

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0400a.9
(321 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
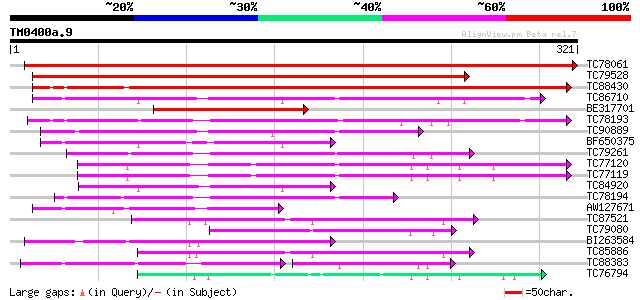
Score E
Sequences producing significant alignments: (bits) Value
TC78061 weakly similar to PIR|T10824|T10824 auxin-induced protei... 448 e-126
TC79528 similar to GP|15808674|gb|AAL06644.1 putative quinone ox... 365 e-101
TC88430 weakly similar to PIR|T10824|T10824 auxin-induced protei... 272 1e-73
TC86710 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_10... 129 2e-30
BE317701 similar to GP|14532287|gb| quinone oxidoreductase-like ... 124 5e-29
TC78193 similar to PIR|T05166|T05166 quinone reductase homolog F... 113 9e-26
TC90889 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_10... 110 6e-25
BF650375 similar to GP|15724256|gb AT4g13010/F25G13_100 {Arabido... 103 1e-22
TC79261 similar to PIR|T49047|T49047 quinone reductase-like prot... 100 6e-22
TC77120 similar to PIR|D96533|D96533 ARP protein [imported] - Ar... 94 9e-20
TC77119 similar to PIR|D96533|D96533 ARP protein [imported] - Ar... 91 6e-19
TC84920 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_10... 91 6e-19
TC78194 similar to PIR|T05166|T05166 quinone reductase homolog F... 87 9e-18
AW127671 weakly similar to GP|15724256|gb AT4g13010/F25G13_100 {... 83 1e-16
TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 75 3e-14
TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogena... 68 5e-12
BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1... 64 6e-11
TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 64 1e-10
TC88383 similar to GP|21592515|gb|AAM64465.1 nuclear receptor bi... 49 1e-08
TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbi... 55 3e-08
>TC78061 weakly similar to PIR|T10824|T10824 auxin-induced protein (clone
MII-3) - mung bean, complete
Length = 1315
Score = 448 bits (1153), Expect = e-126
Identities = 223/313 (71%), Positives = 264/313 (84%)
Frame = +3
Query: 9 IPSQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI 68
IP+ TKAW YS++G +ILK PN P LK DQVLIKV AA+LNPVD KR FK+
Sbjct: 102 IPTHTKAWFYSEHGKASDILKLHPNWSIPQLKDDQVLIKVVAASLNPVDYKRMHALFKET 281
Query: 69 DSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKV 128
D LP VPG+DVAG+V+KVGS+V K KVGDE+YGD+NE L + K +G+L+EYT AEE++
Sbjct: 282 DPHLPIVPGFDVAGIVIKVGSEVVKFKVGDEIYGDINEEGLSNLKILGTLSEYTIAEERL 461
Query: 129 LAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFE 188
LAHKP NLSFIEAAS+PLA+ TAY+GLE AQ SAGKSILVLGGAGGVGS IQLAKHV+
Sbjct: 462 LAHKPKNLSFIEAASIPLAMETAYEGLERAQLSAGKSILVLGGAGGVGSFAIQLAKHVYG 641
Query: 189 ASKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRALKAVKEGG 248
ASKIAAT+STGK+EFLRKLG DLPIDYTKENFE++ EKFDVVYD VG+ DRA+KA+KEGG
Sbjct: 642 ASKIAATSSTGKIEFLRKLGVDLPIDYTKENFEDLPEKFDVVYDGVGEVDRAMKAIKEGG 821
Query: 249 KVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEAFSYLN 308
KVVTI+PPG PPAI F+LTS G++LEKL+PY ESG++KPILD K+P PFS+ +EA+SYL
Sbjct: 822 KVVTIVPPGFPPAIFFVLTSKGSILEKLRPYFESGQLKPILDSKTPVPFSEVIEAYSYLE 1001
Query: 309 TSRATGEVVIYPI 321
TSRATG+VVIYPI
Sbjct: 1002TSRATGKVVIYPI 1040
>TC79528 similar to GP|15808674|gb|AAL06644.1 putative quinone
oxidoreductase {Fragaria x ananassa}, partial (72%)
Length = 834
Score = 365 bits (937), Expect = e-101
Identities = 182/247 (73%), Positives = 209/247 (83%)
Frame = +2
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSPLP 73
KAW YS+YG ++LKFDPNVP P +K DQVLIKV AA+LNP+D KR G FK DSPLP
Sbjct: 92 KAWTYSEYGHSVDVLKFDPNVPIPQVKDDQVLIKVAAASLNPIDYKRMEGAFKASDSPLP 271
Query: 74 TVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKP 133
T PGYDV+GVVVKVGS+VKK KVGDEVYGD+N L++PK +GSLAEYTAAEE +LAHKP
Sbjct: 272 TAPGYDVSGVVVKVGSEVKKFKVGDEVYGDINVKALEYPKVIGSLAEYTAAEEILLAHKP 451
Query: 134 SNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIA 193
NLSF EAASLPL I TAY+GLE FSAGKSILVLGGAGGVG+ VIQLAKHV+ ASK+A
Sbjct: 452 KNLSFAEAASLPLTIETAYEGLERTGFSAGKSILVLGGAGGVGTHVIQLAKHVYGASKVA 631
Query: 194 ATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRALKAVKEGGKVVTI 253
AT+ST KLE L LGADLPIDYTKENFE++ EKFDVV+D VG++++A KA+KEGGKVVTI
Sbjct: 632 ATSSTKKLELLSNLGADLPIDYTKENFEDLPEKFDVVFDTVGETEKAFKALKEGGKVVTI 811
Query: 254 LPPGTPP 260
+PPG PP
Sbjct: 812 VPPGFPP 832
>TC88430 weakly similar to PIR|T10824|T10824 auxin-induced protein (clone
MII-3) - mung bean, partial (41%)
Length = 1450
Score = 272 bits (696), Expect = 1e-73
Identities = 141/307 (45%), Positives = 205/307 (65%), Gaps = 2/307 (0%)
Frame = +2
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSPLP 73
KAW Y +YG +E+LK + P P +Q+L++V AAALNP+DSKR + S P
Sbjct: 35 KAWFYEEYGP-KEVLKLG-DFPIPSPLENQLLVQVYAAALNPIDSKRRMRPI--FPSDFP 202
Query: 74 TVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEI-TLDHPKTVGSLAEYTAAEEKVLAHK 132
VPG D+AGVV+ G VKK +GDEVYG++ + +++ PK +G+LA++ EE ++A K
Sbjct: 203 AVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARK 382
Query: 133 PSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKI 192
P LSF EAASLPLA+ TA +G +T F G+++ V+GGAGGVG+LV+QLAK +F AS +
Sbjct: 383 PKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 562
Query: 193 AATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRALKAVKEGGKVVT 252
++ ST KL+F+++ GAD +DYTK +E++ EKFD +YD VG ++ K+ G +V
Sbjct: 563 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVD 742
Query: 253 IL-PPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEAFSYLNTSR 311
I P A+ LT G +LEKL+PYLE G++K ++DPK + F ++AF Y+ T R
Sbjct: 743 ITWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGR 922
Query: 312 ATGEVVI 318
A G+VV+
Sbjct: 923 AWGKVVV 943
>TC86710 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_100
{Arabidopsis thaliana}, partial (98%)
Length = 1176
Score = 129 bits (324), Expect = 2e-30
Identities = 104/318 (32%), Positives = 150/318 (46%), Gaps = 28/318 (8%)
Frame = +3
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSP-- 71
+A YS YG LK VP P K ++VL+K+EA ++NP+D K G + I P
Sbjct: 60 QALQYSSYGGGVSGLKH-AEVPVPIPKTNEVLLKLEATSINPIDWKIQSGALRAIFLPRK 236
Query: 72 LPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAH 131
P +P DVAG VV+VG QVK K GD+V + + G LAE+ A E + A
Sbjct: 237 FPHIPCTDVAGEVVEVGPQVKDFKSGDKVIAKLTH------QYGGGLAEFAVASESLTAT 398
Query: 132 KPSNLSFIEAASLPLAIITAYQ------GLETAQFSAGKSILVLGGAGGVGSLVIQLAKH 185
+P +S EAA LP+A +TA G++ K++LV +GGVGS +QLAK
Sbjct: 399 RPPEVSAAEAAGLPIAGLTARDALTEIGGIKLDGTGEQKNVLVTAASGGVGSFAVQLAK- 575
Query: 186 VFEASKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRAL---- 241
+ + AT ++F++ LGAD +DY + YDAV +
Sbjct: 576 -LGNNHVTATCGARNIDFVKSLGADEVLDYKTPEGTSLKSPSGKKYDAVIHCTTGIPWST 752
Query: 242 --KAVKEGGKVVTILPP--------------GTPPAIPFLLTSDGAVLEKLQPYLESGKV 285
+ E G VV + P +PF++T LE L ++ GK+
Sbjct: 753 FDPNLSEKGVVVDLTPGPSSMLTFALKKTTFSKKRLVPFVVTVKREGLEHLAQLVKDGKL 932
Query: 286 KPILDPKSPFPFSQTVEA 303
K I+D K FP S+ +A
Sbjct: 933 KTIIDSK--FPLSKAEDA 980
>BE317701 similar to GP|14532287|gb| quinone oxidoreductase-like protein
{Helianthus annuus}, partial (28%)
Length = 269
Score = 124 bits (311), Expect = 5e-29
Identities = 65/88 (73%), Positives = 72/88 (80%)
Frame = +3
Query: 82 GVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEA 141
GVVVKVGS+VK KVGDEVYGDVNE L+ P GSLAEYTA EEK+LA KP+NL F +A
Sbjct: 3 GVVVKVGSEVKDFKVGDEVYGDVNEKALEGPNQFGSLAEYTAVEEKLLALKPTNLDFAQA 182
Query: 142 ASLPLAIITAYQGLETAQFSAGKSILVL 169
ASLPLAI TAY+GLE FS+GKSILVL
Sbjct: 183 ASLPLAIETAYEGLERTGFSSGKSILVL 266
>TC78193 similar to PIR|T05166|T05166 quinone reductase homolog F18E5.200 -
Arabidopsis thaliana, partial (95%)
Length = 1379
Score = 113 bits (283), Expect = 9e-26
Identities = 101/341 (29%), Positives = 165/341 (47%), Gaps = 33/341 (9%)
Frame = +2
Query: 11 SQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDS 70
++ KA V ++ G+ E+L+ V P +K D+VLIKV A ALN D+ + G +
Sbjct: 11 TKMKAIVITKPGE-PEVLQLQ-EVQDPQIKDDEVLIKVHATALNRADTLQRKGAYPPPQG 184
Query: 71 PLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLA 130
P PG + +G++ VG V K K+GD+V + G AE A E +
Sbjct: 185 ASP-YPGLECSGIIEFVGKNVSKWKIGDQVCALL---------AGGGYAEKVAVAEGQVL 334
Query: 131 HKPSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEA 189
P +S +AAS P T + + T++ S G+++L+ GG+ G+G+ IQ+AK++
Sbjct: 335 PVPPGISLKDAASFPEVACTVWSTIFMTSRLSKGETLLIHGGSSGIGTFAIQIAKYL--G 508
Query: 190 SKIAATA-STGKLEFLRKLGADLPIDYTKENF------EEVSEKFDVVYDAVGQS--DRA 240
S++ TA S KL F + +GAD+ I+Y E+F E + DV+ D +G S R
Sbjct: 509 SRVFVTAGSEEKLAFCKSIGADVGINYKTEDFVARVKEETGGQGVDVILDCMGASYYQRN 688
Query: 241 LKAVK-EG---------------------GKVVTILPPGTPPAIPFLLTSDGAVLEK-LQ 277
L ++ +G GK +T+ G P A +EK +
Sbjct: 689 LASLNFDGRLFIIGFQGGVSTELDLRALFGKRLTVQAAGLRSRSPENKAVIVAEVEKNVW 868
Query: 278 PYLESGKVKPILDPKSPFPFSQTVEAFSYLNTSRATGEVVI 318
P + GKVKP++ FP S+ EA + +S+ G++++
Sbjct: 869 PAIAEGKVKPVV--YKSFPLSEAAEAHRLMESSQHIGKILL 985
>TC90889 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_100
{Arabidopsis thaliana}, partial (69%)
Length = 779
Score = 110 bits (276), Expect = 6e-25
Identities = 81/225 (36%), Positives = 113/225 (50%), Gaps = 8/225 (3%)
Frame = +3
Query: 18 YSQY-GDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI-DSPLPTV 75
Y Y G + + ++PTP K ++VLIKVEA+++NPVD K G + + P
Sbjct: 75 YDSYDGGSSGLKHVEVHIPTP--KANEVLIKVEASSINPVDWKIQDGLLRYLLPRKFPFT 248
Query: 76 PGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSN 135
P DVAG VV GSQVK KVGD+V +N + G LAE+ A E + +PS
Sbjct: 249 PCTDVAGEVVDFGSQVKDFKVGDKVIAKLNN------QDGGGLAEFAVASESLTTLRPSE 410
Query: 136 LSFIEAASLPLA------IITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEA 189
+S E A LP+A IT G++ + K+ILV +GGVG +QLAK
Sbjct: 411 VSAAEGAGLPVAGLAAHDAITKMAGIKLDRTGEPKNILVTAASGGVGVYAVQLAK--LGN 584
Query: 190 SKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAV 234
+ + AT +E ++ LGAD +DY + YDAV
Sbjct: 585 NHVTATCGARNIELIKSLGADEVLDYKTPEGASLKSPSGRKYDAV 719
>BF650375 similar to GP|15724256|gb AT4g13010/F25G13_100 {Arabidopsis
thaliana}, partial (55%)
Length = 564
Score = 103 bits (256), Expect = 1e-22
Identities = 72/175 (41%), Positives = 102/175 (58%), Gaps = 8/175 (4%)
Frame = +1
Query: 18 YSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSP--LPTV 75
Y+ YG LK VP P K ++VLIK+EA ++NP+D K G + + P P +
Sbjct: 25 YNSYGGGASGLKH-VEVPIPTPKTNEVLIKLEAISINPLDWKIQQGVLRAMFFPRKFPHI 201
Query: 76 PGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSN 135
P DVAG VV+VG +VK KVGD+V + ++T ++ G LAE+ A E + A +PS
Sbjct: 202 PCTDVAGEVVEVGPRVKDFKVGDKV---IAKLTNEYG---GGLAEFAVASESLTAARPSE 363
Query: 136 LSFIEAASLPLAIITAYQ------GLETAQFSAGKSILVLGGAGGVGSLVIQLAK 184
+S EAA LP+A ITA+ G++ K+ILV +GGVG+ V+QLAK
Sbjct: 364 VSAAEAAGLPIAGITAHDALTKNGGIKLDGTGEQKNILVTAASGGVGAYVVQLAK 528
>TC79261 similar to PIR|T49047|T49047 quinone reductase-like protein -
Arabidopsis thaliana, partial (95%)
Length = 1239
Score = 100 bits (250), Expect = 6e-22
Identities = 77/245 (31%), Positives = 118/245 (47%), Gaps = 14/245 (5%)
Frame = +3
Query: 33 NVPTPHL-K*DQVLIKVEAAALNPVDSKRALGHFKDIDSPLPTVPGYDVAGVVVKVGSQV 91
N P P L V ++++A +LN + + LG +++ LP +PG D +G+V VGS V
Sbjct: 108 NHPIPQLDSPTSVRVRIKATSLNFANYLQILGKYQE-KPHLPFIPGSDFSGIVDSVGSNV 284
Query: 92 KKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITA 151
+VGD V +GS A+Y ++ L P + A +L +A T+
Sbjct: 285 TNFRVGDPVCSFA---------ALGSYAQYLVVDQTQLFRVPEGCDLVAAGALAVAFGTS 437
Query: 152 YQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGAD 210
+ GL AQ +G+ +LVLG AGGVG +Q+ K A IA K++ L+ +G D
Sbjct: 438 HVGLVHRAQLKSGQVLLVLGAAGGVGLAAVQIGK-ACGAIVIAVARGAEKVQLLKSMGVD 614
Query: 211 LPIDYTKENFEEVSEKF---------DVVYDAVGQS--DRALKAVKEGGKVVTI-LPPGT 258
+D EN E ++F DV+YD VG +L+ +K G ++ I G
Sbjct: 615 HVVDLGNENVTESVKEFLKVKRLKGIDVLYDPVGGKLMKESLRLLKWGANILIIGFASGE 794
Query: 259 PPAIP 263
P IP
Sbjct: 795 IPVIP 809
>TC77120 similar to PIR|D96533|D96533 ARP protein [imported] - Arabidopsis
thaliana, partial (93%)
Length = 2203
Score = 93.6 bits (231), Expect = 9e-20
Identities = 89/321 (27%), Positives = 138/321 (42%), Gaps = 41/321 (12%)
Frame = +3
Query: 39 LK*DQVLIKVEAAALNPVDSKRALGHF-----KDIDSPLPTVPGYDVAGVVVKVGSQVKK 93
+K + VL+K+ A +N D + G + K+ + LP G++ G++ VG V
Sbjct: 1059 IKPNHVLVKIIYAGVNASDVNFSSGRYFGGNNKETAARLPFDAGFEAVGIIAAVGDSVTD 1238
Query: 94 LKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQ 153
LKVG T G AE+T K P EA ++ + +TA
Sbjct: 1239 LKVGMPCAF----------MTFGGYAEFTMIPSKYALPVPRPDP--EAVAMLTSGLTASI 1382
Query: 154 GLETA-QFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTG-KLEFLRKLGADL 211
LE A Q +GK +LV AGG G +QLAK + + AT G K + L++LG D
Sbjct: 1383 ALEKAGQMESGKVVLVTAAAGGTGQFAVQLAK--LAGNTVVATCGGGAKAKLLKELGVDR 1556
Query: 212 PIDYTKENFEEVSEK-----FDVVYDAVG--QSDRALKAVKEGGKVVTI----------- 253
IDY E+ + V +K D++Y++VG L A+ G+++ I
Sbjct: 1557 VIDYHSEDIKTVLKKEFPKGIDIIYESVGGDMLKLCLDALAVHGRLIVIGMISQYQGEKG 1736
Query: 254 -LPPGTPPAIPFLLTSDGAV---------------LEKLQPYLESGKVKPILDPKSPFPF 297
P P LL+ AV L++L GK+K +DPK
Sbjct: 1737 WTPSKYPGLCEKLLSKSQAVAGFFLVQYSHMWQEHLDRLFDLYSQGKLKVAVDPKKFIGL 1916
Query: 298 SQTVEAFSYLNTSRATGEVVI 318
+A YL++ ++ G+VV+
Sbjct: 1917 HSVADAVEYLHSGKSAGKVVV 1979
Score = 28.1 bits (61), Expect = 4.7
Identities = 15/32 (46%), Positives = 20/32 (61%), Gaps = 1/32 (3%)
Frame = +3
Query: 154 GLETAQFSAGKSILVLGGAGGVG-SLVIQLAK 184
G + + AG S LV GGA G+G LV+ LA+
Sbjct: 108 GTKRMEIKAGLSALVTGGASGIGKGLVLALAE 203
>TC77119 similar to PIR|D96533|D96533 ARP protein [imported] - Arabidopsis
thaliana, partial (93%)
Length = 2277
Score = 90.9 bits (224), Expect = 6e-19
Identities = 87/321 (27%), Positives = 135/321 (41%), Gaps = 41/321 (12%)
Frame = +1
Query: 39 LK*DQVLIKVEAAALNPVDSKRALGHF-----KDIDSPLPTVPGYDVAGVVVKVGSQVKK 93
+K + VL+K+ A +N D + G + K+ + LP G++ G++ VG V
Sbjct: 1033 VKPNLVLVKIIYAGVNASDVNFSSGRYFGGNNKETTARLPFDAGFEAVGIIAAVGDSVTD 1212
Query: 94 LKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQ 153
LKVG T G AE+T K P E ++ + +TA
Sbjct: 1213 LKVGMPCAF----------MTFGGYAEFTMIPSKYALPVPRPDP--EGVAMLTSGLTASI 1356
Query: 154 GLETA-QFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTG-KLEFLRKLGADL 211
LE A Q +GK +LV AGG G +QLAK + + AT G K + L++LG D
Sbjct: 1357 ALEKAGQMESGKVVLVTAAAGGTGQFAVQLAK--LAGNTVVATCGGGTKAKLLKELGVDR 1530
Query: 212 PIDYTKENFEEVSEK-----FDVVYDAVG--QSDRALKAVKEGGKVVTI----------- 253
IDY E+ + V K D++Y++VG L A+ G+++ I
Sbjct: 1531 VIDYNSEDIKTVLRKEFPKGIDIIYESVGGDMLKLCLDALAVHGRLIVIGMISQYQGEHG 1710
Query: 254 -LPPGTPPAIPFLLTSDGAV---------------LEKLQPYLESGKVKPILDPKSPFPF 297
P P + LL V L++L GK+K +DPK
Sbjct: 1711 WTPSKYPGLLEKLLAKSQTVAGFFLVQYSHFYQEHLDRLFDLYSKGKLKVAVDPKKFIGL 1890
Query: 298 SQTVEAFSYLNTSRATGEVVI 318
+A YL++ ++ G+VV+
Sbjct: 1891 HSVADAVEYLHSGKSVGKVVV 1953
>TC84920 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_100
{Arabidopsis thaliana}, partial (54%)
Length = 563
Score = 90.9 bits (224), Expect = 6e-19
Identities = 61/153 (39%), Positives = 83/153 (53%), Gaps = 8/153 (5%)
Frame = +2
Query: 40 K*DQVLIKVEAAALNPVDSKRALGHFKDIDSP--LPTVPGYDVAGVVVKVGSQVKKLKVG 97
K ++VLIK+EA +NP D K G + I P P P DVAG VV++G QVK KVG
Sbjct: 71 KTNEVLIKLEAIRINPADWKIQTGVLRAIFFPRKFPHTPCTDVAGEVVEIGEQVKDFKVG 250
Query: 98 DEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQ---- 153
D+V +N + G LAE+ A E + A +PS +S EAA LP+A +TA
Sbjct: 251 DKVIAKLNNL------YGGGLAEFAVARESLTAARPSEVSAAEAAGLPIAGLTARDALTE 412
Query: 154 --GLETAQFSAGKSILVLGGAGGVGSLVIQLAK 184
G++ K++LV +GGV +QLAK
Sbjct: 413 IGGVKLDGTGEPKNVLVTAASGGVXVYAVQLAK 511
>TC78194 similar to PIR|T05166|T05166 quinone reductase homolog F18E5.200 -
Arabidopsis thaliana, partial (60%)
Length = 709
Score = 87.0 bits (214), Expect = 9e-18
Identities = 65/197 (32%), Positives = 100/197 (49%), Gaps = 2/197 (1%)
Frame = +1
Query: 26 EILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSPLPTVPGYDVAGVVV 85
E+L+ V P K ++VLI+V A ALN D+ + G F I PG + +G +V
Sbjct: 157 EVLQLQ-EVQDPQPKDNEVLIRVHATALNSADTVQRKG-FYPIPQGASPYPGLECSGTIV 330
Query: 86 KVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLP 145
VG V K K+GD+V + G AE A E + P +S +AAS
Sbjct: 331 CVGINVSKWKIGDQVCALL---------AGGGYAEKVAVPEGQVLPVPPGISLKDAASFT 483
Query: 146 LAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATA-STGKLEF 203
T + + T++ S G+++LV GG+ G+G+ I +AK ++ SK+ TA S KL F
Sbjct: 484 EVACTVWSTIFMTSRLSKGETLLVHGGSSGIGTFAIXIAK--YQGSKVFVTAGSEEKLAF 657
Query: 204 LRKLGADLPIDYTKENF 220
+ +GAD+ +Y +F
Sbjct: 658 CKSIGADVGXNYXTXDF 708
>AW127671 weakly similar to GP|15724256|gb AT4g13010/F25G13_100 {Arabidopsis
thaliana}, partial (48%)
Length = 493
Score = 83.2 bits (204), Expect = 1e-16
Identities = 60/145 (41%), Positives = 77/145 (52%), Gaps = 3/145 (2%)
Frame = +2
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVD---SKRALGHFKDIDS 70
+A Y+ YG LK VP P D+VLIK+EAA++NP D KR L F +
Sbjct: 26 RAVQYNAYGGGSTGLKH-VEVPIPSPSKDEVLIKLEAASINPFDWKVQKRMLWPF--LPP 196
Query: 71 PLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLA 130
P +P DVAG V+ VG VKK K GD+V G V+ P + G LAE+ +E V A
Sbjct: 197 KFPYIPCTDVAGEVMMVGKGVKKFKTGDKVVGLVS------PFSGGGLAEFAVVKESVTA 358
Query: 131 HKPSNLSFIEAASLPLAIITAYQGL 155
P +S E LP+A +TA Q L
Sbjct: 359 SIPQEISASECVGLPVAGLTALQAL 433
>TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (98%)
Length = 1433
Score = 75.1 bits (183), Expect = 3e-14
Identities = 65/228 (28%), Positives = 97/228 (42%), Gaps = 32/228 (14%)
Frame = +2
Query: 70 SPLPTVPGYDVAGVVVKVGSQVKKLKVGDEV---------------------YGDVNEIT 108
S P VPG+++AG+V +VGS+V+K K+GD+V Y T
Sbjct: 257 STYPLVPGHEIAGIVTEVGSKVEKFKIGDKVGVGCLVDSCRACQNCEENLENYCPKQTNT 436
Query: 109 L-----DHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAG 163
D T G ++ A+E + H P L AA L A IT Y L F
Sbjct: 437 YSAKYSDGSITYGGYSDSMVADEHFIVHIPDGLPLESAAPLLCAGITVYSPLR--YFGLD 610
Query: 164 KSILVLG--GAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGAD-LPIDYTKENF 220
K + +G G GG+G L ++ AK + +T+ + E + LGAD I + ++
Sbjct: 611 KPGMNIGIVGLGGLGHLGVKFAKAFGANVTVISTSPNKEKEAIENLGADSFLISHDQDKM 790
Query: 221 EEVSEKFDVVYDAVGQSDRALKAV---KEGGKVVTILPPGTPPAIPFL 265
+ D + D V L V K GK+V + P PP +P +
Sbjct: 791 QAAMGTLDGIIDTVSADHPLLPLVGLLKYHGKLVMVGAPDKPPELPHI 934
>TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogenase-like
protein {Arabidopsis thaliana}, partial (96%)
Length = 1524
Score = 67.8 bits (164), Expect = 5e-12
Identities = 50/150 (33%), Positives = 79/150 (52%), Gaps = 10/150 (6%)
Frame = +3
Query: 114 TVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGA 172
++G LAEY LA P+++ + E+A L A+ TAY + A+ G ++ V+G
Sbjct: 651 SMGGLAEYCVVPANALAVLPNSMPYTESAILGCAVFTAYGAMAHAAEVRPGDTVAVIG-T 827
Query: 173 GGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGADLPIDYTKEN-FEEVSE-----K 226
GGVGS +Q+A+ + IA KLE + LGA I+ KE+ E++ E
Sbjct: 828 GGVGSSCLQIARAFGASDIIAVDVQDEKLEKAKTLGATHTINSAKEDPIEKILEITGGKG 1007
Query: 227 FDVVYDAVGQSD---RALKAVKEGGKVVTI 253
DV +A+G+ + ++VK+GGK V I
Sbjct: 1008VDVAVEALGRPQTFAQCTQSVKDGGKAVMI 1097
>BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1.1.1.1).
{Neurospora crassa}, partial (57%)
Length = 682
Score = 64.3 bits (155), Expect = 6e-11
Identities = 55/195 (28%), Positives = 87/195 (44%), Gaps = 19/195 (9%)
Frame = +2
Query: 9 IPSQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI 68
IP + A V+ + E + P P D+VL+ V+ + + D G + +
Sbjct: 77 IPEKQWAQVFEKTAGPIEYKQIPVQKPGP----DEVLVNVKFSGVCHTDLHAWQGDWP-L 241
Query: 69 DSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEV----------------YGDVN---EITL 109
D+ LP V G++ AGVVV G VK +K+G++V D + E L
Sbjct: 242 DTKLPLVGGHEGAGVVVARGDLVKDVKIGEKVGIKWLNGSCLSCSYCQNADESLCAEALL 421
Query: 110 DHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVL 169
GS +Y A+ +A P + + A IT Y+GL+ + AG+SI ++
Sbjct: 422 SGYTVDGSFQQYAIAKAIHVARIPEECDLESISPILCAGITVYKGLKESGVKAGQSIAIV 601
Query: 170 GGAGGVGSLVIQLAK 184
G GG+GS+ Q K
Sbjct: 602 GAGGGLGSIAXQYCK 646
>TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (94%)
Length = 1395
Score = 63.5 bits (153), Expect = 1e-10
Identities = 58/223 (26%), Positives = 93/223 (41%), Gaps = 32/223 (14%)
Frame = +2
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEV---------------YGDV-----------NE 106
P VPG+++ GVV +VGS+VKK KVGD+V D+ N
Sbjct: 269 PLVPGHELTGVVTEVGSKVKKFKVGDKVGVGYMVDSCRSCENCADDIENYCTKYTQTFNG 448
Query: 107 ITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSI 166
+ D T G ++ A+E + P +L A L A +T Y L F K
Sbjct: 449 KSRDGTITYGGFSDSMVADEHFVIRIPDSLPLDGAGPLLCAGVTVYSPLR--HFGLDKPG 622
Query: 167 LVLG--GAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGAD-LPIDYTKENFEEV 223
+ +G G GG+G + ++ AK + +T+ + E + LGAD + E +
Sbjct: 623 MNIGVVGLGGLGHMAVKFAKAFGAKVTVISTSPKKEKEAIEHLGADSFLVSRDPEQMQAA 802
Query: 224 SEKFDVVYDAVGQSDRALKAV---KEGGKVVTILPPGTPPAIP 263
+ + + D V S + + K GK+V + P +P
Sbjct: 803 TSTLNGIIDTVSASHPVVPLIGLLKSNGKLVMVGAVAKPLELP 931
>TC88383 similar to GP|21592515|gb|AAM64465.1 nuclear receptor binding
factor-like protein {Arabidopsis thaliana}, partial
(90%)
Length = 1313
Score = 48.9 bits (115), Expect(2) = 1e-08
Identities = 40/150 (26%), Positives = 66/150 (43%)
Frame = +3
Query: 7 SNIPSQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFK 66
S + +KA +Y +G + + K ++P +K + + +K+ AA +NP D R G +
Sbjct: 105 SAVSPPSKAIIYESHGQPDAVTKL-VDIPATEVKENDLCVKMLAAPINPSDINRIQGVYP 281
Query: 67 DIDSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEE 126
P P V GY+ G V VGS V GD V + P + G+ Y ++
Sbjct: 282 VRPEP-PAVGGYEGVGEVHSVGSAVTCFSPGDWV--------IPSPPSFGTWQTYIVKDQ 434
Query: 127 KVLAHKPSNLSFIEAASLPLAIITAYQGLE 156
V + AA++ + +TA LE
Sbjct: 435 NVWHKVNKGVPMEYAATITVNPLTALLMLE 524
Score = 27.3 bits (59), Expect(2) = 1e-08
Identities = 28/103 (27%), Positives = 47/103 (45%), Gaps = 11/103 (10%)
Frame = +1
Query: 161 SAGKSILVLGGAGGVGSLVIQLAKH--VFEASKIAATASTGKL-EFLRKLGADLPIDYTK 217
++G +I+ G VG VIQLAK + + I G++ E L+ LGAD ++
Sbjct: 541 NSGDAIVQNGATSMVGQCVIQLAKSRGIHNINIIRDRPGVGEVKERLKDLGADEVFTESE 720
Query: 218 ENFEEVSEKFDVV------YDAVG--QSDRALKAVKEGGKVVT 252
+ V + ++ VG + LK ++ GG +VT
Sbjct: 721 LEVKNVKSLLGGIPEPALGFNCVGGNSASLVLKFLRRGGTMVT 849
>TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbitol
dehydrogenase {Prunus persica}, partial (93%)
Length = 1691
Score = 55.5 bits (132), Expect = 3e-08
Identities = 77/280 (27%), Positives = 112/280 (39%), Gaps = 48/280 (17%)
Frame = +2
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGD--VNEITLDH-----------------PK 113
P V G++ AG++ +VGSQVK L GD V + ++ DH P
Sbjct: 428 PMVIGHECAGIIEEVGSQVKTLVPGDRVAIEPGISCWRCDHCKLGRYNLCPDMKFFATPP 607
Query: 114 TVGSLAEYTAAEEKVLAHKPSNLSFIEAASL-PLAIITAYQGLETAQFSAGKSILVLGGA 172
GSLA + P N+S E A PL++ A ++L++ GA
Sbjct: 608 VHGSLANQIVHPADLCFKLPENVSLEEGAMCEPLSV--GVHACRRANIGPETNVLIM-GA 778
Query: 173 GGVGSLVIQLAKHVFEASKIAAT-ASTGKLEFLRKLGADLPIDYTKENFEEVSEK----- 226
G +G LV L+ F A +I +L + LGAD I N ++V+E+
Sbjct: 779 GPIG-LVTMLSARAFGAPRIVVVDVDDHRLSVAKSLGAD-DIVKVSTNIQDVAEEVKQIH 952
Query: 227 ------FDVVYDAVG---QSDRALKAVKEGGKVVTI-------LPPGTPPAIPFLLTSDG 270
DV +D G AL A + GGKV + P TP A +
Sbjct: 953 NVLGAGVDVTFDCAGFNKTMTTALTATQPGGKVCLVGMGHSEMTVPLTPAAAREVDVVGI 1132
Query: 271 AVLEKLQP----YLESGK--VKPILDPKSPFPFSQTVEAF 304
+ P +L SGK VKP++ + F + EAF
Sbjct: 1133FRYKNTWPLCLEFLRSGKIDVKPLITHRFGFSQKEVEEAF 1252
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,719,489
Number of Sequences: 36976
Number of extensions: 90442
Number of successful extensions: 464
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 430
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 440
length of query: 321
length of database: 9,014,727
effective HSP length: 96
effective length of query: 225
effective length of database: 5,465,031
effective search space: 1229631975
effective search space used: 1229631975
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0400a.9


