGFHTSEAANNVLA 1121
>TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protein
{Arabidopsis thaliana}, partial (60%)
Length = 1398
Score = 199 bits (505), Expect = 2e-51
Identities = 114/318 (35%), Positives = 170/318 (52%), Gaps = 5/318 (1%)
Frame = +1
Query: 30 VPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGF 88
VP +FGDS VD+GNNN + + ++++ PYG D GG P+GRFSNG+ D I+E G
Sbjct: 127 VPAVIVFGDSSVDSGNNNMISTFLKSNFRPYGRDIDGGRPTGRFSNGRIPPDFISEAFGI 306
Query: 89 DDFIPPYVSTSG--DDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVN 146
IP Y+ + DD + GV FASA G T L I +V+ Y+ ++
Sbjct: 307 KSLIPAYLDPAYTIDDFVTGVCFASAGTGYDNATSAILNV-IPLWKEVEFYKEYQDKLKA 483
Query: 147 ILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQ 206
+G E+++ +S+ +Y I LG+ND+L NY+ F + +YT QY D LI ++
Sbjct: 484 HIG-EEKSIEIISEALYIISLGTNDFLGNYY--GFTTLRFRYTISQYQDYLIGIAENFIR 654
Query: 207 TLYNFGARKMVLFGIGQIGCSPNELAQRSQDG-RTCVSDINDANQIFNNKLKSVVDQFNN 265
LY+ GARK+ + G+ +GC P E A G C N FN KL++++ + N
Sbjct: 655 QLYSLGARKLAITGLIPMGCLPLERAINIFGGFHRCYEKYNIVALEFNVKLENMISKLNK 834
Query: 266 QLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQ-TPCENRRE 324
+LP + + N Y +F DII P+ YG + CC G C M C++ +
Sbjct: 835 ELPQLKALSANVYDLFNDIITRPSFYGIEEVEKACCSTGTIEMSYLCNKMNLMTCKDASK 1014
Query: 325 YLFWDAFHPSEAGNVVIA 342
Y+FWDAFHP+E N +I+
Sbjct: 1015YMFWDAFHPTEKTNRIIS 1068
>TC80033 weakly similar to GP|21593518|gb|AAM65485.1 putative GDSL-motif
lipase/hydrolase {Arabidopsis thaliana}, partial (89%)
Length = 1164
Score = 199 bits (505), Expect = 2e-51
Identities = 114/320 (35%), Positives = 173/320 (53%), Gaps = 5/320 (1%)
Frame = +1
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPS-GRFSNGKTTVDVIAELLG 87
+VP +FGDS VD GNNN + ++AR+++ PYG DF GG + GRFSNG+ D IAE G
Sbjct: 124 KVPAIIVFGDSSVDAGNNNFIPTVARSNFQPYGRDFQGGKATGRFSNGRIPTDFIAESFG 303
Query: 88 FDDFIPPYVSTSGD--DILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVV 145
+ +P Y+ + D GV+FASAA G T L I Q++ Y+ +
Sbjct: 304 IKESVPAYLDPKYNISDFATGVSFASAATGYDNATSDVLSV-IPLWKQLEYYKDYQKNLS 480
Query: 146 NILGNEDQAANYLSKCIYSIGLGSNDYLNNYF-MPQFYSTSRQYTTDQYADVLIQAYTEQ 204
+ LG E +A +S+ ++ + +G+ND+L NY+ MP + QYT QY L
Sbjct: 481 SYLG-EAKAKETISESVHLMSMGTNDFLENYYTMP---GRASQYTPQQYQTFLAGIAENF 648
Query: 205 LQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFN 264
++ LY GARK+ L G+ +GC P E CV++ N+ FN+KLK++ + N
Sbjct: 649 IRNLYALGARKISLGGLPPMGCLPLERTTNFMGQNGCVANFNNIALEFNDKLKNITTKLN 828
Query: 265 NQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGR-NNGQITCLPMQTPCENRR 323
+LPD ++++ N Y I II P YGF + + CC G G C +
Sbjct: 829 QELPDMKLVFSNPYYIMLHIIKKPDLYGFESASVACCATGMFEMGYACSRGSMFSCTDAS 1008
Query: 324 EYLFWDAFHPSEAGNVVIAQ 343
+++FWD+FHP+E N ++A+
Sbjct: 1009KFVFWDSFHPTEKTNNIVAK 1068
>BQ141293 weakly similar to GP|20466732|gb| unknown protein {Arabidopsis
thaliana}, partial (24%)
Length = 583
Score = 195 bits (495), Expect = 3e-50
Identities = 92/178 (51%), Positives = 126/178 (70%)
Frame = +2
Query: 8 MVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG 67
+VV ++ + + V QVPC FIFGDSL D+GNNN L + A+++Y PYGIDFP G
Sbjct: 50 LVVLIVFLSANYFKQCVNGKSQVPCVFIFGDSLSDSGNNNNLPTSAKSNYKPYGIDFPMG 229
Query: 68 PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGR 127
P+GRF+NG+T +D+I +LLGF++FIPP+ + SG DIL+GVN+AS AGIR ET G
Sbjct: 230 PTGRFTNGRTAIDIITQLLGFENFIPPFANISGSDILKGVNYASGGAGIRMETYSAKGYA 409
Query: 128 ISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTS 185
IS Q++N++ VSQ+ + LG D+A YL+KC+Y + +GSN Y+NN F P FY TS
Sbjct: 410 ISLGLQLRNHRVIVSQIASRLGGIDKAQQYLNKCLYYVNIGSNXYINNXFXPXFYPTS 583
>TC79258 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(73%)
Length = 1067
Score = 179 bits (454), Expect = 2e-45
Identities = 98/268 (36%), Positives = 151/268 (55%), Gaps = 2/268 (0%)
Frame = +3
Query: 81 VIAELLGFDDFIPPYVSTS--GDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQ 138
+ E LG + +P Y+S G+ +L G NFASA GI +TG Q I Q+ +
Sbjct: 24 IAGERLGLEPSLP-YLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFN 200
Query: 139 STVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLI 198
++ +G E A ++K I I LG ND++NNY++ F + SRQ++ Y LI
Sbjct: 201 QYQQKLSAQIGAEG-AKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLI 377
Query: 199 QAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKS 258
Y + LQ LY+ GAR++++ G G +GC+P ELA +S++G C +++ A ++N +L
Sbjct: 378 SEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNG-DCDAELMRAASLYNPQLVQ 554
Query: 259 VVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTP 318
++ Q N ++ D I +NA+ + D I +P +GF + CCG GR NG C P+
Sbjct: 555 MITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKL 734
Query: 319 CENRREYLFWDAFHPSEAGNVVIAQRAY 346
C NR Y FWDAFHPSE + +I Q+ +
Sbjct: 735 CPNRNLYAFWDAFHPSEKASRIIVQQMF 818
>TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(65%)
Length = 881
Score = 178 bits (451), Expect = 3e-45
Identities = 104/273 (38%), Positives = 170/273 (62%), Gaps = 5/273 (1%)
Frame = +1
Query: 10 VAMLVMVLGLWGGWVGAAPQVP-CYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG- 67
VA++++V+G G +V +P + +FGDSLVDNGNNN L + ARAD PYGID+
Sbjct: 55 VALVILVVG--GIFVHEIEAIPRTFLVFGDSLVDNGNNNYLATTARADAPPYGIDYQPSH 228
Query: 68 -PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTS--GDDILRGVNFASAAAGIREETGQQL 124
P+GRFSNG D+I++ LG + +P Y+S G+ +L G NFASA GI +TG Q
Sbjct: 229 RPTGRFSNGYNIPDIISQKLGAEPTLP-YLSPELRGEKLLVGANFASAGIGILNDTGIQF 405
Query: 125 GGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYST 184
I Q + +Q S++ ++G QA + +++ + I +G ND++NNY++ + +
Sbjct: 406 INIIRMYRQYEYFQEYQSRLSALIG-ASQAKSRVNQALVLITVGGNDFVNNYYLVPYSAR 582
Query: 185 SRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSD 244
SRQY +Y LI Y + LQ LY+ GAR++++ G G +GC P+E+AQR ++G+ C ++
Sbjct: 583 SRQYPLPEYVKYLISEYQKLLQKLYDLGARRVLVTGTGPMGCVPSEIAQRGRNGQ-CSTE 759
Query: 245 INDANQIFNNKLKSVVDQFNNQLPDARVIYINA 277
+ A+ +FN +L++++ N ++ R ++I A
Sbjct: 760 LQRASSLFNPQLENMLLGLNKKI--GRDVFIAA 852
>TC87519 weakly similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(76%)
Length = 1084
Score = 160 bits (406), Expect = 6e-40
Identities = 101/305 (33%), Positives = 161/305 (52%), Gaps = 5/305 (1%)
Frame = +2
Query: 63 DFPGG-PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTS--GDDILRGVNFASAAAGIREE 119
DFP P+GRFSNG D+I++ LG +P Y+S G +L G NFASA GI +
Sbjct: 2 DFPTRQPTGRFSNGLNVPDLISKELGSSPPLP-YLSPKLRGHRMLNGANFASAGIGILND 178
Query: 120 TGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMP 179
TG Q I Q+ ++ +V +++G ++ A ++ + I G ND++NNY++
Sbjct: 179 TGFQFIEVIRMYKQLDFFEEYQKRVSDLIGKKE-AKKLINGALILITCGGNDFVNNYYLV 355
Query: 180 QFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGR 239
SRQY +Y L+ Y + L+ LY+ GAR++++ G G +GC+P LA DG
Sbjct: 356 PNSLRSRQYALPEYVTYLLSEYKKILRRLYHLGARRVLVSGTGPMGCAPAALAIGGTDGE 535
Query: 240 TCVSDINDANQIFNNKLKSVVDQFNNQLPD--ARVIYINAYGIFQDIIASPATYGFSNTN 297
C ++ A ++N KL ++ + N Q+ V+ I+A +F + F +
Sbjct: 536 -CAPELQLAASLYNPKLVQLITELNQQIGSDVFSVLNIDALSLFGN--------EFKTSK 688
Query: 298 EGCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPI 357
CCG G NG C + C+NR ++LFWDAFHPSE N +I ++ + S YP+
Sbjct: 689 VACCGQGPYNGIGLCTLASSICQNRDDHLFWDAFHPSERANKMIVKQIMTG-STDVIYPM 865
Query: 358 DIKRL 362
++ +
Sbjct: 866 NLSTI 880
>TC89608 similar to PIR|T01143|T01143 probable GDSL-motif lipase/hydrolase
[imported] - Arabidopsis thaliana, partial (70%)
Length = 959
Score = 159 bits (403), Expect = 1e-39
Identities = 87/271 (32%), Positives = 141/271 (51%), Gaps = 2/271 (0%)
Frame = +2
Query: 97 STSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAAN 156
+ +G IL GVN+AS GI TG+ RI Q+ + T Q+ +LG
Sbjct: 17 NATGKSILYGVNYASGGGGILNATGRIFVNRIGMDIQIDYFTITRKQIDKLLGQSKARDF 196
Query: 157 YLSKCIYSIGLGSNDYLNNYFMPQFYSTSR-QYTTDQYADVLIQAYTEQLQTLYNFGARK 215
+ K I+SI +G+ND+LNNY +P +R + D + D +I + QL LY ARK
Sbjct: 197 IMKKSIFSITVGANDFLNNYLLPVLSVGARISQSPDAFVDDMINHFRGQLTRLYKMDARK 376
Query: 216 MVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYI 275
V+ +G IGC P + + CV N +N +LK ++ + N+ LP A +
Sbjct: 377 FVIGNVGPIGCIPYQKTINQLNEDECVDLANKLAIQYNGRLKDMLAELNDNLPGATFVLA 556
Query: 276 NAYGIFQDIIASPATYGFSNTNEGCCG-VGRNNGQITCLPMQTPCENRREYLFWDAFHPS 334
N Y + ++I + YGF+ ++ CCG G+ G I C P C +R +++FWD +HPS
Sbjct: 557 NVYDLVMELIKNYDKYGFTTSSRACCGNGGQFAGIIPCGPTSCICNDRYKHVFWDPYHPS 736
Query: 335 EAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
EA N++IA++ + P+++++L +
Sbjct: 737 EAANIIIAKQLLDGDKRYIS-PVNLRQLRDL 826
>TC86678 similar to PIR|E96579|E96579 hypothetical protein T18A20.15
[imported] - Arabidopsis thaliana, partial (43%)
Length = 1341
Score = 159 bits (403), Expect = 1e-39
Identities = 95/321 (29%), Positives = 159/321 (48%), Gaps = 7/321 (2%)
Frame = +2
Query: 34 FIFGDSLVDNGNNNALRSLA--RADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFDDF 91
FIFGDS +D GNNN + + +A+++PYG + P+GRFS+G+ D IAE +
Sbjct: 188 FIFGDSFLDAGNNNYINTTTFDQANFLPYGETYFNFPTGRFSDGRLISDFIAEYVNIP-L 364
Query: 92 IPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNE 151
+PP++ + GVNFAS AG ET Q G I F Q N++ + + + LG+
Sbjct: 365 VPPFLQPDNNKYYNGVNFASGGAGALVETFQ--GSVIPFKTQAINFKKVTTWLRHKLGSS 538
Query: 152 DQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNF 211
D + LS +Y +GSNDYL+ + + Y+ +Y ++I +T ++ ++
Sbjct: 539 D-SKTLLSNAVYMFSIGSNDYLSPFLTNS--DVLKHYSHTEYVAMVIGNFTSTIKEIHKR 709
Query: 212 GARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPDAR 271
GA+K V+ + +GC P +SQ +C+ +++ I N L V+ + QL +
Sbjct: 710 GAKKFVILNLPPLGCLPGTRIIQSQGKGSCLEELSSLASIHNQALYEVLLELQKQLRGFK 889
Query: 272 VIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTP-----CENRREYL 326
+ +I P YGF CCG G G+ +C + C+ E +
Sbjct: 890 FSLYDFNSDLSHMINHPLKYGFKEGKSACCGSGPFRGEYSCGGKRGEKHFELCDKPNESV 1069
Query: 327 FWDAFHPSEAGNVVIAQRAYS 347
FWD++H +E+ +A + +S
Sbjct: 1070FWDSYHLTESAYKQLAAQMWS 1132
>AW775568 similar to GP|20466732|gb unknown protein {Arabidopsis thaliana},
partial (43%)
Length = 668
Score = 159 bits (401), Expect = 2e-39
Identities = 78/171 (45%), Positives = 116/171 (67%), Gaps = 2/171 (1%)
Frame = +3
Query: 197 LIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRT-CVSDINDANQIFNNK 255
L+ Y QL L++ GARK+++ +GQIGC P EL + + + T C IN+A Q FN+
Sbjct: 3 LLHYYARQLSQLHSLGARKVIVTAVGQIGCIPYELGRINGNSSTGCNDKINNAIQYFNSG 182
Query: 256 LKSVVDQFNN-QLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLP 314
LK +V N QLP A+ ++++ Y D+ + + GF ++GCCGVGRNNGQITCLP
Sbjct: 183 LKQLVQNINGGQLPGAKFVFLDFYQSSADLALNGKSMGFDVVDKGCCGVGRNNGQITCLP 362
Query: 315 MQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
+Q CE+R +YLFWDAFHP+E N+++A+ +YS+QS + PI+I++LA +
Sbjct: 363 LQQVCEDRGKYLFWDAFHPTELANILLAKASYSSQSYTS--PINIQQLAML 509
>BI273040 similar to GP|6721157|gb| putative GDSL-motif lipase/acylhydrolase
{Arabidopsis thaliana}, partial (60%)
Length = 692
Score = 156 bits (394), Expect = 1e-38
Identities = 88/216 (40%), Positives = 129/216 (58%), Gaps = 5/216 (2%)
Frame = +2
Query: 28 PQVPC--YFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAE 84
PQV +F+FGDSLVDNGNNN L + ARAD PYGID+P +GRFSNG D+I+E
Sbjct: 29 PQVEARAFFVFGDSLVDNGNNNYLATTARADSYPYGIDYPTHRATGRFSNGLNMPDLISE 208
Query: 85 LLGFDDFIPPYVST--SGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVS 142
+G +P Y+S +G+ +L G NFASA GI +TG Q I + Q+Q ++
Sbjct: 209 RIGSQPTLP-YLSPELNGEALLVGANFASAGIGILNDTGIQFFNIIRITRQLQYFEQYQQ 385
Query: 143 QVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYT 202
+V ++G E++ +++ +Y + LG ND++NNYF+ F + SRQ+ Y LI Y
Sbjct: 386 RVSALIG-EEETVRLVNEALYLMTLGGNDFVNNYFLVPFSARSRQFXLPDYVVYLISEYR 562
Query: 203 EQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDG 238
+ L LY GAR++++ G +GC P L ++G
Sbjct: 563 KILARLYELGARRVLVTGTXPLGCVPXXLXXHXKNG 670
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,406,015
Number of Sequences: 36976
Number of extensions: 135679
Number of successful extensions: 824
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 701
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 706
length of query: 365
length of database: 9,014,727
effective HSP length: 97
effective length of query: 268
effective length of database: 5,428,055
effective search space: 1454718740
effective search space used: 1454718740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0404.11


