

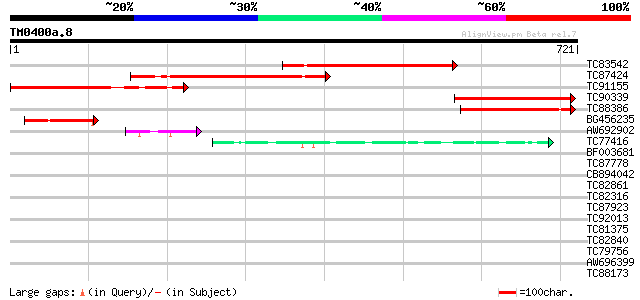
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0400a.8
(721 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83542 similar to GP|19387245|gb|AAL87157.1 putative kinesin li... 375 e-104
TC87424 similar to GP|19699309|gb|AAL91265.1 AT4g10840/F25I24_50... 272 3e-73
TC91155 similar to GP|9294312|dbj|BAB01483.1 contains similarity... 247 1e-65
TC90339 similar to GP|19387245|gb|AAL87157.1 putative kinesin li... 216 2e-56
TC88386 similar to GP|19699309|gb|AAL91265.1 AT4g10840/F25I24_50... 198 5e-51
BG456235 weakly similar to GP|9802538|gb|A F17L21.29 {Arabidopsi... 92 7e-19
AW692902 similar to GP|19699309|gb| AT4g10840/F25I24_50 {Arabido... 62 6e-10
TC77416 similar to GP|18139887|gb|AAL60196.1 O-linked N-acetyl g... 55 7e-08
BF003681 similar to GP|17380952|gb putative kinesin light chain ... 40 0.004
TC87778 similar to GP|5103827|gb|AAD39657.1| ESTs gb|F20110 and ... 38 0.012
CB894042 similar to PIR|T49093|T4 hypothetical protein F4F15.250... 38 0.016
TC82861 similar to GP|6630450|gb|AAF19538.1| F23N19.10 {Arabidop... 32 0.66
TC82316 similar to GP|19807751|gb|AAL99323.1 hypothetical protei... 32 0.66
TC87923 85p protein 31 1.9
TC92013 homologue to GP|11595557|emb|CAC18142. related to c-modu... 31 1.9
TC81375 weakly similar to GP|21537367|gb|AAM61708.1 unknown {Ara... 30 3.3
TC82840 similar to GP|2073450|emb|CAA73365.1 Krm protein {Lotus ... 29 5.6
TC79756 29 5.6
AW696399 homologue to PIR|T06370|T0 probable DNA (cytosine-5-)-m... 29 7.3
TC88173 similar to GP|4512677|gb|AAD21731.1| unknown protein {Ar... 29 7.3
>TC83542 similar to GP|19387245|gb|AAL87157.1 putative kinesin light chain
gene {Oryza sativa (japonica cultivar-group)}, partial
(30%)
Length = 665
Score = 375 bits (963), Expect = e-104
Identities = 191/223 (85%), Positives = 205/223 (91%)
Frame = +1
Query: 347 VAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALE 406
+AEAHVQALQFDEA++LCQMAL+IH+GNG AS EE+ADRRLMGLI DSKGDYE+ALE
Sbjct: 1 LAEAHVQALQFDEAKKLCQMALDIHKGNG--LPASFEEAADRRLMGLICDSKGDYESALE 174
Query: 407 HYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNV 466
HYVLAS+ M+ NGQE DVASVD SIGDAY +LARYDEA+FSYQKALTVFKSTKGENHP V
Sbjct: 175 HYVLASVVMAENGQELDVASVDCSIGDAYSSLARYDEAIFSYQKALTVFKSTKGENHPTV 354
Query: 467 ASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDL 526
ASV+VRLADLYNKIGKFKDSK+YCENALRIFGK KPGI EEIANGLIDVAAIYQSMNDL
Sbjct: 355 ASVFVRLADLYNKIGKFKDSKTYCENALRIFGKKKPGISLEEIANGLIDVAAIYQSMNDL 534
Query: 527 EKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSD 569
EKGLKLLKKALKIY + PGQ STIAGIEAQMGVMYYMLGNYSD
Sbjct: 535 EKGLKLLKKALKIYNNVPGQHSTIAGIEAQMGVMYYMLGNYSD 663
>TC87424 similar to GP|19699309|gb|AAL91265.1 AT4g10840/F25I24_50
{Arabidopsis thaliana}, partial (32%)
Length = 1014
Score = 272 bits (696), Expect = 3e-73
Identities = 143/255 (56%), Positives = 183/255 (71%), Gaps = 1/255 (0%)
Frame = +2
Query: 154 SAKSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIEERIATGLDNPDLGP 213
S+K+ +S ++P + S RS +K S P ++ E LDNPDLGP
Sbjct: 248 SSKTPTQSPLPNKKPPSPSPSSRSKKKTSETP-------TTNSLLSE---ASLDNPDLGP 397
Query: 214 FLLKQTRDMISSGENPQKALDLALRALKSFEMCA-DGKPSLELVMCLHVLATIYCNLGQY 272
FLLK RD I+SG+ P KALD A+RA KSFE CA +G+PSL+L M LHVLA IYC+L ++
Sbjct: 398 FLLKVARDTIASGDGPAKALDYAIRASKSFERCAVEGEPSLDLAMSLHVLAAIYCSLNRF 577
Query: 273 NEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQV 332
EA+P+LER+I +P +E G DHALA F+G MQLGDT++M+G ++ SIL Y GL+IQ Q
Sbjct: 578 EEAVPVLERAIHVPDVERGADHALAAFSGYMQLGDTFSMLGQVDKSILMYDQGLQIQIQT 757
Query: 333 LGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMG 392
LGETDPR GETCRY+AEA+VQA+QFD+AE LC LEIHR + AS+EE+ADRRLM
Sbjct: 758 LGETDPRVGETCRYLAEANVQAMQFDKAEELCNKTLEIHR--AHSEPASLEEAADRRLMA 931
Query: 393 LIYDSKGDYEAALEH 407
LI ++KGDYE+ALEH
Sbjct: 932 LICEAKGDYESALEH 976
>TC91155 similar to GP|9294312|dbj|BAB01483.1 contains similarity to kinesin
light chain~gene_id:K24A2.5 {Arabidopsis thaliana},
partial (8%)
Length = 746
Score = 247 bits (631), Expect = 1e-65
Identities = 156/234 (66%), Positives = 171/234 (72%), Gaps = 7/234 (2%)
Frame = +1
Query: 1 MPERAMDELNMNIAGEGPSGSYTPHKDNFN-QQASPRSTLSPRSIQSSDSIDLAIDGVVD 59
MP AM+E NMN A E S SYTPHKDNF QQASPRSTLSPRSIQS DSIDLAIDGVVD
Sbjct: 124 MPTIAMEEFNMNNATEEASESYTPHKDNFTAQQASPRSTLSPRSIQS-DSIDLAIDGVVD 300
Query: 60 TSIEQLYYNVCEMRSSDQSPSRASFYSYGEESRIDSELGHLVG--GILEITKEVV-TENK 116
TSIEQLY NV EMRSSDQSPSRAS YSYGEESRIDSELGHLVG G LEI KE+V TENK
Sbjct: 301 TSIEQLYTNVYEMRSSDQSPSRASLYSYGEESRIDSELGHLVGYIGDLEIRKEIVTTENK 480
Query: 117 EESNGNAAEKDIVSCGKEATKKDNNQSHSPSSRIIEGSAKSTPKSRNLRERPSTDKRSER 176
EE N +A EK IV S S+RI EGS+KS KSR+L KR+E+
Sbjct: 481 EEPNVDAIEKVIV---------------SSSTRIGEGSSKSAAKSRSLH------KRNEK 597
Query: 177 SSRKGSLY--PMRKHRSLALR-GIIEERIATGLDNPDLGPFLLKQTRDMISSGE 227
+RKG+ + MRKH+ L L+ GI++E LDNPDLGPFLLK TRDMISSGE
Sbjct: 598 GTRKGNGFYNMMRKHKRLGLKDGIVDE-----LDNPDLGPFLLKNTRDMISSGE 744
>TC90339 similar to GP|19387245|gb|AAL87157.1 putative kinesin light chain
gene {Oryza sativa (japonica cultivar-group)}, partial
(19%)
Length = 779
Score = 216 bits (550), Expect = 2e-56
Identities = 109/154 (70%), Positives = 128/154 (82%)
Frame = +2
Query: 566 NYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEK 625
NY++SYN K A+AK RA GEKKS+ FGIALNQ+GLACVQRYA+ EA +LFEEA+++LE
Sbjct: 2 NYTESYNTLKKAIAKLRAIGEKKSSFFGIALNQLGLACVQRYALREAIELFEEAKSVLEH 181
Query: 626 EYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAE 685
E G YHP+TLGVYSNLAGTYDA+GR+DDAI LE VV MREEKLGTANPDVDDEKRRL+E
Sbjct: 182 ELGPYHPETLGVYSNLAGTYDAIGRLDDAILTLEHVVSMREEKLGTANPDVDDEKRRLSE 361
Query: 686 LLKEAGRGRNRKSKRSLETLLDANSRLIKNNSIK 719
LLKEAG+ R+RK RSLE L D ++ + N IK
Sbjct: 362 LLKEAGKVRSRK-VRSLENLFDGDAHTLNNLVIK 460
>TC88386 similar to GP|19699309|gb|AAL91265.1 AT4g10840/F25I24_50
{Arabidopsis thaliana}, partial (21%)
Length = 736
Score = 198 bits (504), Expect = 5e-51
Identities = 104/146 (71%), Positives = 120/146 (81%)
Frame = +2
Query: 574 FKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPD 633
F+SAV K R SGE+KSA FG+ LNQMGLACVQ + I+EAA+LFEEAR ILE+E G H D
Sbjct: 14 FESAVLKLRTSGERKSAFFGVVLNQMGLACVQLFKIDEAAELFEEARGILEQECGPCHQD 193
Query: 634 TLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGRG 693
TLGVYSNLA TYDAMGRV DAIEILE+V+ +REEKLG ANPD +DEKRRLAELLKEAG+
Sbjct: 194 TLGVYSNLAATYDAMGRVGDAIEILEYVLKLREEKLGIANPDFEDEKRRLAELLKEAGKT 373
Query: 694 RNRKSKRSLETLLDANSRLIKNNSIK 719
R+RK+K SLE L+D NS+ K S K
Sbjct: 374 RDRKAK-SLENLIDPNSKRTKKESNK 448
>BG456235 weakly similar to GP|9802538|gb|A F17L21.29 {Arabidopsis thaliana},
partial (7%)
Length = 538
Score = 92.0 bits (227), Expect = 7e-19
Identities = 55/97 (56%), Positives = 68/97 (69%), Gaps = 3/97 (3%)
Frame = +3
Query: 19 SGSYTPHKDNFNQQASPRSTLSPRSIQSSDSIDLAIDGVVDTSIEQLYYNVCEMRSSDQS 78
+G+ +P K + SPRS++SP+ QS + DGVV+ SIEQLY NVC+M+SSDQS
Sbjct: 129 NGNRSPTKASLTPIKSPRSSMSPQRRQSVGP-NFQDDGVVEPSIEQLYENVCDMQSSDQS 305
Query: 79 PSRASFYSYGEESRIDSELGHLVGG---ILEITKEVV 112
PSR SF S G+ESRIDSEL HLVGG LEI +E V
Sbjct: 306 PSRQSFGSDGDESRIDSELHHLVGGRMRELEIMEEEV 416
>AW692902 similar to GP|19699309|gb| AT4g10840/F25I24_50 {Arabidopsis
thaliana}, partial (5%)
Length = 601
Score = 62.4 bits (150), Expect = 6e-10
Identities = 39/103 (37%), Positives = 60/103 (57%), Gaps = 7/103 (6%)
Frame = +3
Query: 148 SRIIEGSAKSTPKSRN----LRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIEERIA 203
S + E + K+ P+ ++ LRE P + + + R ++S A+R I++ I
Sbjct: 273 STMTELTVKTPPQMQSFGVFLREPPQNESTDDDNHR---------NQSPAMRKTIKKTII 425
Query: 204 ---TGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSF 243
+ L+NPDLGPFLLK RD I+SGENP+KALD A+ ++ F
Sbjct: 426 IDESSLENPDLGPFLLKMARDTIASGENPEKALDYAIPGIQVF 554
>TC77416 similar to GP|18139887|gb|AAL60196.1 O-linked N-acetyl glucosamine
transferase {Arabidopsis thaliana}, partial (94%)
Length = 3465
Score = 55.5 bits (132), Expect = 7e-08
Identities = 96/459 (20%), Positives = 173/459 (36%), Gaps = 26/459 (5%)
Frame = +2
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCM-QLGDTYAMMGNIEN 317
L +L IY L ++ + E ++ I + H FA C + + + GNI+
Sbjct: 500 LLLLGAIYYQLHDFDMCVAKNEEALRI------EPH----FAECYGNMANAWKEKGNIDL 649
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEI------- 370
+I +Y +E++ P F + +A A+++ + EA + C+ AL I
Sbjct: 650 AIRYYLIAIELR--------PNFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDA 805
Query: 371 HRGNGNGSSAS--VEES----------------ADRRLMGLIYDSKGDYEAALEHYVLAS 412
H GN A V+E+ A L GL +S GD+ AL++Y
Sbjct: 806 HSNLGNLMKAQGLVQEAYSCYLEALRIQPTFAIAWSNLAGLFMES-GDFNRALQYY---- 970
Query: 413 MAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVR 472
A + ++G+ Y AL EA+ YQ AL + PN Y
Sbjct: 971 --KEAVKLKPSFPDAYLNLGNVYKALGMPQEAIACYQHAL--------QTRPNYGMAYGN 1120
Query: 473 LADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKL 532
LA ++ + G+ + + + A+ + + N L DV + E+ ++
Sbjct: 1121LASIHYEQGQLDMAILHYKQAIACDPRFLEAY--NNLGNALKDVGRV-------EEAIQC 1273
Query: 533 LKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALF 592
+ L + + P + LGN +N+ +A + ++A+ + L
Sbjct: 1274YNQCLSLQPNHPQALTN--------------LGNIYMEWNMVAAAASYYKATLNVTTGL- 1408
Query: 593 GIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVD 652
N + + Q+ +A + E I P N TY +GRV
Sbjct: 1409SAPYNNLAIIYKQQGNYADAISCYNEVLRI--------DPLAADGLVNRGNTYKEIGRVS 1564
Query: 653 DAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAG 691
DAI+ +++ + T P + + LA K++G
Sbjct: 1565DAIQ--DYIRAI------TVRPTMAEAHANLASAYKDSG 1657
>BF003681 similar to GP|17380952|gb putative kinesin light chain {Arabidopsis
thaliana}, partial (23%)
Length = 462
Score = 39.7 bits (91), Expect = 0.004
Identities = 38/146 (26%), Positives = 66/146 (45%), Gaps = 5/146 (3%)
Frame = +2
Query: 210 DLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVLATIYCNL 269
+LG LK + GE+P+KAL+ A RALK FE + K S M L ++ ++ L
Sbjct: 26 ELGLAALKIGLKLDREGEDPEKALEFANRALKVFE--NEKKDSFPYAMSLQLMGSLNYGL 199
Query: 270 GQYNEAIPILERSIDI--PVLEDG---QDHALAKFAGCMQLGDTYAMMGNIENSILFYTA 324
++++++ L ++ + + +G D A +L + MG E ++
Sbjct: 200 NRFSDSLGYLNKANRVLGRLENEGVCVDDIRPVLHAVQFELANVKTAMGRREEALENLRK 379
Query: 325 GLEIQGQVLGETDPRFGETCRYVAEA 350
LEI+ E G+ +AEA
Sbjct: 380 CLEIKEMTFDEDSEELGKGNMDLAEA 457
>TC87778 similar to GP|5103827|gb|AAD39657.1| ESTs gb|F20110 and gb|F20109
come from this gene. {Arabidopsis thaliana}, partial
(24%)
Length = 1350
Score = 38.1 bits (87), Expect = 0.012
Identities = 31/129 (24%), Positives = 55/129 (42%)
Frame = +2
Query: 410 LASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASV 469
LA M ++ AS S + +++A QKAL + + G +HP+
Sbjct: 557 LAKMMTVCGPYHRNTASAYSLLAVVLYHTGDFNQATIYQQKALDINERELGLDHPDTMKS 736
Query: 470 YVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKG 529
Y L+ Y ++ + + Y AL + G+ A I+VA + + M ++
Sbjct: 737 YGDLSVFYYRLQHIELALKYVNRALFLL-HFTCGLSHPNTAATYINVAMMEEGMGNVHVA 913
Query: 530 LKLLKKALK 538
L+ L +ALK
Sbjct: 914 LRYLHEALK 940
>CB894042 similar to PIR|T49093|T4 hypothetical protein F4F15.250 -
Arabidopsis thaliana, partial (14%)
Length = 653
Score = 37.7 bits (86), Expect = 0.016
Identities = 15/46 (32%), Positives = 28/46 (60%)
Frame = +2
Query: 450 KALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALR 495
+AL + + G +HP+VA+ ++ +A +Y IGK + Y + AL+
Sbjct: 8 RALLLLSLSSGPDHPDVAATFINVAMMYQDIGKMNTALRYLQEALK 145
Score = 33.9 bits (76), Expect = 0.23
Identities = 22/80 (27%), Positives = 38/80 (47%)
Frame = +2
Query: 410 LASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASV 469
L +++S+ DVA+ ++ Y + + + A+ Q+AL + GE H A
Sbjct: 14 LLLLSLSSGPDHPDVAATFINVAMMYQDIGKMNTALRYLQEALKKNERLLGEEHIQTAVC 193
Query: 470 YVRLADLYNKIGKFKDSKSY 489
Y LA +N +G FK S +
Sbjct: 194 YHALAIAFNCMGAFKLSHQH 253
Score = 29.6 bits (65), Expect = 4.3
Identities = 22/93 (23%), Positives = 42/93 (44%)
Frame = +2
Query: 508 EIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNY 567
++A I+VA +YQ + + L+ L++ALK G++ Q V Y+ L
Sbjct: 53 DVAATFINVAMMYQDIGKMNTALRYLQEALKKNERLLGEE------HIQTAVCYHALA-- 208
Query: 568 SDSYNIFKSAVAKFRASGEKKSALFGIALNQMG 600
I + + F+ S + + + I + Q+G
Sbjct: 209 -----IAFNCMGAFKLSHQHEKKTYDILVKQLG 292
>TC82861 similar to GP|6630450|gb|AAF19538.1| F23N19.10 {Arabidopsis
thaliana}, partial (29%)
Length = 750
Score = 32.3 bits (72), Expect = 0.66
Identities = 19/63 (30%), Positives = 37/63 (58%), Gaps = 1/63 (1%)
Frame = +2
Query: 393 LIYDSKGDYEAALEHYVLA-SMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKA 451
++Y ++ A+L++Y A + A + D + S +G A+L L++YD+AV +Y+K
Sbjct: 191 VLYSNRSAAYASLQNYTDALTDAKKTVELKPDWSKGYSRLGAAHLGLSQYDDAVSAYKKG 370
Query: 452 LTV 454
L +
Sbjct: 371 LEI 379
>TC82316 similar to GP|19807751|gb|AAL99323.1 hypothetical protein
{Dictyostelium discoideum}, partial (1%)
Length = 632
Score = 32.3 bits (72), Expect = 0.66
Identities = 29/116 (25%), Positives = 48/116 (41%), Gaps = 12/116 (10%)
Frame = +1
Query: 63 EQLYYNVCEMRSSDQSPSRASFYSYGEESRIDSELGHLVGGILEITKEVVTENKEESNGN 122
E+ ++C++ +S SP R F +YG++ D + +L EE N +
Sbjct: 100 EEETLSLCDLPNSTISPQRGDFSNYGKDHHDDDDDDNLFEFF-----------SEEFNTS 246
Query: 123 AAEKDIVSCGKEATKKD-----NNQ-------SHSPSSRIIEGSAKSTPKSRNLRE 166
+I+ CGK KD +NQ S+S + + GS + RN E
Sbjct: 247 TIHDNIIFCGKLIPFKDHQYVPHNQKNCAKPTSNSKAMKSSNGSIANLKSKRNEEE 414
>TC87923 85p protein
Length = 2793
Score = 30.8 bits (68), Expect = 1.9
Identities = 12/32 (37%), Positives = 20/32 (62%)
Frame = +2
Query: 491 ENALRIFGKSKPGIPPEEIANGLIDVAAIYQS 522
ENA + K KP + +ANG++++ A+Y S
Sbjct: 365 ENANPLENKEKPSVYRRALANGIVEILAVYSS 460
>TC92013 homologue to GP|11595557|emb|CAC18142. related to c-module-binding
factor {Neurospora crassa}, partial (2%)
Length = 1437
Score = 30.8 bits (68), Expect = 1.9
Identities = 15/48 (31%), Positives = 24/48 (49%)
Frame = -2
Query: 2 PERAMDELNMNIAGEGPSGSYTPHKDNFNQQASPRSTLSPRSIQSSDS 49
P A E +++ P S + H+D+ N++ SP S+ SSDS
Sbjct: 599 PTEAAKEATPDLSPPAPRASRSLHRDDANEEEVIAPAASPSSVSSSDS 456
>TC81375 weakly similar to GP|21537367|gb|AAM61708.1 unknown {Arabidopsis
thaliana}, partial (30%)
Length = 797
Score = 30.0 bits (66), Expect = 3.3
Identities = 26/83 (31%), Positives = 40/83 (47%)
Frame = +1
Query: 627 YGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAEL 686
Y ++HPD L V++ LA T G++ A E+L+ + R ++ + + LAE
Sbjct: 37 YLEWHPDILTVWA-LAKT----GQISKAEELLKSLK-KRISRMTKKKQQIMQKGMVLAEA 198
Query: 687 LKEAGRGRNRKSKRSLETLLDAN 709
L GRG N+ L DAN
Sbjct: 199 LYAYGRGNNKHGVELLGPDFDAN 267
>TC82840 similar to GP|2073450|emb|CAA73365.1 Krm protein {Lotus japonicus},
partial (83%)
Length = 719
Score = 29.3 bits (64), Expect = 5.6
Identities = 13/49 (26%), Positives = 25/49 (50%)
Frame = +1
Query: 313 GNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAE 361
G ++++ + A ++ + GE DP +C +AE + FD+AE
Sbjct: 418 GKLDDAERLFVAAIKEAKEGFGEQDPHVASSCNNLAELYRVKKAFDKAE 564
>TC79756
Length = 765
Score = 29.3 bits (64), Expect = 5.6
Identities = 21/62 (33%), Positives = 33/62 (52%)
Frame = +1
Query: 230 QKALDLALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLE 289
QK LD A++AL + A KP ++ + L N G NEA + +R I++ +L+
Sbjct: 118 QKKLDSAIQAL-DLMVKAQCKPDGKIY---YTLLKSVANEGMVNEANDLHQRLIELKILK 285
Query: 290 DG 291
DG
Sbjct: 286 DG 291
>AW696399 homologue to PIR|T06370|T0 probable DNA
(cytosine-5-)-methyltransferase (EC 2.1.1.37) - garden
pea, partial (11%)
Length = 553
Score = 28.9 bits (63), Expect = 7.3
Identities = 16/55 (29%), Positives = 29/55 (52%)
Frame = +2
Query: 666 EEKLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRLIKNNSIKV 720
EEK D +DE +LA LL+E +++K +++ + +N IK N ++
Sbjct: 41 EEKKDQMAVDEEDEDAKLARLLQEEEYWKSKKQRKNPRSSSSSNKFYIKINEDEI 205
>TC88173 similar to GP|4512677|gb|AAD21731.1| unknown protein {Arabidopsis
thaliana}, partial (19%)
Length = 898
Score = 28.9 bits (63), Expect = 7.3
Identities = 28/109 (25%), Positives = 48/109 (43%), Gaps = 11/109 (10%)
Frame = +1
Query: 359 EAERLCQMALE-------IHRGNGNGSSASVEESADRRLMGLI----YDSKGDYEAALEH 407
EAE+ Q+ L+ I G S S+ + + G + S+ D + +L
Sbjct: 352 EAEKQIQLQLQAVPNKNIIRNGRRKSGSKSLSDLEFEEVKGFMDLGFVFSEEDKDTSLVS 531
Query: 408 YVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFK 456
+ + NG+E+DV+ V S+I YL+ A D+ +K L +K
Sbjct: 532 IIPGLQRLGKNGEEEDVSDV-SAIQRPYLSEAWEDQKRRKKEKPLMKWK 675
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.132 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,912,766
Number of Sequences: 36976
Number of extensions: 194216
Number of successful extensions: 802
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 777
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 793
length of query: 721
length of database: 9,014,727
effective HSP length: 103
effective length of query: 618
effective length of database: 5,206,199
effective search space: 3217430982
effective search space used: 3217430982
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0400a.8


