

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
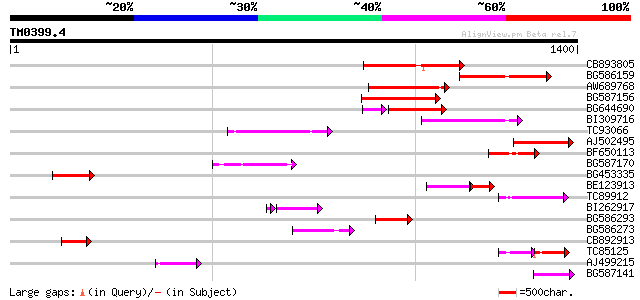
Query= TM0399.4
(1400 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 211 1e-54
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 204 3e-52
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 189 8e-48
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 177 2e-44
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 139 2e-42
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 169 9e-42
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 168 1e-41
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 156 6e-38
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 124 3e-28
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 120 3e-27
BG453335 weakly similar to GP|18071369|g putative gag-pol polypr... 119 6e-27
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 88 5e-25
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 112 8e-25
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 96 3e-21
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 92 1e-18
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 86 1e-16
CB892913 similar to GP|1769897|emb lectin receptor kinase {Arabi... 80 7e-15
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 74 3e-13
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 73 9e-13
BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440... 71 3e-12
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 211 bits (538), Expect = 1e-54
Identities = 106/259 (40%), Positives = 163/259 (62%), Gaps = 10/259 (3%)
Frame = +3
Query: 873 LLADANPVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNP 932
L ++P FEEA+K++ WR +M E+ + ERN TW+L DL + I +KW+FK KLN
Sbjct: 24 LTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNE 203
Query: 933 DGSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWD---LWQLDVK 989
+G I K+KARLV +G+ Q+ G+DY+EVFAPVAR +T+RM++ALA+ D + +
Sbjct: 204 NGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KA 383
Query: 990 SAFLNGPLEEEVYITQPPGFEIKGSEHKVL-------KLRKALYGLKQAPRAWNKRIDTF 1042
+ + + + + I H+V+ ++++ALYGLKQAPRAW RI+ +
Sbjct: 384 HSCMEN*MRKFLLI-----------NHRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAY 530
Query: 1043 LSQTGFHKCSVEHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTD 1102
++ GF KC EH ++VK + G +L++ LYVDDL+ G+ + + K+S+ EF M+D
Sbjct: 531 FTKEGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSD 710
Query: 1103 LGKLSYFLGIEFVQTGEGI 1121
LGK+ YFLG+E Q +GI
Sbjct: 711 LGKMHYFLGVEVTQNEKGI 767
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 204 bits (518), Expect = 3e-52
Identities = 100/225 (44%), Positives = 144/225 (63%)
Frame = +1
Query: 1112 IEFVQTGEGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQL 1171
+E +Q EGI + QRKY+ ++L+RF + N + P+ KL E +VD+T ++Q+
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 1172 VGCLRFICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQ 1231
VGCL ++ +R ++ Y + L+SRFM+CP + H+ A KR+LRYL GT N G+ +
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMY------- 339
Query: 1232 KEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAA 1291
K +G+ L AYTDSD+ GD DR+ST GYVF +SWSSKKQ V LST +AE+IAA
Sbjct: 340 KRNGSEKLEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAA 519
Query: 1292 CSAACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHG 1336
ACQ +W++ +LE+LG ++ + DN S I L+KNPV HG
Sbjct: 520 AFCACQSVWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLHG 654
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 189 bits (479), Expect = 8e-48
Identities = 96/200 (48%), Positives = 135/200 (67%)
Frame = +1
Query: 886 IKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISKHKARLVV 945
+ + W AMK E ++ NKTWDLV LP +K I KWV++VK NPDGS++K KARLV
Sbjct: 82 LSDPRWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVA 261
Query: 946 RGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLEEEVYITQ 1005
+GF Q G DY+E F+PV + T+R+I+ +A W++ Q+D+ +AFLNG L+EEVY++Q
Sbjct: 262 KGFSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQ 441
Query: 1006 PPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVYVKSCDSG 1065
P GFE ++ V KL K+LYGLKQAPRAW + + + Q GF K + + + + +G
Sbjct: 442 PQGFE-AANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYN-QNG 615
Query: 1066 GVLLLCLYVDDLLITGSSYS 1085
+ L +YVDD+LITGSS S
Sbjct: 616 ACIYLXIYVDDILITGSSAS 675
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 177 bits (450), Expect = 2e-44
Identities = 86/199 (43%), Positives = 126/199 (63%), Gaps = 4/199 (2%)
Frame = -1
Query: 870 HMALLADAN----PVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWV 925
H A + + N P +EEA+++K W+ ++ E ++ +N TW +LP K +S +W+
Sbjct: 597 HCAFMVNLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWI 418
Query: 926 FKVKLNPDGSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQ 985
F +K DGSI + K RLV RGF G DY E FAPVA+L T+R++++LA W LWQ
Sbjct: 417 FTIKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQ 238
Query: 986 LDVKSAFLNGPLEEEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQ 1045
+DVK+AFL G LE+EVY+ PPG E VL+L+KA+YGLKQ+PRAW ++ T L+
Sbjct: 237 MDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNG 58
Query: 1046 TGFHKCSVEHGVYVKSCDS 1064
GF K ++H ++ + S
Sbjct: 57 RGFRKSELDHTLFTLTTPS 1
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 139 bits (351), Expect(2) = 2e-42
Identities = 64/142 (45%), Positives = 106/142 (74%)
Frame = -2
Query: 936 ISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNG 995
I+++K++LVV+G+ Q+ G+DY E F+PVAR+E +R+++A A++ + L+Q+DVKSAF+NG
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 996 PLEEEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEH 1055
L+EEV++ QPPGFE + V +L K LYGLKQAPRAW +R+ FL + GF + +++
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 1056 GVYVKSCDSGGVLLLCLYVDDL 1077
+++ + +L++ +YVDD+
Sbjct: 64 TLFLLKRE-*ELLIIQVYVDDI 2
Score = 52.8 bits (125), Expect(2) = 2e-42
Identities = 24/59 (40%), Positives = 35/59 (58%)
Frame = -3
Query: 872 ALLADANPVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKL 930
A ++ P +EA+++ W +M+EEL ER+K W LV P KT I +WVF+ KL
Sbjct: 606 AFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNKL 430
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 169 bits (427), Expect = 9e-42
Identities = 96/249 (38%), Positives = 147/249 (58%)
Frame = +2
Query: 1017 KVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVYVKSCDSGGVLLLCLYVDD 1076
KV +L+K++YGLKQA R W ++ L G+ + S + ++ K DS LL +YVDD
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLL-VYVDD 193
Query: 1077 LLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFVQTGEGILMHQRKYILEVLKRF 1136
+++ G+ S+IQ VK L + F++ DLG L YFLG+E ++ +GIL++QRKY LE+L+
Sbjct: 194 IVLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDS 373
Query: 1137 NLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVGCLRFICHSRLEISYGVGLVSRFM 1196
L+ TP + +LKL + D T +R+L+G L ++ +R +IS+ V +S+F+
Sbjct: 374 GNLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFV 553
Query: 1197 SCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLHLVAYTDSDWCGDQVDRRS 1256
S P+Q H AA R+L+YLK P G+F+ NL L ++ DSDW R+S
Sbjct: 554 SKPQQVHYQAAIRVLQYLKTAPAKGLFY-------SATSNLKLSSFADSDWATCPTTRKS 712
Query: 1257 TMGYVFFFG 1265
GY F G
Sbjct: 713 VTGYWVFLG 739
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 168 bits (426), Expect = 1e-41
Identities = 98/268 (36%), Positives = 155/268 (57%), Gaps = 7/268 (2%)
Frame = +1
Query: 537 CENCLV-SKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIGGNKYFASFIDEYSRKM 595
C++ L + + FS+ T R+ +LD I+SD+ GP + S GG +Y + ID++ RK+
Sbjct: 22 CKHLLFFGNRKKVSFSTATH-RTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKV 198
Query: 596 WVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVT 655
WVY L+ K+E F FK ++ + + Q+G+++K L TD E+CS++ + FC +GI T
Sbjct: 199 WVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKT 378
Query: 656 APYTPQHNGVAERRNRTVLNMVRSMLKGKSLPH--RFCGEAVMTAVYVLNLCPTKSVDSQ 713
P PQ NGVAER RT+L R ML L + EA TA +++N P ++D +
Sbjct: 379 IPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFK 558
Query: 714 VPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGYSS-TGAYKLY--N 770
VPE +WSG +LRIFGC + + D KL ++ IF+ Y+S + Y+L+ +
Sbjct: 559 VPEDIWSGNLVDYSNLRIFGCPAYALVND---GKLAPRAGECIFLSYASESKGYRLWCSD 729
Query: 771 PRTSQVEFSRDVVFEEHSAW-KGKETMV 797
P++ ++ SRDV F E + GK++ V
Sbjct: 730 PKSQKLILSRDVTFNEDALLSSGKQSFV 813
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 156 bits (394), Expect = 6e-38
Identities = 72/147 (48%), Positives = 105/147 (70%)
Frame = +2
Query: 1245 SDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQGLWIQAL 1304
SDW GD R+ST GY F G ISWSSKKQ VA ST EAEYIA+ S A Q +W++ +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1305 LEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKEKIKLQHCGT 1364
LE + + + ++ DNKSAI L+KNPV HGRSKHI+ ++H +R+ ++++++ +++C T
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 1365 DLQIADVFTKPLKADRFKTLKKMMNVL 1391
+ +IAD+FTKPLK + F LKKM+ ++
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGMM 442
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 124 bits (310), Expect = 3e-28
Identities = 64/127 (50%), Positives = 84/127 (65%)
Frame = +1
Query: 1182 RLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLHLVA 1241
R +I Y V ++S+FM PR+ HL AA RILRY++GT +G+ FP S+ + L+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYE----LIC 288
Query: 1242 YTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQGLWI 1301
Y+DSDWCGD RRST GYVF F A ISW +KKQ ALS+ EAEYIA A Q LW+
Sbjct: 289 YSDSDWCGD---RRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWL 459
Query: 1302 QALLEEL 1308
++++EL
Sbjct: 460 DSVIKEL 480
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 120 bits (302), Expect = 3e-27
Identities = 71/207 (34%), Positives = 109/207 (52%)
Frame = -3
Query: 502 LWHQRFGHLNFKDLSSLKSKDMVHGLSQIKLPSKVCENCLVSKQPRAPFSSFTPTRSTAV 561
LWH R GH + + L+ + L + +K CE C++ K + F T T
Sbjct: 584 LWHARLGHPHGRALNLM--------LPGVVFENKNCEACILGKHCKNVFPR-TSTVYENC 432
Query: 562 LDVIYSDVCGPFETASIGGNKYFASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQS 621
D+IY+D+ + S +KYF +FIDE S+ W+ L+ +K V FK F+
Sbjct: 431 FDLIYTDLWTA-PSLSRDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHY 255
Query: 622 GRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSML 681
IK+LR+D GGEY S + + +GILH+ + PYTPQ NGVA+R+N+ ++ + RS++
Sbjct: 254 HAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEVARSLM 75
Query: 682 KGKSLPHRFCGEAVMTAVYVLNLCPTK 708
+ V TA Y++N PTK
Sbjct: 74 FQAN---------VSTACYLINWIPTK 21
>BG453335 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (11%)
Length = 660
Score = 119 bits (299), Expect = 6e-27
Identities = 56/104 (53%), Positives = 86/104 (81%)
Frame = +1
Query: 106 EKLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRTL 165
+K++++KLQ++RR+F+L+QME + IAD+ +++I++ N +KA GE +TDQ IVEK++RTL
Sbjct: 250 DKVEQIKLQSLRRKFKLMQMEDEKKIADYISKLINVVNQIKAYGEVVTDQQIVEKIMRTL 429
Query: 166 TPKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KS 209
+P+FD +VVAI ESK ++ LKIEEL+ SLEAH+ + ERSS +S
Sbjct: 430 SPRFDFIVVAIHESKDVKTLKIEELKSSLEAHKLMVYERSSERS 561
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 88.2 bits (217), Expect(2) = 5e-25
Identities = 41/118 (34%), Positives = 67/118 (56%)
Frame = +1
Query: 1030 QAPRAWNKRIDTFLSQTGFHKCSVEHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQA 1089
Q+PR W R + + G+ +C +H +++K + +L +YVDD+ +TG I+
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 1090 VKRSLNNEFEMTDLGKLSYFLGIEFVQTGEGILMHQRKYILEVLKRFNLLSCNPAETP 1147
+K L EFE+ DLG L YFLG+E + +G + QRKY+L++LK ++ C P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 46.2 bits (108), Expect(2) = 5e-25
Identities = 22/58 (37%), Positives = 37/58 (62%)
Frame = +2
Query: 1139 LSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVGCLRFICHSRLEISYGVGLVSRFM 1196
L P+ETP++ +KLG + VD +++LVG L ++ H+R +IS+ V +S+FM
Sbjct: 329 LDVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 112 bits (281), Expect = 8e-25
Identities = 64/175 (36%), Positives = 102/175 (57%)
Frame = +1
Query: 1206 AAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFG 1265
A K +L+YL + + + K +Q+ED L Y D+D+ G+ R+S G+VF
Sbjct: 4 ALKWVLKYLNESLKSSLKYTK--AAQEEDA---LEGYVDADYAGNVDTRKSLSGFVFTLY 168
Query: 1266 KAPISWSSKKQAAVALSTCEAEYIAACSAACQGLWIQALLEELGLKTDEAVQLMVDNKSA 1325
ISW + +Q+ V LST +AEYIA +W++ ++ ELG+ T E V++ D++SA
Sbjct: 169 GTTISWKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGI-TQEYVKIHCDSQSA 345
Query: 1326 IDLAKNPVSHGRSKHIETKYHFLRDQVSKEKIKLQHCGTDLQIADVFTKPLKADR 1380
I LA + V H R+KHI+ + HF+RD + ++I ++ ++ ADVFTK K R
Sbjct: 346 IHLANHQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTKLKKHAR 510
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 96.3 bits (238), Expect(2) = 3e-21
Identities = 49/115 (42%), Positives = 66/115 (56%), Gaps = 1/115 (0%)
Frame = +1
Query: 658 YTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQVPEA 717
+TPQ NGVAER NRT+L R+MLK + F EAV TA YV+N P+ +D + P
Sbjct: 82 HTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPME 261
Query: 718 VWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGYS-STGAYKLYNP 771
+W G+ L +FGC + Q R KLD KS IF+GY+ + Y L++P
Sbjct: 262 MWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWDP 426
Score = 25.0 bits (53), Expect(2) = 3e-21
Identities = 9/22 (40%), Positives = 13/22 (58%)
Frame = +3
Query: 634 GEYCSNEMSNFCEENGILHEVT 655
GEY E FC++ GI+ + T
Sbjct: 9 GEYVDGEFLAFCKQEGIVRQFT 74
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 92.4 bits (228), Expect = 1e-18
Identities = 41/92 (44%), Positives = 66/92 (71%)
Frame = +2
Query: 904 RNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISKHKARLVVRGFMQRGGLDYSEVFAPV 963
+ +T LV P PI ++W++K+K N DG++ K+KARLV +G++++ G+D+ EVFAPV
Sbjct: 47 QKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPV 226
Query: 964 ARLETVRMIVALASWKNWDLWQLDVKSAFLNG 995
R+ET+ +++ALA+ + +DVK AFLNG
Sbjct: 227 VRIETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 85.5 bits (210), Expect = 1e-16
Identities = 52/155 (33%), Positives = 81/155 (51%), Gaps = 2/155 (1%)
Frame = -2
Query: 698 AVYVLNLCPTKSVDSQVPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIF 757
A Y++N PT+ + Q P V + RKPS+ ++R+FGCLC+ +P + R KL+ +S +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 758 IGYSST-GAYKLYNPRTSQVEFSRDVVF-EEHSAWKGKETMVVNDSMQRVNLDLDHDDSE 815
IGYS+T YK Y+P +V SRDV F EE ++ K + D D
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDLRDLT---------SDKA 372
Query: 816 GIESAVVDVPGTSQNQNQIQVHNPRPIRTKTLPAR 850
G+ +++ G NQ+Q + +P + P R
Sbjct: 371 GVLRVILEGLGIKMNQDQ-STRSRQPEESSNEPRR 270
>CB892913 similar to GP|1769897|emb lectin receptor kinase {Arabidopsis
thaliana}, partial (35%)
Length = 837
Score = 79.7 bits (195), Expect = 7e-15
Identities = 36/72 (50%), Positives = 56/72 (77%)
Frame = +1
Query: 129 ESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRTLTPKFDHVVVAIEESKKLENLKIE 188
E + +F+R+IS++N +K EK+ D I+EK+LR+L PKF+H+V IEE+K LE + IE
Sbjct: 472 ERVRVYFSRVISISNQLKRNSEKLEDVRIMEKILRSLDPKFEHIVEKIEETKDLETMTIE 651
Query: 189 ELQGSLEAHEQR 200
+LQGSL+A+E++
Sbjct: 652 KLQGSLQAYEEK 687
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 74.3 bits (181), Expect = 3e-13
Identities = 35/88 (39%), Positives = 57/88 (64%), Gaps = 1/88 (1%)
Frame = +3
Query: 1295 AC-QGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVS 1353
AC + +W+Q L+EELG K E + + D++SA+ +A+NP H R+KHI +YHF+R+ V
Sbjct: 240 ACKEAIWMQRLMEELGHK-QEQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVE 416
Query: 1354 KEKIKLQHCGTDLQIADVFTKPLKADRF 1381
+ + +Q T+ +AD TK + D+F
Sbjct: 417 EGSVDMQKIHTNDNLADAMTKSINTDKF 500
Score = 70.9 bits (172), Expect = 3e-12
Identities = 43/99 (43%), Positives = 51/99 (51%), Gaps = 6/99 (6%)
Frame = +1
Query: 1208 KRILRYLKGTPNHGVFFPKHLPSQKEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKA 1267
KRI+RY+KGT V F L + Y DSD+ GD R+ST GYVF
Sbjct: 4 KRIMRYIKGTSGVAVCFG--------GSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGG 159
Query: 1268 PISWSSKKQAAVALSTCEAEYIAACSA------ACQGLW 1300
+SW SK Q VALST EAEY+AA C+G W
Sbjct: 160 AVSWLSKLQTVVALSTTEAEYMAAYLKHARKLFGCKG*W 276
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 72.8 bits (177), Expect = 9e-13
Identities = 39/115 (33%), Positives = 65/115 (55%)
Frame = +3
Query: 360 TKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVRFADDRFISAEGIGDVLVKRE 419
TK+ Y W +DSGC++HMT +++ L+++ S+VR + I EGIG VLVK
Sbjct: 87 TKQPSKY---WLIDSGCTHHMTHDRDLFKELNKSTISKVRMLNGAHIEVEGIGTVLVKSH 257
Query: 420 DGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFDPNERKIMKVPL 474
G IS VLY P + +L+S+ QLL K + + + + D N +++++ +
Sbjct: 258 SGYK-QISNVLYAPKLNQSLLSVPQLLTKGYKVLFEHEKCVIKDQNNKEVLRAQM 419
>BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440.1 [imported]
- Arabidopsis thaliana, partial (20%)
Length = 731
Score = 70.9 bits (172), Expect = 3e-12
Identities = 35/101 (34%), Positives = 60/101 (58%)
Frame = +3
Query: 1293 SAACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQV 1352
+AA Q +W+Q LL E+ + E V + +DN+S I L +NPV HGR HI +YHF+R+ V
Sbjct: 123 TAARQAMWLQDLLSEVTWEPCEEVVIRIDNQSVIALTRNPVFHGRGNHIHKRYHFIRECV 302
Query: 1353 SKEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNVLVL 1393
+++++H + A + TK L F+ ++ + ++ L
Sbjct: 303 ENGQVEVEHVPGEKHRAYI*TKALGRIIFREIRYYIGMIDL 425
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,192,186
Number of Sequences: 36976
Number of extensions: 591789
Number of successful extensions: 2901
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 2803
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2875
length of query: 1400
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1292
effective length of database: 5,021,319
effective search space: 6487544148
effective search space used: 6487544148
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0399.4


